Abstract
Transient and long-term shortages of fresh water are major adverse environmental factors that cause dramatic reductions in crop production and distribution globally. In this study, we isolated a full-length CaSRP1 (Capsicum annuum stress-related protein 1) cDNA, which was rapidly induced by dehydration in hot pepper plants. The predicted CaSRP1 protein sequence exhibited significant amino acid identity to putative stress-related proteins and the small rubber particle protein (SRPP) found in rubber trees (Hevea brasiliensis). To study the cellular functions of CaSRP1, transgenic Arabidopsis plants (35S:CaSRP1) that constitutively expressed the CaSRP1 gene were constructed. Overexpression of CaSRP1 resulted in enhanced root and shoot growth and earlier bolting in the transgenic plants relative to wild-type plants. In addition, 35S:CaSRP1 overexpressors exhibited enhanced tolerance to drought stress as compared to the control plants. These results suggest that CaSRP1 plays dual functions as a positive factor for tissue growth and development and for drought-defensive responses. A possible cellular function of SRPP homologs in non-rubber-producing plants in relation to drought stress tolerance is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to their sessile nature, plants are constantly faced with various adverse environmental factors, including drought, high salt, heavy metals, cold, heat shock, and ozone, during their whole life span. These abiotic stresses are a limiting factor for the growth and development of crop plants. Water deficiency causes dramatic reduction of crop production globally (Boyer 1982; Cushman and Bohnert 2000). Urbanization due to growing populations and industrialization, as well as the recent global warming, has reduced the fresh water availability for crop plants world wide (Vorosmarty et al. 2000; Jackson et al. 2001; Kerr 2007). Worldwide water consumption has been continuously increasing and, thus, the decreasing availability of fresh water may pose a future threat to humans and higher plants (Jackson et al. 2001; Hightower and Pierce 2008; Yoo et al. 2009). Plants have diverse defense strategies to enhance their tolerance to transient and long-term water shortages by triggering signaling network pathways and inducing stress-responsive genes. The cellular and genetic defense mechanisms in response to water stress have been widely documented (Shinozaki and Yamaguchi-Shinozaki 1996; Bray 1997; Ishitani et al. 1997; Zhu 2002; Bohnert et al. 2006; Shinozaki and Yamaguchi-Shinozaki 2007; Vij and Tyagi 2007). However, for stress tolerance or sensitivity, our knowledge concerning the biological functions of stress-related genes in higher plants is still rudimentary. Therefore, it is important to study the functions of stress-responsive genes to increase the productivity and distribution of crop plants.
Hot pepper (Capsicum annuum L.), belonging to the solanaceous species, is a widely cultivated crop for its commercially important hot-tasting fruits. The response of hot pepper plants to water-deficit stress is not well defined. Previously, we isolated and characterized a broad spectrum of genes induced by dehydration in hot peppers using subtractive hybridization and differential-display polymerase chain reaction (PCR) methods (Park et al. 2003; Hong and Kim 2005). Among these cDNAs, Ca-LEAL1 (Capsicum annuum late embryogenesis-abundant-like protein 1) (Park et al. 2003), Ca-DREBLP1 (Capsicum annuum dehydration-responsive element binding-factor-like protein 1) (Hong and Kim 2005), CaPUB1 (Capsicum annuum putative U-box protein 1) (Cho et al. 2006a), CaXTHs (Capsicum annuum xyloglucan endotransglucosylase/hydrolase homologs) (Cho et al. 2006b), and CaRma1H1 (Capsicum annuum RING membrane-anchor ubiquitin ligase homolog 1) (Lee et al. 2009) were rapidly induced by drought stress and may be functionally involved in drought stress responses as positive or negative regulators in hot pepper plants.
Ca-DSR5 (Capsicum annuum drought stress-responsive 5) (GenBank accession No. CK327621) is one of the previously identified partial genes, the mRNA of which was induced in response to dehydration in hot pepper roots (Hong and Kim 2005). The derived partial Ca-DSR5 protein displayed a significant homology to the putative stress-related proteins from soybean and Arabidopsis and, thus, was renamed CaSRP1 (Capsicum annuum stress-related protein 1). In this study, we isolated a full-length CaSRP1 cDNA clone (GenBank accession No. GU373985). The predicted full-length CaSRP1 protein also showed amino acid sequence identity with the small rubber particle proteins (SRPPs) found in rubber trees. To study the cellular functions of CaSRP1, transgenic Arabidopsis plants (35S:CaSRP1) that constitutively expressed the CaSRP1 gene were constructed. We present results indicating that overexpression of CaSRP1 resulted in enhanced growth of roots and shoots in the transgenic Arabidopsis plants relative to that of wild-type plants. This enhanced growth resulted in an earlier bolting phenotype. In addition, 35S:CaSRP1 plants exhibited markedly enhanced tolerance to dehydration stress as compared to the control plants. CaSRP1 may play a cellular role in relation to drought stress tolerance.
Materials and methods
Plant materials
Dried hot pepper (Capsicum annuum cv. Pukang) and Arabidopsis thaliana ecotype Columbia (Col-0) seeds were soaked for 10 min in 30% sodium hypochlorite solution (bleach) and rinsed extensively with sterilized water as previously described by Cho et al. (2006b). Seedlings were grown on MS medium that contained 1% sucrose, 12 μg/ml B5 vitamin, and 0.8% agar (pH 5.7) in a 25°C growth chamber under continuous light conditions.
Application of dehydration stress
Hot pepper plants were grown in pots for 2 weeks in a 25°C growth chamber with a 16-h-light/8-h-dark photoperiod. Whole hot pepper plants were then exposed to a stream of air in a clean bench for varying times (0, 30, and 90 min) as described by Kilian et al. (2007). During this incubation time, the plants gradually lost their fresh weight.
Isolation of a full-length cDNA clone of CaSRP1
Full-length CaSRP1 cDNA was PCR amplified from a λ-uni-Zap II cDNA library constructed from water-stressed leaves of hot pepper plants (Cho et al. 2006b). Vector-specific primers corresponding to the T3 promoter (Table 1) were used to generate the sense oligonucleotide. The antisense oligonucleotide was designed from the sequences corresponding to the 3′ end of the partial Ca-DSR5 cDNA (Table 1). PCRs used high-fidelity Ex-Taq DNA polymerase (Takara, Otsu, Shiga, Japan) and consisted of 40 amplification cycles with an annealing temperature of 52°C for 30 s and an elongation temperature of 68°C for 1 min. PCR products were introduced into the pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into DH5α E. coli. The cDNA inserts from the plasmids of 12 resulting colonies were sequenced.
RNA isolation
Total RNA from hot pepper and transgenic Arabidopsis plants were extracted using RNAiso Plus reagent as per the manufacturer’s instructions (Takara). Isolated RNA was treated with DNase I to remove possible residual DNA contamination as per the manufacturer’s instructions (Promega).
Reverse transcriptase-polymerase chain reaction (RT-PCR)
RT-PCR was carried out in a total volume of 50 μl containing 1 μl of the first strand cDNA reaction products, 1 μM of gene-specific primers (Table 1), 10 mM Tris (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 200 μM deoxynucleotides, and 2.5 units of high-fidelity Ex-Taq polymerase (Takara) as previously described (Jun et al. 2008). Amplification consisted of 27 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C in an automatic thermal cycler (Perkin-Elmer/Cetus, Norwalk, CT, USA). PCR products were separated on 1.0% agarose gels and visualized under UV light.
Generation of 35S:CaSRP1 transgenic Arabidopsis plants
The full-length pCaSRP1 cDNA was introduced into the corresponding sites of the binary vector pBI121. The resulting 35S:CaSRP1 fusion gene was transferred to Agrobacterium tumefaciens strain GV3101 by electroporation. Agrobacterium cells containing the 35S:CaSRP1 construct were transformed into Arabidopsis plants (Col-0) by means of the floral-dip method (Zhang et al. 2006). T1 seeds were collected from regenerated T0 plants. T1 seeds were germinated on 0.5× Murashige and Skoog (MS) medium with 30 μg/ml kanamycin. Homozygous T3 lines were obtained by subsequent self-fertilization and used in phenotypic analyses. The presence and expression level of the CaSRP1 gene were confirmed by genomic Southern blotting and RT-PCR, respectively.
Measurements of dimensional parameters of roots, leaves, and petioles in wild-type and transgenic Arabidopsis seedlings
Root and leaf growth of wild-type and transgenic Arabidopsis plants were monitored as described by Seo et al. (2008) with minor modifications. To examine root growth, vertically oriented agar plates (0.8% select agar; Life Technology, Rockville, MD, USA) were incubated at 22°C under continuous light for 1–7 days. During incubation, the advancing root tips were monitored with the image-analyzing program SCIONIMAGE (Scion Corp., Frederic, MD, USA). Leaves and petioles of 2-week-old wild-type and transgenic Arabidopsis plants were removed from the plants. The surface areas, lengths, and widths were also determined using SCIONIMAGE software.
Microscopy
Five-day-old wild-type and transgenic Arabidopsis leaves were cleared in a chloral hydrate solution as described previously (Kwon et al. 2009). The leaf cell layers were visualized by light-field microscopy (BX51 fluorescence microscope, Olympus, Japan). For the longitudinal root sections, 5-day-old roots were stained with propidium iodide and their images were obtained using a confocal microscope (LSM510 META; Carl Zeiss Inc.) as described by Seo et al. (2008).
Survival rate determination of wild-type and transgenic Arabidopsis plants after drought stress
Three-week-old 35S:CaSRP1 transgenic and 25-day-old wild-type plants, which had been grown under normal growth conditions, were subjected to drought stress by withholding water for 7 days. The plants were then re-watered and their phenotypes were examined after 3 days. Survival was defined as the ability to resume growth when returned to normal conditions following water stress (Cho et al. 2008).
Chlorophyll measurement
Chlorophylls were extracted and analyzed from drought stress-treated wild-type and CaSRP1-overexpressing leaves using 80% acetone as described previously (Bae et al. 2009). The remaining leaf extracts were dried at 105°C for 16 h and weights of the dried extracts were measured (Welti et al. 2002). The content of chlorophyll a + b was expressed as mg/g DW.
Results
Isolation and characterization of full-length CaSRP1 cDNA
In our previous study, a partial hot pepper Ca-DSR5 (Capsicum annuum drought stress-responsive 5) gene, which was rapidly induced by drought stress in root tissue, was identified (Hong and Kim 2005). The derived partial Ca-DSR5 protein showed homology to the putative stress-related proteins from soybean (PvSRP) and Arabidopsis. Thus, we referred to this gene as CaSRP1 for Capsicum annuum stress-related protein 1 and proceeded to isolate its full-length cDNA. Total recombinant lambda DNA was obtained from the λ-uni-Zap II cDNA library constructed from water-stressed leaves of hot pepper plants (Cho et al. 2006b). The entire coding region of CaSRP1 was PCR amplified using the lambda DNA as a template with primers corresponding to the 5′-end of the library vector sequence and the 3′-untranslated region of the partial Ca-DSR5 cDNA clone (Table 1).
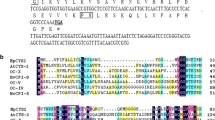
Figure 1a shows the restriction enzyme map of pCaSRP1. The pCa-SRP1 clone (GenBank accession No. GU373985) is 978 bp long, consisting of a 68-bp 5′-untranslated region, a 684-bp coding region encoding 228 amino acids, and a 226-bp 3′-untranslated region. The predicted molecular mass and calculated isoelectric point of CaSRP1 are 24.9 kDa and 9.27, respectively. A database search showed that CaSRP1 is 60.5 and 58.5% identical to grape and poplar proteins, respectively, the cellular functions of which are not yet known (Fig. 1b, c). In addition, CaSRP1 is 51.8–28.3% homologous to the putative stress-related proteins in Arabidopsis (At3g05500, At2g47780, and At1g67360), soybean (PvSRP), and alfalfa. Functions of these proteins are also unknown. Interestingly, CaSRP1 shares a significant degree of sequence identity (~46%) with the small rubber particle proteins (SRPP) of rubber tree (Hevea brasiliensis) and the guayule homolog of SRPP (GHS) from Parthenium argentatum Gray, both of which produce natural rubber (cis-1, 4-polyisoprene) (Oh et al. 1999; Sookmark et al. 2002; Kim et al. 2004). Hevea brasiliensis currently supplies most of the commercially used high molecular weight natural rubber, while Parthenium argentatum Gray is a potential rubber tree for an alternative source of natural rubber (Mooibroek and Cornish 2000).
Sequence analysis of hot pepper CaSRP1. a Schematic structure of CaSRP1 cDNA clone (Genbank accession No. GU373985). The gray bar represents the coding region and the black lines indicate 5′- and 3′-untranslated regions. Restriction sites for EcoRI, PstI, NcoI, and SpeI are indicated. b Multiple alignments of 10 SRP1 homologs. The predicted amino acid sequence of CaSRP1 is compared with those of proteins from poplar (Genbank accession No. XP_002319520), grape (Genbank accession No. XP_002283697), Arabidopsis (Genbank accession Nos. At3g05500, At2g47780, and At1g67360), alfalfa (Genbank accession No. ABD28680), soybean (PvSRP; Genbank accession No. AAB00555), rubber tree (HbSRPP; Genbank accession No. AAC82355), and Parthenium argentatum Gray (GHS; Genbank accession No. AAQ11374). Amino acid residues identical in at least six of the ten sequences are shaded. Amino acid sequences conserved in all ten proteins are indicated in black. c Phylogenetic relationship of the ten SRP homologs from hot pepper, poplar, grape, Arabidopsis, alfalfa, rubber tree, and Parthenium argentatum Gray
Drought induction of the CaSRP1 gene in hot pepper leaves
The CaSRP1 gene was induced rapidly in roots of hot pepper plants in response to dehydration (Hong and Kim 2005). To examine whether CaSRP1 is also up-regulated in leaf tissue in response to dehydration, we monitored the steady-state level of CaSRP1 transcripts in water-stressed leaves. For drought stress treatment, light-grown 2-week-old hot pepper plants were exposed to a stream of air in a clean bench for increasing times (0, 30, and 90 min) (Kilian et al. 2007). Total RNA was then isolated from the treated leaves and subjected to RT-PCR using gene-specific primers (Table 1). Figure 2 indicates that the low basal-level of CaSRP1 transcripts already began to increase after 30 min of dehydration. mRNA levels were continuously elevated for at least 90 min. The CaLEAL1 gene, which encodes abiotic stress-induced late embryogenesis-abundant-like protein 1 (Park et al. 2003), was chosen for the RT-PCR experiment as a positive control for water deficit. The amount of CaLEAL1 mRNA was also elevated concomitantly in response to water loss. In contrast, the expression of the hot pepper actin gene (CaACT) remained unchanged during incubation. These results, along with those of Hong and Kim (2005), indicate that CaSRP1 is induced by dehydration in both roots and leaves (Fig. 2) of hot pepper plants.
Induction of CaSRP1 gene expression in hot pepper leaves in response to dehydration. Light-grown 2-week-old hot pepper plants were subjected to dehydration stress by exposing whole plants to a stream of air in a clean bench for increasing times (0, 30, and 90 min). Total RNA was isolated from leaves and analyzed by RT-PCR using gene-specific primers designed for CaSRP1, CaLEAL1 (positive control), and CaACT (negative control)
CaSRP1-overexpressing transgenic Arabidopsis plants grew more rapidly than control plants
The structure (Fig. 1) and expression profile (Fig. 2) of the CaSRP1 gene raises the possibility that CaSRP1 may be involved in cellular responses to drought stress in hot pepper plants. Thus, we wanted to investigate the cellular functions of CaSRP1 with the aid of the transgenic approach. It was previously reported that construction of transgenic hot pepper plants was extremely difficult (Seo et al. 2008). Specifically, transformation and regeneration yields were too low to obtain sufficient independent transgenic lines. Alternatively, hot pepper genes appeared to be fully functional in heterologous Arabidopsis cells (Cho et al. 2006a, b; Seo et al. 2008; Lee et al. 2009). We indeed failed to obtain transgenic hot pepper plants that constitutively expressed the CaSRP1 gene. Thus, in this study, we generated transgenic Arabidopsis plants that overexpressed CaSRP1 under the control of the 35S CaMV promoter by means of Agrobacterium-mediated transformation. Several independent primary transformants were identified due to kanamycin resistance. Transgenic plants were subsequently regenerated and used for phenotype analyses. The presence and expression levels of transgenes were elucidated by RT-PCR. Figure 3a indicates that several independent T3 transgenic lines (#1, #2, #3, #4, and #7) possessed varied amounts of the CaSRP1 transcripts under normal growth conditions. Transgenic line #3 contained the highest level of CaSRP1 mRNA and line #4 the lowest level (Fig. 3a).
Phenotypic characterizations of T3 35S:CaSRP1 transgenic Arabidopsis plants. a Expression analysis of the CaSRP1 transgene. The presence and expression levels of CaSRP1 were examined by RT-PCR in wild-type and various independent T3 35S:CaSRP1 transgenic lines (#1, #2, #3, #4, and #7). The actin gene (AtACT8) was used as a loading control. b Morphological comparisons of light-grown 4-day-old wild-type and T3 35S:CaSRP1 transgenic seedlings (lines #1, #2, #3, and #7). c Differences in growth patterns of early roots in wild-type and CaSRP1-overexpressing seedlings 1–7 days after germination. Error bars indicate mean ± SD (n = 70). d Morphological comparisons of light-grown 2-week-old wild-type and 35S:CaSRP1 transgenic plants (lines #1, #2, #3, and #7). e Determination of the dimensional parameters of second leaves 2 weeks after germination. The second leaves were detached from wild-type and transgenic lines, scanned for image analysis using SCIONIMAGE program, and their blade widths and lengths, petiole lengths, and blade areas were calculated. Error bars indicate mean ± SD (n = 35)
In pursuing the phenotypic analysis, we first found that light-grown 4-day-old CaSRP1-overexpressing seedlings contained considerably longer (1.2- to 1.3-fold) roots relative to the wild-type roots under normal growth conditions (Fig. 3b). The lengths of transgenic and wild-type roots were similar in very early seedlings (1 day after germination), but began to differ 2 days after germination. Thereafter, the differences in root length became greater during the period 5–7 days after germination (Fig. 3c). In addition, although the morphology of transgenic leaves appeared to be somewhat similar in 5- to 7-day-old-seedlings (Fig. 3a), the 2-week-old transgenic leaves were markedly larger than those of wild-type Arabidopsis plants (Fig. 3d). Both blade length and width of the second leaf were increased 1.3- to 2.1-fold and 1.2- to 1.8-fold, respectively, in transgenic plants as compared to the wild-type plants (Fig. 3e). The petiole lengths were also increased to a similar extent as the blade length and width (Fig. 3e). An increase in blade length was paralleled with that in blade width, resulting in the uniformly larger shape in 35S:CaSRP1 lines. Consequently, the leaf-blade area of the independent transgenic CaSRP1-overexpressing lines was 1.6- to 3.6-fold greater than that of wild-type leaves.
In addition, the 35S:CaSRP1 plants bolted earlier than wild-type plants. The CaSRP1-overexpressing transgenic lines bolted 20–21 days after germination, whereas control plants bolted approximately 25 days after germination (Fig. 4a). At 28 days after germination, the inflorescence length of CaSRP1 overexpressors was from 7.7 to 12.9 cm depending on the transgenic lines, while that of control plants averaged 1.0 cm (Fig. 4b). Taken together, these results suggest that the ectopic expression of the hot pepper-originated CaSRP1 gene caused faster growth in heterologous Arabidopsis plants that is coupled with enhanced development of both vegetative and reproductive organs.
CaSRP1 overexpressors bolted earlier than did wild-type plants. a Growth morphology of wild-type and 35S:CaSRP1 transgenic Arabidopsis plants grown under normal growth conditions 28 days after germination. b Differences in growth patterns of inflorescence stems of wild-type and transgenic Arabidopsis plants 20–28 days after germination. Error bars indicate mean ± SD (n = 80)
Faster-growing phenotypes of 35S:CaSRP1 transgenic plants correlated with enhanced cell cycle progression
Based on the aforementioned results of phenotypic analyses, we hypothesized that CaSRP1 was associated with a subset of cell and tissue development. To examine whether the elevation of leaf and root growth in 35S:CaSRP1 transgenic plants was due to changes in cell expansion and/or proliferation pathways, we investigated expression profiles of genes that were intimately tied with cell division and elongation, respectively. The D-type cyclin AtCYCD3 (Riou-Khamlichi et al. 1999) and cell cycle-dependent kinase-related gene AtCDC2b (Yoshizumi et al. 1999) were selected as marker genes for cell cycling activity. The expansin gene AtEXP5 (Li et al. 2002, GenBank accession No. NM_113824) was included as a marker for cell elongation. Total RNA was extracted from light-grown 7-day-old wild-type and 35S:CaSRP1 whole seedlings (lines #2 and #7) and analyzed by RT-PCR with gene-specific primers (Table 1). The results showed that mRNA levels of AtCYCD3 and AtCDC2b were markedly enhanced in the CaSRP1-overexpressor lines #2 and #7 (Fig. 5a). In contrast, the amount of AtEXP5 transcript was nearly identical in transgenic and control seedlings (Fig. 5a). Thus, these results allowed the suggestion that the faster-growing phenotypes of 35S:CaSRP1 transgenic plants correlated with enhanced expression of cell cycle progression genes, rather than elongation-related genes.
Faster-growing phenotypes of 35S:CaSRP1 transgenic plants correlated with enhanced cell cycle progression. a Comparisons of the expression levels of cell division- and cell elongation-associated genes in wild-type (WT) and CaSRP1-overexpressing transgenic seedlings (lines #2 and #7). The steady-state mRNA levels of D-type cyclin AtCYCD3, cell cycle-dependent kinase-related gene AtCDC2b, and the expansin gene AtEXP5 were elucidated by RT-PCR using gene-specific primers in light-grown 1-week-old wild-type and 35S:CaSRP1 seedlings. The actin gene (AtACT8) was used as a loading control. b Longitudinal views of root tips from 5-day-old wild-type and CaSRP1-overexpressing transgenic seedlings (lines #2 and #7). Root sections were stained with propidium iodide and analyzed by confocal microscopy. Vertical lines indicate the proximal meristem. Arrows indicate the transition zone between the proximal meristem and elongation-differentiation zones. The transition zone is more clearly shown in the insets. Cell numbers in the proximal meristem are shown in the right panel. PM, proximal meristem. Scale bars 20 μm c Light microscope demonstrating the epidermal and palisade mesophyll cells from the cotyledons of 5-day-old light-grown wild-type and CaSRP1 overexpressors (lines #2 and #7). Scale bars 20 μm
The cellular phenotype of 35S:CaSRP1 transgenic plants were next examined by confocal and light microscopy. As shown in Fig. 5b, the proximal meristem (PM) of CaSRP1-overexpressing root tips (lines #2 and #7) contain 32 ± 2 cells in average, while the average number of PM cells in wild-type root tips was 27 ± 1. In addition, both epidermal and palisade mesophyll cells in 35S:CaSRP1 leaves were smaller and more populated compared to those in wild-type plants (Fig. 5c). Overall, these results indicate that the overexpression of CaSRP1 resulted in the increased cell numbers in both root and leaves in transgenic Arabidopsis plants. Based on these results, we concluded that the faster-growing phenotypes of 35S:CaSRP1 transgenic plants were associated with increased cell cycle progression.
Overexpression of CaSRP1 conferred increased drought tolerance
Because CaSRP1 is rapidly induced by dehydration in hot pepper, we considered the possibility that, in addition to a subset of cell and tissue development, CaSRP1 also participates in drought stress responses. To test this possibility, we first examined the cut rosette water loss (CRWL) rate (Bouchabke et al. 2008) of 35S:CaSRP1 plants. The rosette leaves were detached from 3-week-old wild-type and transgenic plants, placed on open-lid Petri dishes, and incubated for different time periods (0–4 h) at room temperature under dim light. Decreases in fresh weight were monitored. As shown in Fig. 6a, detached rosette leaves from 35S:CaSRP1 plants (lines #1, #2, #3, and #7) lost water more slowly than those from wild-type plants. After 1 h of incubation, CRWL rates of wild-type and CaSRP1-overexpressing leaves were already distinguishable, with that of the transgenic leaves being significantly lower than that of control leaves. After 4 h of incubation, the difference became clearer; the average fresh weight of wild-type leaves was reduced to about 27% of the starting weights, while the fresh weight of the transgenic leaves was reduced to only 42–51% depending on the independent lines (Fig. 6a).
Overexpression of CaSRP1 conferred increased tolerance against drought stress in transgenic Arabidopsis plants. a Measurement of cut rosette water loss (CRWL) rates. The rosette leaves were detached from light-grown 3-week-old wild-type and 35S:CaSRP1 transgenic plants (lines #1, #2, #3, and #7), placed on open-lid Petri dishes and incubated for 0–4 h at room temperature. Decreases in fresh weights were determined. Water loss is expressed as the percentage of initial fresh weight of the detached leaves. Error bars indicate mean ± SD (n = 20). b Survival rates of wild-type and 35S:CaSRP1 transgenic plants after drought stress. Light-grown 25-day-old wild-type and 3-week-old transgenic plants were further grown for 7 days without watering. Plants were then re-watered and their survival rates were determined after 3 days. The survival rate was considered as the ability of plants to continue growing after return to normal water conditions following a 7-day drought stress. The survival rates are as follows: wild type, 9.8% (11 out of 112 plants); transgenic line #1, 27.3% (27 out of 99 plants); transgenic line #2, 73.0% (65 out of 89 plants); transgenic line #3, 60.2% (50 out of 83 plants); and transgenic line #7, 87.1% (108 out of 124 plants). c Amount (mg/g DW) of chlorophyll a + chlorophyll b in leaves of wild-type and CaSRP1-overexpressing transgenic lines (#1, #2, #3, and #7). The experiments were independently repeated three times, and the error bars indicate SD
As a next experiment, the response of transgenic lines to drought stress was examined at the whole plant level. Wild-type and 35S:CaSRP1 Arabidopsis plants were grown for 25 days and 3 weeks, respectively, in pots under normal growth conditions. Because 35S:CaSRP1 transgenics grew faster (Fig. 3) and bolted 4 days earlier in average than control plants (Fig. 4), 3-week-old 35S:CaSRP1 and 25-day-old wild-type plants were used in drought stress experiment. The soil was then allowed to dry without watering for 7 days. After this drought condition, wild-type plants appeared to be severely wilted, their leaves were rolling, and they ceased to grow (Fig. 6b). After re-watering for 3 days, most of the wild-type plants were unable to re-grow and died with a survival rate of 9.8% (11 out of 112 plants). In contrast, significant numbers of 35S:CaSRP1 leaves remained turgid and green after withholding water for 7 days. At 3 days of re-watering, many CaSRP1 overexpressors continued to grow and their survival rates were between 27.3 and 87.1% depending on the transgenic lines [#1; 27.3% (27 out of 99 plants), #2; 73.0% (65 out of 89 plants), #3; 60.2% (50 out of 83 plants), and #7; 87.1% (108 out of 124 plants)] (Fig. 6b). Thus, these results are in agreement with the slower CRWL rate of 35S:CaSRP1 leaves as compared to the wild-type leaves (Fig. 6a) and indicate that CaSRP1-overexpressing transgenic plants were more tolerant to water deficits than were the control plants.
In addition, CaSRP1-overexpressing leaves contained higher levels of chlorophyll after drought stress. As shown in Fig. 6c, transgenic leaves had 15.5 ± 0.4 to 23.1 ± 0.4 mg/g DW of chlorophyll a + chlorophyll b, depending on the independent transgenic lines (#1, #2, #3, and #7), whereas wild-type leaves contained 13.1 ± 0.5 mg/g DW of chlorophyll a + chlorophyll b. This suggests that the photosynthetic capacity of CaSRP1 overexpressors is significantly higher than that of wild-type plants during and after severe dehydration stress. Overall, our data in Figs. 3, 4, and 6 are consistent with the notion that CaSRP1 is involved not only in a subset of cell and tissue development, but also in the drought stress response of transgenic Arabidopsis plants. On the other hand, the phenotypes of the independent 35S:CaSRP1 lines were not necessarily paralleled with the levels of CaSRP1 transcripts (Fig. 3a), suggesting that CaSRP1 protein levels may be subject to control in transgenic Arabidopsis plants.
Discussion
In this study, we isolated a full-length CaSRP1 cDNA clone from water-stressed hot pepper plants. The derived full-length CaSRP1 protein exhibited significant amino acid sequence identity not only to putative stress-related proteins, but also to SRPP from rubber trees (Fig. 1). Natural rubber is cis-1, 4-polyisoprene produced through the mevalonate pathway in the cytosolic fractions (latex) of latex vessel tissues in rubber trees (Oh et al. 1999; Sookmark et al. 2002; Kim et al. 2004; Chow et al. 2007). Hot pepper as well as Arabidopsis does not produce natural rubber, and, hence, it was unexpected that a hot pepper water stress-induced gene shared sequence homology with rubber biosynthetic genes.
These results prompted us to construct transgenic Arabidopsis plants that constitutively expressed the CaSRP1 gene (Fig. 3a). 35S:CaSRP1 transgenic plants displayed unique phenotypes as compared to wild-type plants as evidenced by the following observations. First, CaSRP1-overexpressing plants grew more rapidly with longer roots and larger leaves and, thus, bolted earlier with markedly prolonged inflorescences (Figs. 3, 4). This faster-growing phenotype may be associated with increased cell proliferating activity (Fig. 5). Secondly, 35S:CaSRP1 plants were highly tolerant to drought stress relative to the control plants. Under our experimental conditions, the majority of wild-type plants failed to re-grow after 7 days of dehydration, whereas 27.3–87.1% of CaSRP1 overexpressors were healthy and could continue to grow before and after drought stress (Fig. 6). Thus, these results invoke a working hypothesis in which CaSRP1 participates in a subset of tissue development and in response to water deficiency in transgenic Arabidopsis plants.
These phenotypes of the 35S:CaSRP1 plants are quite intriguing because the normal response of plants to abiotic stresses is growth arrest, rather than promotion (Rohde et al. 1999; Xiong and Zhu 2001). Growth retardation may be a natural adaptive procedure, which allows the plants to prepare their defensive mechanisms against the stresses. In addition, overexpression of water stress-induced genes often results in reduced growth under normal growth conditions (Yamaguchi-Shinozaki and Shinozaki 1994; Sakuma et al. 2006; Mlynarova et al. 2007; Qin et al. 2008), which is opposite to that of the 35S:CaSRP1 plants. The faster-growing phenotype of 35S:CaSRP1 plants is somewhat reminiscent of 35S:CaPUB1 transgenic plants that overexpress drought stress-induced hot pepper U-box E3 ubiquitin ligase in terms of longer roots and early bolting with increased cell division activity (Cho et al. 2006a). However, the faster-growing 35S:CaPUB1 plants displayed hypersensitivity to dehydration stress, which was in sharp contrast to the 35S:CaSRP1 plants. Thus, the unique phenotypes of CaSRP1 lend strong support to a model in which CaSRP1 may play dual functions: a positive factor in tissue growth and development and in defensive responses against water deficit.
SRPP, originally known as a latex allergin, is a protein tightly bound to a small rubber particle in the latex of rubber trees (Hevea brasiliensis). The bark tissue of rubber trees is constantly stripped, known as a tapping process, to collect rubber latex. Thus, plugging of latex vessels is essential for rubber trees to prevent the loss of their cytoplasmic components, such as primary metabolites, and to prevent pathogen infection of the latex vessel tissues (Wititsuwannakul et al. 2008b). In the process of latex vessel plugging, hevein or Hevea latex lectin (HLL) interacts with rubber particle (RP) protein to form a rubber latex coagulum (Gidrol et al. 1994; Wititsuwannakul et al. 2008a). Recently, SRPP was purified as an RP glycoprotein that bound HLL and was, therefore, termed HLL-binding protein (HLLBP) (Wititsuwannakul et al. 2008c). Interaction between an N-acetylglucosamine moiety in SRPP and HLL may modulate the degree of latex coagulation in response to tapping and mechanical wounding.
Given that SRPP is involved in stress responses in rubber trees, SRPP homologs in non-rubber-producing plants may play defensive roles against abiotic stresses. This proposal is supported by our present results that overexpression of CaSRP1 confers strong resistance to drought stress in transgenic Arabidopsis plants (Fig. 6). It would be of keen interest whether CaSRP1 and homologs of SRPPs are indeed glycosylated in hot pepper and transgenic Arabidopsis plants as is SRPP in rubber trees (Wititsuwannakul et al. 2008c). Thus, more detailed cellular and biochemical characterizations of SRPP homologs are required in higher plants other than rubber trees. The Arabidopsis genome contains at least three SRPP homologs (putative stress-related proteins: At3g05500, At2g47780, and At1g67360) (Fig. 1). Currently, we have obtained T-DNA-inserted knockout mutants of each gene and are producing transgenic Arabidopsis plants that overexpress each homolog. Phenotypic analysis of these plants will provide a more conclusive view about the SRPP homologs in higher plants.
In conclusion, CaSRP1 and SRPP homologs were seemingly unrelated proteins found in multiple plant species. However, the data presented here illustrate the relatedness of these proteins in both sequence and function. Better characterization of SRPP homologs may lead to crops with improved tolerance toward drought stress.
Abbreviations
- CaSRP1:
-
Capsicum annuum stress-related protein 1
- DW:
-
Dry weight
- GHS:
-
Guayule homolog of SRPP
- PCR:
-
Polymerase chain reaction
- SRPP:
-
Small rubber particle protein
References
Bae H, Choi SM, Yang SW, Pai H-S, Kim WT (2009) Suppression of the ER-localized AAA ATPase NgCDC48 inhibits tobacco growth and development. Mol Cells 28:57–65
Bohnert HJ, Gong Q, Li P, Ma S (2006) Unraveling abiotic stress tolerance mechanisms—getting genomics going. Curr Opin Plant Biol 9:180–188
Bouchabke O, Chang F, Simon M, Voisin R, Pelletier G, Durand-Tardif M (2008) Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought responses. PLoS ONE 3:e1705
Boyer JS (1982) Plant productivity and environment. Science 218:443–448
Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54
Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk YY, Kim J, Pai HS, Kim WT (2006a) Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol 142:1664–1682
Cho SK, Kim JE, Park J-A, Eom TJ, Kim WT (2006b) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580:3136–3144
Cho SK, Ryu MY, Song C, Kwak JM, Kim WT (2008) Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20:1899–1914
Chow K-S, Wan K-L, Isa MNM, Bahari A, Tan S-H, Harikrishna K, Yeang H-Y (2007) Insight into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J Exp Bot 58:2429–2440
Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3:117–124
Gidrol X, Chrestin H, Tan H-L, Kush A (1994) Hevein, a lectin-like protein from Hevea brasiliensis (rubber tree) is involved in the coagulation of latex. J Biol Chem 269:9278–9283
Hightower M, Pierce AS (2008) The energy challenge. Nature 452:285–286
Hong JP, Kim WT (2005) Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv. Pukang). Planta 220:875–888
Ishitani M, Xiong LM, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9:1935–1949
Jackson RB, Carpenter SR, Dahm CN, McKnight DM, Naiman RJ, Postel SL, Running SW (2001) Water in a changing world. Ecol Appl 11:1027–1045
Jun S-S, Choi HJ, Lee HY, Hong Y-N (2008) Differential protection of photosynthetic capacity in trehalose- and LEA protein-producing transgenic plants under abiotic stresses. J Plant Biol 51:327–336
Kerr RA (2007) How urgent is climate change? Science 318:230–231
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363
Kim IJ, Ryu SB, Kwak YS, Kang H (2004) A novel cDNA from Parthenium argentatum Gray enhances the rubber biosynthetic activity in vitro. J Exp Bot 55:377–385
Kwon SH, Lee BH, Kim EY, Seo YS, Lee S, Kim WT, Song JT, Kim JH (2009) Overexpression of a Brassica rapa NGATHA gene in Arabidopsis thaliana negatively affects cell proliferation during leaf and root development. Plant Cell Physiol 50:2162–2173
Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21:622–641
Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ (2002) Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128:854–864
Mlynarova L, Nap J-P, Bisseling T (2007) The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J 51:874–885
Mooibroek H, Cornish K (2000) Alternative sources of natural rubber—mini review. Appl Microbiol Biotech 53:355–365
Oh SK, Kang H, Shin DH, Yang J, Chow K-S, Yeang HY, Wagner B, Breiteneder H, Han K-H (1999) Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J Biol Chem 274:17132–17138
Park JA, Cho SK, Kim JE, Chung HS, Hong JP, Hwang B, Hong CB, Kim WT (2003) Isolation of cDNAs differentially expressed in response to drought stress and characterization of the Ca-LEAL1 gene encoding a new family of atypical LEA-like protein homologue in hot pepper (Capsicum annuum L. cv. Pukang). Plant Sci 165:471–481
Qin F, Sakuma Y, Tran L-SP, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K-I, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20:1693–1707
Riou-Khamlichi C, Huntly R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283:1541–1544
Rohde A, van Montagu M, Boerjan W (1999) The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environ 22:261–270
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309
Seo YS, Kim EY, Mang HG, Kim WT (2008) Heterologous expression and biochemical and cellular characterization of CaPLA1 encoding a hot pepper phospholipase A1 homolog. Plant J 53:895–908
Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotech 7:161–167
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Sookmark U, Pujade-Renaud V, Chrestin H, Lacote R, Naiyanetr C, Seguin M, Romruensukharom P, Narangajavana J (2002) Characterization of polypeptides accumulated in the latex cytosol of rubber trees affected by the tapping panel dryness syndrome. Plant Cell Physiol 43:1323–1333
Vij S, Tyagi AK (2007) Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol J 5:361–380
Vorosmarty CJ, Green P, Salisbury J, Lammers RB (2000) Global water resources: vulnerability from climate change and population growth. Science 289:284–288
Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X (2002) Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277:31994–32002
Wititsuwannakul R, Pasitkul P, Jewtragoon P, Wititsuwannakul D (2008a) Hevea latex lectin binding protein in C-serum as an anti-latex coagulating factor and its role in a proposed new model for latex coagulation. Phytochemistry 69:656–662
Wititsuwannakul R, Pasitkul P, Kanokwiroon K, Wititsuwannakul D (2008b) A role for a Hevea latex lectin-like protein in mediating rubber particle aggregation and latex coagulation. Phytochemistry 69:339–347
Wititsuwannakul R, Rukseree K, Kanokwiroon K, Wititsuwannakul D (2008c) A rubber particle protein specific for Hevea latex lectin binding protein involved in latex coagulation. Phytochemistry 69:1111–1118
Xiong L, Zhu JK (2001) Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol Plant 112:152–166
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Yoo CY, Pence HE, Hasegawa PM, Mickelbart MV (2009) Regulation of transpiration to improve crop water use. Crit Rev Plant Sci 28:410–431
Yoshizumi T, Nagata N, Shimaga H, Matsui M (1999) An Arabidopsis cell cycle-dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11:1883–1895
Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
This work was supported by grants from the Technology Development Program for Agriculture and Forestry (Project No. 309017-5 funded by the Ministry for Agriculture, Forestry and Fisheries, Republic of Korea), the National Research Foundation (Project No. 2009-0078317 funded by the Ministry of Education, Science, and Technology, Republic of Korea), and the BioGreen 21 Program (funded by the Rural Development Administration).
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported here has been deposited in the GenBank database under accession number GU373985 (CaSRP1).
Rights and permissions
About this article
Cite this article
Kim, E.Y., Seo, Y.S., Lee, H. et al. Constitutive expression of CaSRP1, a hot pepper small rubber particle protein homolog, resulted in fast growth and improved drought tolerance in transgenic Arabidopsis plants. Planta 232, 71–83 (2010). https://doi.org/10.1007/s00425-010-1149-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1149-2