Abstract
The number of different anthocyanin molecules potentially produced by Arabidopsis thaliana and which anthocyanin molecule is the first product of anthocyanidin modification remain unknown. To accelerate the understanding of these questions, we investigated anthocyanin biosynthesis in rosette leaves of both pap1-D and wild-type (WT) A. thaliana plants grown in nine growth conditions, which were composed of three light intensities (low light, middle light, and high light) and three media derived from MS medium (medium-1, 2, and 3). These nine growth conditions differentially affected the levels of anthocyanins and pigmentation patterns of rosette leaves, which were closely related to the diversification levels of cyanin structures. The combined growth conditions of high light and either medium-2 or medium-1 induced the most molecular diversity of anthocyanin structures in rosette leaves of pap1-D plants. Twenty cyanin molecules, including five that were previously unknown, were characterized by HPLC-ESI-MS and HPLC-TOF-MS analyses. We detected that the A. thaliana anthocyanin molecule A11 was most likely the first cyanin derived from the multiple modification steps of cyanidin. In addition, in the same growth condition, rosette leaves of pap1-D plants produced much higher levels and more diverse molecular profiling of cyanins than those of WT plants. The transcript levels of PAP1, PAL1, CHS, DFR, and ANS cDNAs were much higher in pap1-D rosette leaves than in WT ones. Furthermore, on the same agar-solidified medium, an enhancement of light intensity increased levels and molecular diversity of cyanins in both pap1-D and WT rosette leaves. In the same light intensity condition, the responses of anthocyanin levels and profiling to medium alternation were different between pap1-D and WT plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanins are the most abundant plant flavonoid pigments prevalent in vascular plants. Their biosynthetic pathway starting from phenylalanine has been extensively studied in numerous plants (Grotewold 2006; Holton and Cornish 1995; Springob et al. 2003; Winkel-Shirley 2001). From phenylalanine to anthocyanidins, the biosynthetic pathway is catalyzed by multiple enzymes, the number of which varies according to plant species and to the structures of core anthocyanidins (Holton and Cornish 1995; Springob et al. 2003). In general, these enzymes include phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumaroyl: CoA-ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′,5′-hydroxylase (F3′5′H), dihydroflavonol reductase (DFR) and anthocyanidin synthase (ANS, also called leucoanthocyanidin dioxygenase, LDOX). Variations in the presence, substrate recognition, and number of homologs for each anthocyanin biosynthetic enzyme occur in different plant species. The enzyme flavonoid 3′,5′-hydroxylase is absent in some plant species, such as A. thaliana (Fig. 1). The genome of A. thaliana has four PAL and four 4CL gene members (Olsen et al. 2008; Rowan et al. 2009). The Medicago truncatula genome has two DFR gene members, DFR1 and DFR2 (Xie et al. 2004). Based on substrate specificity, DFR can be a dihydrokaempferol reductase (DHK), a dihydroquercetin reductase (DHQ) and/or a dihydromyricetin reductase (DHM). The two DFR homologs of M. truncatula show all three activities to utilize dihydrokaempferol, dihydroquercetin, and dihydromyricetin as substrates (Xie et al. 2004), while Petunia hybrid lacks DHK activity (Meyer et al. 1987). ANS also can function as cyanidin, pelargonidin, or/and delphinidin synthase.
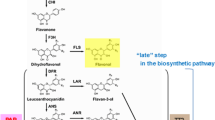
The biosynthetic pathway of anthocyanins in A. thaliana. Enzymes include: PAL phenylalanine ammonia lyase, C4H cinnamate 4-hydroxylase, 4CL 4-coumaroyl: CoA-ligase, CHS chalcone synthase, CHI chalcone isomerase, F3H flavanone 3-hydroxylase, F3′H flavonoid 3′-hydroxylase, DFR dihydroflavonol reductase, ANS anthocyanidin synthase (also called leucoanthocyanidin dioxygenase: LDOX), FLS flavonol synthase, ANR anthocyanidin reductase and 3-GT 3-glucosyltransferase. Transcription factors include: PAP1 and 2: production of anthocyanin pigment 1 and 2, TT1, 2, 8, 16 transparent testa 1, 2, 8, and 16, TTG1 transparent testa glabra 1, MYB 12, 113, and 114 R2R3 MYB 12, 113, and 114. Transporters include TT12 and TT19, transparent testa 12 and 19
Most of the anthocyanin pathway genes in A. thaliana have been cloned and functionally characterized by different approaches. The pathway is summarized in Fig. 1, which shows the properties of anthocyanin biosynthesis in A. thaliana. Based on the current genome sequence information (available on TAIR website, http://www.arabidopsis.org), the PAL gene family includes four members; PAL1 (AT2G37040), PAL2 (AT3G53260), PAL3 (AT5G0423) and PAL4 (AT3G10340). The 4CL gene family has 4 members: 4CL1 (AT1G51680), 4CL2 (AT3G21240), 4CL3 (AT1G65060) and 4CL5 (AT3G21230). Other pathway gene families have been reported to include only one member, i.e., C4H (AT2G30490), CHS (AT5G13930), CHI (AT3G55120), F3H (AT3G51240), F3′H (AT3G51240), DFR (AT5G42800) and ANS/LDOX (AT4G22880) (Rowan et al. 2009). As described above, the A. thaliana genome lacks the F3′5′H gene. 5-O- and 3-O-glycosyltransferase genes and other modification genes of cyanidin are not listed here (Tohge et al. 2005). With the ongoing functional characterization of A. thaliana proteins, more genes are likely to be identified from the genome.
The exact number of anthocyanin molecules that A. thaliana can produce remains unknown. Recently, a few excellent works have identified nearly 21 anthocyanin molecules (including isomers) from this model plant (Bloor and Abrahams 2002; Nakabayashi et al. 2009; Rowan et al. 2009; Tohge et al. 2005). Most of these anthocyanins were isolated from leaves of the production of anthocyanin pigment 1-Dominant (pap1-D) mutants (Tohge et al. 2005). Changes of growth conditions increase the number of anthocyanins in the pap1-D mutants (Rowan et al. 2009). Based on all structures identified so far, A. thaliana anthocyanins are cyanins derived from cyanidin. It is unclear whether other types of anthocyanins (i.e. pelargonins and delphinins) are produced in A. thaliana. The recently identified 21 cyanins are derived from 3-O- and 5-O-glycosylation of cyanidin (Fig. 2) (Rowan et al. 2009; Tohge et al. 2005). Addition of a coumaroyl, sinapoyl, malonyl, or xylosyl group to glucose groups at 2′′-O, 6′′-O, and 6′′′-O increases the structural diversities of cyanins (Fig. 2) (Rowan et al. 2009; Tohge et al. 2005). Other modifications also increase the diversity of cyanin molecules in A. thaliana. For example, a cyanin acyl transferase that stabilizes the coloration in a neutral pH condition was recently identified in this model plant (Luo et al. 2007).
Assigned structures of cyanins are derived from cyanidin via glycosylation at 3-OH and 5-OH in pap1-D and WT plants (Bloor and Abrahams 2002; Rowan et al. 2009; Tohge et al. 2005). A1: R1 –xylosyl, R2 –H, R3 –H (MW: 743) A2: R1 –xylosyl, R2 –H, R3 –malonyl (MW: 829) A3: R1 –xylosyl, R2 –p-coumaroyl, R3 –H (MW: 889) A4: R1 –sinapoyl-xylosyl, R2 –H, R3 –H (MW: 949) A5: R1 –xylosyl, R2 –p-coumaroyl, R3 –malonyl (MW: 975) A6: R1 –xylosyl, R2 –p-coumaroyl-Glc, R3 –H (MW: 1,051) A7: R1 –sinapoyl-xylosyl, R2 –p-coumaroyl, R3 –H (MW: 1,095) A8: R1 –xylosyl, R2 –p-coumaroyl-Glc, R3 –malonyl (MW: 1,137) A9: R1 –sinapoyl-xylosyl, R2 –p-coumaroyl, R3 –malonyl (MW: 1,181) A10: R1 –sinapoyl-xylosyl, R2 –p-coumaroyl-Glc, R3 –H (MW: 1,257) A11: R1 –sinapoyl-xylosyl, R2 –p-coumaroyl-Glc, R3 –malonyl (MW: 1,343) A12: R1 –H, R2 –p-coumaroyl-Glc, R3 malonyl (MW: 1,005) A13: R1 –sinapoyl-hexosyl, R2 –p-coumaroyl-Glc, R3 –malonyl (MW: 1,373) A14: R1 –sinapoyl-xylosyl, R2 –p-coumaroyl-Glc, R3 – methyl-malonyl (MW: 1,357)
PAP1 is an R2R3-MYB factor that is essential for regulating anthocyanin biosynthesis in A. thaliana. This gene was identified by T-DNA activation tagging. The insertion of 4-enhancers of the 35S promoter immediately near the PAP1 gene activated its expression, which resulted in purple pigmentation of A. thaliana (Borevitz et al. 2000). The over expression of the A. thaliana PAP1 gene in tobacco created an anthocyanin pathway leading to formation of new anthocyanins in the transgenic plants (Borevitz et al. 2000; Xie et al. 2006). Furthermore, an ectopic expression of this gene increased anthocyanins in tomato (Zuluaga et al. 2008). Anthocyanin biosynthesis in A. thaliana is highly regulated by multiple regulatory genes. To date, functional genomics approaches have identified several regulatory gene families, which directly or indirectly control the biosyntheses of anthocyanins and procyanidins in A. thaliana (Fig. 1) (Dixon et al. 2005; Grotewold 2006; Lepiniec et al. 2006; Springob et al. 2003). These regulatory protein families include MYB (TT2, PAP1/PAP2, MYB113, MYB114 and MYBL2); bHLH (TT8, GL3 and EGL3); WD40 (TTG1); WKRY (TTG2); WIP (TT1); Homeodomain (ANL2); and bMADS (TT16) (Dixon et al. 2005). In addition, two transcription factors, which are Phytochrome-Interacting Factor 3 (PIF3), a member of bHLH family) and HY5 (Long Hypocotyl5, a member of bZIP factors) involved in light signal transduction, regulates anthocyanin biosynthesis particularly under red and far-red light conditions (Ang and Deng 1994; Kim et al. 2003; Ni et al. 1998; Shin et al. 2007). In vitro binding and in vivo CHIP experiments demonstrated that the two transcription factors regulated anthocyanin biosynthesis via binding promoters of several pathway genes in a complementary manner and likely formed a complex (Shin et al. 2007; Song et al. 2008).
Multiple reports have shown that anthocyanin biosynthesis in A. thaliana can be induced by different abiotic and biotic factors that either induce or increase the expression of the PAP1 gene (Lea et al. 2007; Lillo et al. 2008; Rowan et al. 2009; Teng et al. 2005). Transcription of the PAP1 gene strongly responds to all environmental stimulations investigated so far (Lillo et al. 2008). Nitrogen deficiency increased the mRNA levels of the PAP1 gene in both wild-type and pap1-D seedlings sixfold (Lea et al. 2007). Deprivation or a low concentration of phosphorus in growth media resulted in an increase of anthocyanins in seedlings and increased the transcript level of the PAP1 gene and its homolog PAP2 (Lillo et al. 2008; Misson et al. 2005; Morcuende et al. 2007; Muller et al. 2007). Induction of anthocyanin by sucrose also correlated positively with PAP1 transcript levels in seedlings (Lillo et al. 2008; Solfanelli et al. 2006; Teng et al. 2005).
Other factors that control the regulatory function of PAP1 remain unknown. To study these factors under controlled conditions, we established two types of epigenetically stable cells lines in vitro from the same transgenic tobacco leaf (Zhou et al. 2008). RT-PCR analysis showed that the two cell lines highly expressed the PAP1 transgene in a similar level. However, the anthocyanin biosynthesis in the two cell lines was obviously different. One cell line is phenotypically red because of extremely high production of anthocyanins resulting from the high expression of the PAP1 transgene. The other one is phenotypically white due to low production or lack of anthocyanins in spite of the high expression of the PAP1 transgene. These data demonstrates that the regulatory function of the PAP1 gene in anthocyanin biosynthesis is dependent upon cell types. More importantly, the regulatory function of the PAP1 gene on anthocyanin biosynthesis was regulated by auxins, nitrogen nutrients, and light conditions (Zhou et al. 2008). Another excellent recent study also reported that a high temperature and low light intensity growth condition led to less production of cyanins in leaves, although the transcript level of the PAP1 gene was greatly increased, suggesting that the regulatory function of PAP1 required the involvement of other factors (Rowan et al. 2009).
The recent work completed by Rowan et al. (2009) excellently showed that anthocyanin biosynthesis activated by overexpression of the PAP1 gene was controlled by changes of light and temperature conditions. To further understand the regulation of anthocyanin biosynthesis by other environmental factors, we here report features of the anthocyanin biosynthesis in rosette leaves of both pap1-D and wild-type plants grown in nine growth conditions consisting of three light intensities in combination with three media containing different nitrogen nutrient concentrations. Both HPLC-ESI-MS and HPLC-TOF-MS were employed to characterize cyanin molecules. Semi-quantitative RT-PCR analysis was carried to characterize the transcript levels of the PAP1 gene and four pathway genes. Our study suggests that the biosynthesis and diversity of cyanin molecules regulated by the transcription of the PAP1 gene is strongly dependent upon these growth conditions.
Materials and methods
Chemicals
Pelargonidin, cyanidin and delphinidin standards were purchased from Indofine Company (Hillsborough, NJ, USA). Hydrochloric acid (36.5–38%) was purchased from BDH (cat#: BHH3028-2.5L, West Chester, PA, 19380, USA). Acetonitrile (LC-MS grade) was purchased from EMD (cat#: AX0156-1, Gibbstown, NJ 08027, USA). Acetic acid (HPLC grade, cat#: 9515-03) and methanol (LC-MS grade, cat#: 9830-03) were purchased from J. T. Baker (Phillipsburg, NJ 08865, USA). All macronutrients, micronutrients and organic nutrients used in MS medium (Murashige and Skoog 1962), sucrose and phytoagar were purchased from Plant Media Company (Dublin, OH, USA).
Medium preparation
Medium-1 (M-1) was basal MS medium (with full strength of 20 mM NH4NO3 and 18.8 mM KNO3 (Murashige and Skoog 1962). Medium-2 (M-2) was MS medium with 10 mM NH4NO3 and 9.4 mM KNO3 (also called as 1/2 strength). Medium-3 (M-3) was MS medium with 9.4 mM KNO3 but 0 mM NH4NO3 (also called 1/2 strength KNO3 and no NH4NO3).
All media used in this study contained 3% sucrose and were solidified with 0.8% phytoagar (g/100 ml). All media were adjusted to pH 5.8 and then autoclaved for 15 min. Twenty milliliter of medium was poured into one petri dish (15 × 100 mm, height × diameter in size) and then cooled down to room temperature for both seed germination and seedling growth.
Light conditions
Three light conditions were used in this study. Low light (LL) condition was 16/8 h (light/dark) of lighting with a light intensity of 50 μmol m−2 s−1. Middle light (ML) condition was 24 h of continuous lighting with a light intensity of 200 μmol m−2 s−1. High light (HL) condition was 24 h continuous lighting with a light intensity of 500 μmol m−2 s−1. The light resource was a mixture of fluorescent and incandescent lights.
Growth conditions, seed germination, seedling growth, and harvest of leaves
Seeds of the A. thaliana production of anthocyanin pigment1-Dominant (pap1-D) mutant were purchased from ABRC for this study. Seeds of wild-type A. thaliana ecotype Columbia (Col-0) were harvested by us as controls. Seeds were cold treated for 5 days in a 4°C refrigerator. Surfaces of seeds were sterilized by using 70% alcohol for 1 min followed by three washes with autoclaved MiniQ water. For further sterilization, these surface-cleaned seeds were treated with 10% Clorox (a regular bleach product) for 8 min and then washed 6 times with autoclaved MiniQ water.
Ninety-six seeds were equally planted in eight petri dishes containing agar-solidified medium and tested for each growth condition (Table 1). Seed germination and seedling growth were compared in nine conditions (Table 1). The temperature used in all growth conditions was 22 ± 0.5°C. The numbers of seed germinating and surviving seedlings were recorded for each growth condition. The rates of seed germination, seedling survival, and flowering were calculated. Rosette leaves of 20-day-old plants grown in each condition were harvested into a 15 ml plastic tube and immediately frozen in the liquid nitrogen and then stored at −80°C until analysis. This experiment was repeated twice.
Reverse transcription-polymerase chain reaction
Frozen rosette leaves were ground into a fine powder in liquid nitrogen. We used 0.1 g powder and Qiagen RNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) to isolate total RNA and then utilized on-column DNase treatment (Qiagen product) to remove DNA from RNA samples by following this manufacturer’s protocols. For each sample, four μg of total RNA was used as a template to synthesize the first strand cDNA. We used Ready-to-Go T-Primed First-Strand Kit (Amersham Biosciences, Piscataway, NJ, USA) to carry out reverse transcription and obtained the first strand cDNA by following this manufacturer’s protocol.
Gene specific primers were designed to amplify cDNA fragments of PAL1, CHS, DFR, ANS, PAP1 and ACTIN genes (Table 2). Polymerase chain reaction (PCR) of 25 cycles was performed on a thermocycler (Mastercycler®-Ep, Eppendorf). The primers and thermal gradient programs for each gene are included in Table 2. PCR products were examined by 0.8% agarose gel electrophoresis and imaged with a Bio-Rad imaging system.
The expression level of each gene was normalized by using ACTIN transcriptional level as a reference in a semi-quantitative analysis on QuantityOne gel image software (Bio-Rad) (Supplementary data).
Extraction of anthocyanin and absorbance measurements
One hundred mg powder of leaves was suspended in 1 ml extraction solvent (0.5% HCl in methanol 100%) in a 1.5 ml extraction tube. The tube was vigorously vortexed for 30 s, sonicated for 15 min and then centrifuged for 15 min at 10,000 rpm. The supernatant was transferred into a new 2.0 ml tube. Another 1 ml extraction solvent was added to the extraction tube to re-suspend the pellet and the extraction steps were repeated. The two methanol extractions were pooled into the 2.0 ml tube.
To remove chlorophyll in the extraction, 500 μl out of the 2 ml methanol extraction was mixed with 500 μl MiniQ H2O and 300 μl chloroform in a 1.5 ml tube. The mixture was vortexed vigorously for 30 s and centrifuged at the speed of 10,000 rpm for 5 min, then the upper water–methanol phase containing red pigment was pipetted into a new tube. The bottom chloroform phase containing chlorophyll was disposed into a waste container. These steps were repeated once and the upper red phase was stored at −20°C for anthocyanin analysis. For each sample, three replicates were conducted in this experiment.
The absorbance (ABS) of methanol–water phase extracts was recorded at the wavelength of 530 nm on a HEλIOSγ UV–Visible spectrophotometer. The extraction buffer was used as a control blank. A volume of 600 μl methanol–water phase extract was measured to obtain an ABS value. Authentic standard cyanidin was used to establish a standard curve. Total anthocyanin content in rosette leaves was estimated as cyanin equivalent (μg/g) according to the standard curve.
Hydrolysis of anthocyanins
Two hundred microliters of anthocyanin extraction was added into 800 μl of n-butanol: HCl (95:5, v/v) solvent contained in a 1.5 ml tube. This mixture was boiled for 1 h. After the sample was cooled down to room temperature, it was dried off with flow nitrogen gas. The remaining residue was suspended in 100 μl of 0.1% HCl-methanol. The sample was centrifuged at 10,000 rpm for 15 min, and then the supernatant was transferred to a new 1.5 ml tube for HPLC-ESI-MS and HPLC-TOF-MS analyses.
HPLC-UV-ESI-MS analysis of anthocyanins
The analysis of anthocyanin profiles was carried out by using high performance liquid chromatography/mass spectrometry on 2,010 eV LC/UV/ESI/MS instrument (Shimadzu). Samples were separated on an analytical column of Eclipse XDB-C18 (250 mm × 4.6 mm, 5 μm, Agilent). The mobile phase solvents were composed of 1% acetic acid in water (solvent A: 1% HPLC grade acetic acid in LC-MS grade water) and 100% acetonitrile (solvent B) (LC-MS grade). The column was treated with a gradient solvent system that was designed to separate metabolites and was composed of ratios of solvent A to B: 90:10 (0–5 min), 90:10 to 88:12 (5–10 min), 88:12 to 80:20 (10–20 min), 80:20 to 75:25 (20–30 min), 75:25 to 65:35 (30–35 min), 65:35 to 60:40 (35–40 min), 60:40 to 50:50 (40–55 min) and 50:50 to 10:90 (55–60 min). The gradient step was followed by a 10 min column wash with 10% of solvent B. The flow rate was 0.4 ml/min and the injection volume was 20 μl. The UV spectrum was recorded from 190 to 800 nm. The total ion chromatograms of positive electrospray ionization were recorded from 0 to 60 min by mass spectrum detector and the mass spectrum was scanned and stored from m/z of 120–1,600 at a speed of 1,000 amu per second. Pelargonidin, cyanidin and delphinidin were used as authentic standards.
HPLC-TOF-MS analysis of anthocyanins
For structural characterization of anthocyanins, samples were analyzed on an Agilent Technologies (Santa Clara, CA, USA) 6210 time-of-flight LC/MS. The HPLC gradient program, the separation column and the flow rate were the same as described above. The injection volume of samples was 10 μl. The drying gas flow was set to 12 l/min, and the nebulizer pressure was set to 35 psi.
Statistical analysis
To compare the effects of the nine growth conditions on anthocyanin biosynthesis, we used student’s T test to statistically compare contents of total anthocyanins and peak area values of the individual anthocyanin molecules detected by LC and MS analyses.
Results
Different anthocyanin pigmentation patterns in rosette leaves of pap1-D and WT plants
Under the same growth conditions, rosette leaves, stems, and roots of pap1-D plants consistently showed much more intense anthocyanin pigmentation than those of WT plants (Fig. 3a). For example, in the LLM-1 (50 μmol m−2 s−1 of light intensity and medium-1) condition, WT plants completely lacked anthocyanin pigmentation in all tissues, whereas pap1-D plants accumulated obvious red pigment on petioles of rosette leaves and roots. In the condition of MLM-1 (200 μmol m−2 s−1 of light intensity and medium-1) condition, WT plants showed only weak pink pigmentation in petioles of rosette leaves (Fig. 3a-D), whereas pap1-D plants were very purple (Fig. 3a-M).
Impacts of nine growth conditions on anthocyanin pigmentation and five gene expression levels in rosette leaves of pap1-D and WT plants. a Phenotypes of WT (A–I) and pap1-D (J–R) plants grown in nine conditions. Bar 0.5 cm. Growth conditions: A and J: LLM-1 (low light: 50 μmol m−2 s−1 of light intensity and medium-1: full strength of basal MS medium), B and K: LLM-2 (low night and medium-2: ½ strength of NH4NO3 and KNO3 in MS medium), C and L: LLM-3 (low light and medium-3: 0 NH4NO3 and ½ strength of KNO3 in MS medium), D and M: MLM-1 (middle light: 200 μmol m−2 s−1 of light intensity and medium-1), E and N: MLM-2, F and O: MLM-3, G and P: HLM-1 (high light: 500 μmol m−2 s −1 of light intensity and medium-1), H and Q: HLM-2, I and R: HLM-3. b Gel images show the effects of three light intensities on transcriptional levels of five genes in rosette leaves of both wild-type (WT) (a-B, E, I) and pap1-D plants (a-K, N, Q) grown on agar-solidified MS medium-2. Due to poor growth of wild-type plants in the condition of HLM-2, total RNA isolated from rosette leaves of WT plants (a-I) grown in the condition of HLM-3 were used as a control. c Gel images show the effects of three media on transcript levels of five genes in rosette leaves of both WT (a-D, E) and pap1-D (a-M, N, O) plants grown in the ML condition. RT-PCR analysis for WT plants grown in the MLM-3 condition (a-F) is not shown due to low yield and poor quality of total RNA from rosette leaves. Five genes tested include PAP1: Production of Anthocyanin Pigment1; PAL1: Phenylalanine Ammonia-Lyase 1; CHS: Chalcone Synthase; CHI: Chalcone Isomerase; DFR: Dihydroflavonol Reductase; and ANS: Anthocyanidin Synthase. (Conditions N and E are used in b and c to help compare band intensity in different growth conditions directly and easily)
Enhancing light intensity increased red/purple pigmentation in rosette leaves of both pap1-D and WT plants on any given agar medium. For example, on medium-2, the HL condition induced the most intense pigmentation in rosette leaves of pap1-D plants (Fig. 3a-Q), followed by ML and then LL conditions (Fig. 3a-N, K). The same trend of pigmentation patterns was also observed in rosette leaves of WT plants grown in three light conditions, in the order of HL (Fig. 3a-H), ML (Fig. 3a-E), and then LL (Fig. 3a-B). In addition, on medium-1 and -3, the trends of light intensity effects on anthocyanin pigmentation patterns in rosette leaves were similar to those observed on medium-2 (Fig. 3a).
Under the same light conditions, the pigmentation patterns in rosette leaves of pap1-D and WT responded differently to medium changes. Decreasing total nitrogen nutrient concentration (NH4NO3 and KNO3) in medium (from medium-1 to medium-3) led to an obvious reduction in anthocyanin pigmentation intensity in rosette leaves of pap1-D plants (Fig. 3a, J–L, M–O, Q, R). In contrast, three obvious types of pigmentation pattern trends were observed in rosette leaves of WT plants under the same conditions. The reduction of total nitrogen (from medium-1 to medium-3) (1) induced the formation of anthocyanin 11 (A11) (Fig. 7a-A–C), although red pigmentation patterns were not obviously enhanced (Fig. 3a-A–C) and total levels of anthocyanins measured at 530 nm did not show significant increases under LL (Fig. 4); (2) decreased anthocyanin pigmentation under ML (Fig. 3a-D–F) and (3) increased intensity of pigmentation (from M-1 to M-2) and then decreased anthocyanin pigmentation (from M-2 to M3) in HL condition (Fig. 3a-G–I).
Total levels of anthocyanins in rosette leaves of pap1-D and WT plants
In the nine growth conditions tested, the levels of anthocyanins were in the range of 0.18–1.5 mg/g FW (fresh weight) in the pap1-D rosette leaves and in the range of 0–0.6 mg/g FW in the WT rosette leaves (Fig. 4). In the conditions of HLM-1 or HLM-2, the anthocyanin levels reached the maximum values of nearly 1.5 mg/g FW in rosette leaves of pap1-D plants (Fig. 4).
Under growth conditions identical to those shown in Fig. 3a, the total levels of anthocyanins were significantly higher in pap1-D rosette leaves than in WT ones (Fig. 4). In LL growth conditions, total levels of anthocyanins in pap1-D rosette leaves were approximately 20–40-fold higher than those in WT rosette leaves across all growth media. It was very interesting that when light intensity was increased, the difference in anthocyanin levels between pap1-D and WT rosette leaves was obviously reduced. In all ML growth conditions, levels of anthocyanins in pap1-D rosette leaves were nearly 5–9-fold more than the ones in WT rosette leaves. In the HL growth conditions, levels of anthocyanins in pap1-D rosette leaves were only 2–5-fold more than the ones in WT rosette leaves.
Transcript levels of PAP1 and four pathway genes in pap1-D and WT plants
Total RNA was isolated from rosette leaves of 20-day-old pap1-D plants grown in 8 growth conditions and 20-day-old WT plants grown in six conditions. We could not obtain good quality RNA from both pap1-D and WT plants grown under the HLM-1 conditions due to unhealthy or dead rosette leaves (Fig. 3a-G, P). Consequently, RT-PCR analysis could not be performed to evaluate gene expression levels in either pap1-D or WT rosette leaves for these conditions. In addition, we could not isolate good quality RNA from WT plants grown under either conditions of HLM-2 (Fig. 3a-H) or MLM-3 (Fig. 3a-F) to perform RT-PCR analysis. Expression levels of PAP1 and four pathway genes (PAL1, CHS, DFR and ANS) were examined by semi-quantitative RT-PCR analysis to evaluate their transcriptional responses to different conditions. ACTIN was used as a housekeeping gene control to evaluate mRNA quality and normalize expression levels of these five specific genes across different samples.
RT-PCR analysis showed that the transcriptional levels of PAP1 were much higher in rosette leaves of pap1-D plants than in those of WT plants in all growth conditions tested (Fig. 3b, c; S-Fig. 1 a, b). Under the same growth conditions, the transcriptional levels of CHS, DFR and ANS were obviously higher in rosette leaves of pap1-D plants than in those of WT plants (Fig. 3b, c; S-Fig. 1a, b). For PAL1, except for in the LLM-1 growth condition, its expression levels were higher in rosette leaves of pap1-D than in those of WT plants (Fig. 3b, c; S-Fig. 1a, b).
Semi-quantitative RT-PCR analysis and gel images showed that three light conditions differentially affected gene expression in rosette leaves of plants grown on the same medium. On medium-2, in pap1-D rosette leaves, we observed that the expression levels of the five genes were higher in the ML (200 μmol m−2 s−1 of light intensity) condition than in both HL (500 μmol m−2 s−1 of light intensity) and LL (50 μmol m−2 s−1 of light intensity) conditions (S-Fig. 1a). In WT rosette leaves, on medium-2, we could only compare the effects of LL and ML conditions on gene transcripts, due to low yield and poor quality of total RNA isolated from samples harvested under the HL condition. Gel images and semi-quantitative analysis showed that the expression levels of PAP1, DFR, and ANS genes were higher under the ML condition than under the LL condition; the expression level of CHS was similar under two conditions; in contrast, the expression level of PAL1 was slightly lower under the ML condition than under the LL condition (Fig. 3b; S-Fig. 1 a). To show the effect of HL on gene expression, we included the HLM-3 condition to compare with the MLM-2 as well as the LLM-2 conditions and found that the transcriptional levels of the PAP1, CHS, DFR, and ANS were higher in the former condition than in both of the latter conditions (Fig. 3b; S-Fig. 1a).
Under the same light condition, the three media differentially affected transcription of the five genes in rosette leaves of both pap1-D and WT plants. We chose the ML condition (200 μmol m−2 s−1 of light intensity) to compare effects of these three lighting conditions on gene expression. In rosette leaves of pap1-D plants, the transcriptional levels of the five genes were the highest on medium-2, followed by medium-1 and then medium-3 (Fig. 3c; S-Fig. 1 c). In rosette leaves of WT plants, RT-PCR analysis was carried out for plants grown on medium-1 and medium-2. The transcriptional levels of the five genes were higher on medium-2 than on medium-1 (Fig. 3c; S-Fig. 1b).
Cellular localization of anthocyanins
To examine the cellular localization of anthocyanins, cross sections were made of leaves from pap1-D and WT plants grown in HL conditions. Microscopic observation showed that anthocyanins were mainly accumulated in the epidermis of both adaxial and abaxial sides of leaves, in the outermost layer of mesophyll cells (Fig. 5a), and in the vascular tissues (Fig. 5d). Unlike PAP1 transgenic tobacco trichomes described previously (Xie et al. 2006), the trichomes on rosette leaves of A. thaliana plants did not contain anthocyanins (Fig. 5c). In WT rosette leaves, only epidermal cells showed a very weak red coloration, but none of the other types of cells exhibited red pigmentation (Fig. 5b).
Levels of anthocyanins in rosette leaves of 20-day-old pap1-D and WT plants grown in nine conditions consisting of three different light intensities (50, 200, and 500 μM m−2 s−1) in combination with three different media (M-1, M-2, and M-3). LLM-1 low light: 50 μmol m−2 s −1 of light intensity and medium-1: full strength of basal MS medium; LLM-2 low night and medium-2: ½ strength of NH4NO3 and KNO3 in MS medium; LLM-3 low light and medium-3: 0 NH4NO3 and ½ strength of KNO3 in MS medium; MLM-1 middle light: 200 μmol m−2 s−1 of light intensity and medium-1; MLM-2 middle light and medium-2; MLM-3 middle light and medium-3; HLM-1 high light: 500 μmol m−2 s −1 of light intensity and medium-1; HLM-2 high light and medium-2; HLM-3 high light and medium-3; x no significant difference in these three measurements, y no significant difference between the two measurements
Molecular properties of anthocyanins
HPLC-UV-ESI-MS was used to profile anthocyanins in plants grown under all nine conditions. The growth condition of HLM-2 (500 μmol m−2 s−1 of light intensity and medium-2) maximized the peak numbers of anthocyanins extracted from rosette leaves of both pap1-D (Fig. 6a) and WT plants. Thus, we used HPLC-UV-ESI-MS and HPLC-TOF-MS to analyze samples from this growth condition to characterize most of the anthocyanins detected.
Microscopic images show the cellular localization of cyanins in pap1-D and WT leaf blades grown in the condition of HLM-2 (500 μM m−2 s−1 of light intensity and medium-2: ½ strength of both NH4NO3 and KNO3 in MS medium) (Fig. 3a-Q). a A cross section of a pap1-D leaf, b a cross section of a WT leaf, c leaf trichomes from rosette leaves of pap1-D plants, d a cross section from veins showing red vascular tissues in a pap1-D leaf. V vascular tissues, bar length 0.5 mm (c) and 50 μm (a, b, d)
Twenty peaks were identified in methanolic extraction by HPLC profiling at 530 nm (Fig. 6a). Anthocyanins were characterized on the basis of HPLC profiles, UV absorbance, MS fragments and previous records (Table 3, S-Fig. 2a, b). Cyanins including A2–A11, A13, cis-A5, cis-A7 and cis-A9 that were detected in pap1-D plants by two previous studies (Tohge et al. 2005; Rowan et al. 2009) were confirmed to be produced in pap1-D plants by our study (Table 3). However, we could not detect A1, m/z = 743 [M]+ or A12, m/z = 1,005 [M]+ in our materials.
Five new anthocyanins were identified in pap1-D plants, and four of them are likely methyl esters of the reported anthocyanins in pap1-D. In our current study, we give these five anthocyanins names: A14, m/z = 1,357 [M]+; A15, m/z = 1,195 [M]+; A16, m/z = 989 [M]+; A17, m/z = 1151 [M]+; and A18, m/z = 1,035 [M]+. Based on these MS analyses, we predicted that A14 was a methyl ester of A11, m/z = 1,343 [M]+; A15 was a methyl ester of A9, m/z = 1,181 [M]+; A16 was a methyl ester of A5, m/z = 975 [M]+; and A17 was a methyl ester of A8, m/z = 1,137 [M]+.
Cyanidin is the major anthocyanidin in A. thaliana
Butanol: HCl hydrolysis of anthocyanin extraction was carried out via boiling for 1 h to release core anthocyanidins. HPLC analysis showed that cyanidin formed the predominant red chromophore molecule in rosette leaves of both pap1-D and WT plants (Fig. 6b). MS analyses confirmed its mass (m/z = 287 [M]+) and chemical formula as that of cyanidin. A second minor peak was also detected at retention time 38 min from this hydrolysis (Fig. 6b). The visible wavelength of its maximum absorbance was 522 nm. Single quadrupole MS analysis showed that the second peak had three major masses including 282, 329, and 595 (m/z) [M]+. But HPLC-TOF-MS analysis could not give us a correct chemical formula; thus its structure could not be predicted in this study. Neither delphinidin nor pelargonidin was found from this hydrolysis of anthocyanin extraction.
Responses of cyanin levels and profiles to medium alternation in the same light condition
In each light condition, we compared the effects of the three media (medium-1: 20 mM NH4NO3 and 18.8 mM KNO3; medium-2: 10 mM NH4NO3 and 9.4 mM KNO3; and medium-3: 9.4 mM KNO3 and 0 mM NH4NO3) on the biosynthesis of cyanins. Changes in the cyanin levels and profiles were observed in rosette leaves of both pap1-D and WT plants according to the media used.
First, in the LL (50 μmol m−2 s−1 of light intensity) condition, responses of cyanin levels to the three media were different between pap1-D and WT rosette leaves (Fig. 4). Progressing from medium-1 to medium-3, the cyanin levels were slightly increased in rosette leaves of WT plants (Fig. 4) but significantly reduced in leaves of pap1-D plants (Fig. 4). These two cyanin level trends were confirmed by HPLC analysis. On agar medium-1, in rosette leaves of WT plants, none of cyanins was detected at 530 nm (Fig. 6a-A); on agar medium-2 and medium-3, A11 formed the only peak detected at 530 nm (Fig. 7a-B, C). In contrast, from the rosette leaves of pap1-D plants, on medium-1, 11 peaks of cyanins, including A5, A8, A9, A10, A11 and A14, were detected by HPLC (Fig. 7b-J). The area values of these 11 cyanin peaks were reduced in samples from medium-2 and 3 (Fig. 7b-K, L).
HPLC profiles of cyanins and cyanidins extracted from rosette leaves of plants. a An HPLC cyanin profile shows 20 cyanin peaks detected at 530 nm from HCl-methanol extraction of rosette leaves of pap1-D seedlings grown in the condition of HLM-2 (500 μmol m−2 s−1 of light intensity and medium-2). b HPLC profiles show anthocyanidins released by butanol-HCl hydrolysis of cyanin extracts from rosette leaves of 20-day-old pap1-D (b-1) and WT (b-2) plants and standards (b-3)
Second, in the ML (200 μmol m−2 s−1 of light intensity) condition, the correlation of cyanin levels to medium composition was similar in rosette leaves of pap1-D and WT plants. Progressing from medium-1 to medium 3, a dramatic reduction trend of cyanin levels was observed in rosette leaves (Fig. 4). In comparison with medium-1 (Fig. 4), medium-2 and medium-3 led to nearly 1.3- and 6-fold reductions, respectively, in the total cyanin levels in rosette leaves of pap1-D plants, and nearly 1.5- and 5-fold decreases in the total cyanin levels in rosette leaves of WT plants (Fig. 4). To support these observations, we analyzed cyanin profiles by HPLC (Fig. 7a-D–F; b-M–O). Five peaks of cyanins including A10, A11 and A14 were detected from rosette leaves of WT plants on agar medium-1, but the peak area values of the five cyanins were dramatically reduced on agar medium-2 (Fig. 7a-E, F), and two of them were not detected on medium-3 (Fig. 7a-F). Likewise, nearly 16 peaks of cyanins were detected from rosette leaves of pap1-D plants on medium-1 (Fig. 7b-M); however, the area values of the 16 cyanins were all reduced on medium 2 (Fig. 7b-N) and several cyanins were not detected on medium-3 (Fig. 7b-O).
Third, in the HL (500 μmol m−2 s−1 of light intensity) condition, pap1-D and WT plants showed changes in cyanin levels with changes in media. On the one hand, in WT rosette leaves, the maximum level of total anthocyanins was obtained on medium-2, followed by on medium-1 and then on medium-3 (Fig. 4). HPLC profiling analysis showed nearly 11 cyanin peaks including A5, A8, A9, A10, A11, and A14 in rosette leaves of WT plants on medium-2 (Fig. 7a-H); however, these peak areas were dramatically reduced on medium-1 and -3 (Fig. 7a-G, I). On the other hand, in pap1-D rosette leaves, cyanin levels were similar in plants grown on both medium-1 and -2, significantly higher than on medium-3 (Fig. 4). HPLC profile analysis showed 20 peaks of cyanins in rosette leaves of pap1-D plants on medium-1 and -2 (Figs. 6a; 7b-P, Q). However, the peak area values of all detected cyanins were reduced on medium-3 (Fig. 7b-R).
An enhancement of light intensity increases cyanin levels and profiles on the same medium
On medium-1, in comparison with LL (Fig. 4), ML and HL increased the total levels of cyanins in rosette leaves nearly 6- and 9-fold, respectively, from pap1-D plants and nearly 22- and 46-fold from WT plants (Fig. 4). These increases in cyanin levels likely resulted from induction and diversification of the new cyanin molecules that were confirmed by HPLC profiling analysis. First, in WT rosette leaves, in the LL condition, not a single cyanin peak was detected at 530 nm (Fig. 7a-A); in the ML condition, three major cyanins, A10, A11, and A14, were identified (Fig. 7a-E); in the HL condition, 10 cyanin peaks, including A5, A8, A9, A10, A11, and A14 molecules, were identified, and these peak areas were significantly increased in comparison with LL and ML conditions (Fig. 7a-G). Second, in pap1-D rosette leaves, in the LL condition, at least 10 peaks of cyanins, including A5, A8, A9, A10, A11, and A14 molecules, were detected at 530 nm (Fig. 7b-J); in comparison with the LL condition (Fig. 7b-J), the ML condition significantly increased the peak areas of all detected cyanins (Fig. 7b-M); in comparison with both the LL and ML conditions, the HL condition maximized the peak areas of all detected cyanins (Fig. 7b-P) and increased the peak numbers up to 20 (Fig. 6a).
On medium-2, in comparison with LL, ML and HL increased the total levels of cyanins in rosette leaves nearly 6- and 11-fold, respectively, from pap1-D plants and nearly 20- and 139-fold from WT plants (Fig. 4). These increases of cyanin levels also likely resulted from both the formation of new cyanins and the increase of individual cyanin levels, as demonstrated by the HPLC profile analysis. First, in rosette leaves of WT plants, under the LL condition, A11 was the only detected cyanin molecule (Fig. 7a-B); in the ML condition, A10, A11, and A14 were detected (Fig. 6b-E); in the HL condition, six major cyanins, A5, A8, A9, A10, A11, and A14, were detected along with several other minor peaks at 530 nm (Fig. 7a-H). In addition, in comparison with the LL condition (Fig. 7a-B), both ML and HL conditions increased the peak area values of A11 dramatically (Fig. 7a-E, H). Second, in rosette leaves of pap1-D plants, in comparison with LL (Fig. 7b-K), both ML and HL increased both peak numbers and areas of cyanins (Fig. 7b-N, Q). In the HL condition, 20 anthocyanin peaks were identified by HPLC analysis (Fig. 6a).
On medium-3, in comparison with LL, ML and HL increased the total levels of cyanins in rosette leaves nearly 1.5- and 9-fold, respectively, from pap1-D plants, and nearly 3- and 30-fold from WT plants. The enhancement of light intensity increased both peak areas and peak numbers of cyanins in rosette leaves of both pap1-D (Fig. 7b-L, O, R) and WT (Fig. 7a-C, F, I) plants.
Discussion
Is the structure diversification of A. thaliana cyanins a programmed consequence?
Many questions relating to the modification and molecular diversity of anthocyanins are unanswered. One of the most important ones is whether such multi-step modifications of anthocyanins are synergistically programmed at the transcriptional level for genes encoding enzymes. Specifically, the initial anthocyanin molecule derived from structural modification in different plant species remains unclear. One challenge is the isolation of each anthocyanin molecule in the pathway and ordering them within it. Intensive studies of A. thaliana cyanins lay a foundation to understand the cyanidin modification process. The biosynthesis of cyanins in A. thaliana is very well understood from the results of numerous studies (Borevitz et al. 2000; Gonzalez et al. 2008; Kubasek et al. 1992; Kubo et al. 1999; Lloyd et al. 1992; Pourcel et al. 2009; Rowan et al. 2009). Recent efforts have resolved 21 cyanin peaks, mainly from pap1-D plants. Thirteen of these have been structurally identified as resulting from glycosylation at 3- and 5-OH sites with further additions of sinapoyl, xylosyl, p-coumaroyl, malonyl, etc., groups (Nakabayashi et al. 2009; Rowan et al. 2009; Tohge et al. 2005). In our current report, we have detected 5 new cyanin peaks, increasing the number of known cyanin molecules to 26 in total. In addition, more cyanin molecules are likely to be produced under different growth conditions.
Growth of plants under nine conditions allowed us to investigate the first cyanin formed in rosette leaves. Rowan et al. (2009) found that the cyanin A11was the most abundant molecule and its level was altered by changes of temperature and light conditions. In our study, we found that the molecule A11 most likely was the first cyanin directly resulting from the modification of cyanidin in WT rosette leaves in the growth conditions tested. A11 (Fig. 2) is cyanidin 3-O-[2′′-O-(6′′′-O-(sinapoyl) xylosyl-) 6′′-O-(p-O-(glucosyl)-p-coumaroyl) glucoside] 5-O-(6′′′′-O-malonyl) glucoside, which is predicted to be the product of 7 enzymatic reactions, including three glycosylation reactions and the additions of xylosyl, sinapoyl, coumaroyl, and malonyl groups. We found that in the condition of LLM-1 (50 μmol m−2 s−1 of light intensity and medium-1), rosette leaves of 20-day-old WT plants lacked cyanins. Growth conditions significantly affected cyanin profiles and molecular diversity in WT rosette leaves (Fig. 7a). In the LL condition, when total nitrogen concentration used in the growth medium was reduced twofold in medium-2 or even more in medium-3, a small peak of A11 was the only cyanin detected from the extraction of WT rosette leaves (Fig. 7a-B, C). When WT plants were grown in the condition of MLM-3 (200 μmol m−2 s−1 of light intensity and medium-3), cyanins A10, A11, and A14 were detected at 530 nm, among which A11 formed the main cyanin molecule (Fig. 7a-F). Based on peak area values, A14 was the one likely formed after A11 by cells, then A10 (Fig. 7a-F). In addition, A11 was one of the major cyanin molecules in leaves of pap1-D plants grown in all of nine growth conditions (Fig. 7b-J–R). For example, on medium-1, in pap1-D rosette leaves, A11 accounted for 51.2, 59.4, and 39.4%, respectively, of total cyanin levels in the HL, ML, and LL conditions. In addition, in WT rosette leaves, A11 constituted 62.5 and 67.5% of total cyanin levels in the HL and ML conditions, respectively. In support of our current findings, A11 was found to be the only cyanin in WT leaves grown on an agar-solidified MS medium (Tohge et al. 2005). This A11 cyanin is derived from glycosylation of both 3- and 5-OH of cyanidin and is catalyzed by both flavonoid-3-O-glucosyltransferase and anthocyanin-5-O-glucosyltransferase, which are encoded by UGT78D2 and UGT75C1, respectively (Tohge et al. 2005). Mutation of either these two genes led to the lack of or reduction in A11 levels in the leaves of mutants (Tohge et al. 2005). It is very interesting that A11 has not been found in pap1-D roots (Tohge et al. 2005), indicating that the first cyanin molecule derived from cyanidin modification differs by tissue.
HPLC profiles at 530 nm show major cyanins extracted from rosette leaves of WT (a) and pap1-D (b) plants grown in nine different conditions. A5, A8, A9, A10, A11, and A14 are structurally assigned cyanin molecules. GC Growth conditions, LLM-1 low light: 50 μmol m−2 s −1 of light intensity and medium-1; full strength of basal MS medium; LLM-2 low night and medium-2: ½ strength of NH4NO3 and KNO3 in MS medium; LLM-3 low light and medium-3: 0 NH4NO3 and ½ strength of KNO3 in MS medium; MLM-1 middle light: 200 μmol m−2 s−1 of light intensity and medium-1; MLM-2 middle light and medium-2; MLM-3 middle light and medium-3; HLM-1 high light: 500 μmol m−2 s−1 of light intensity and medium-1; HLM-2 high light and medium-2; HLM-3 high light and medium-3
Properties of PAP1’s regulation on cyanin biosynthesis in A. thaliana
The regulation of PAP1 on cyanin biosynthesis is dependent upon tissue specificity, cell types, environmental conditions, etc. In rosette leaves of pap1-D plants, although the transcriptional levels of the PAP1 gene were uniformly increased (Fig. 3b, c), the cyanins were only localized in epidermal cells, the outmost layers of mesophyll cells, and vascular tissues (Fig. 5). A knockout of the PAP1 gene in A. thaliana led to the loss of cyanin pigmentation in rosette leaves and other vegetative tissues but did not affect procyanidins in seeds, indicating that cyanin biosynthesis in seed coats was likely independent of PAP1 regulation (Gonzalez et al. 2008). Our previous transgenic study demonstrated that anthocyanin accumulation was localized in epidermal and hypodermal cells, trichomes and parenchyma cells in vascular bundles of PAP1-transgenic tobacco plants, although the transgene was expressed in all tissues (Xie et al. 2006). Furthermore, anthocyanin biosynthesis activated by PAP1 transgene expression was demonstrated to be dependent upon transgenic cell types (Zhou et al. 2008). Two types (red and white) of cell lines were established from one single leaf explant of a PAP1-transgenic plant. Although the expression levels of the PAP1 transgene were similar in both cell lines, the production of anthocyanins was much higher in red cells than in white ones (Zhou et al. 2008). Recently, Rowan et al. (2009) demonstrated that environmental factors, e.g., low light and high temperature, could negatively control cyanin biosynthesis activated by PAP1 gene expression (Rowan et al. 2009). In our current report we confirm that the over expression of the PAP1 gene essentially activates the cyanin pathway and increases levels of cyanins in rosette leaves (Figs. 3, 5); however, we found that the levels of cyanins were not positively correlated with the expression levels of PAP1 in certain growth conditions. For example, on agar-solidified medium-2, the expression level of the PAP1 gene in rosette leaves of pap1-D plants in the ML condition was nearly 3-fold higher than in the HL condition (S-Fig. 1 a, Fig. 3b). However, the level of cyanins in rosette leaves was nearly twofold less in the ML condition than in the HL condition (Fig. 4). In addition, in the same ML condition, the transcript level of the PAP1 gene in rosette leaves of pap1-D plants on medium-2 was nearly twofold higher than on medium-1 (Fig. 3c, S-Fig. 1 b), but the level of cyanins was lower on medium-2 than on medium-1 (Fig. 4). Similar results were also obtained for WT plants in other growth conditions, e.g., the ML condition (Figs. 3c and 4; S-Fig. 1b). Although mechanisms behind these observations are unknown, we suggest that the lack of a positive correlation between the expression levels of PAP1 and the levels of total cyanins to a probable extent results from variable transcript levels of multiple genes. In particular, we hypothesize that in addition to PAP1 expression, cyanin biosynthesis in these growth conditions consisting of different combinations of light and medium is likely regulated by both other genes expression particularly those encoding proteins to form the MBW complexes and the ratios of carbon/nitrogen discussed bellow. In addition, in these growth conditions, the expression (if any) of PIF3 and HY5 (Ang and Deng 1994; Kim et al. 2003; Ni et al. 1998; Shin et al. 2007) likely regulates cyanin biosynthesis. Further comparative studies on cyanin biosynthesis in pap1, pap2, pap1-D, pap2-D, hy5, pif3, and other mutants and wild-type plants will enhance the understanding of regulation mechanisms in these growth conditions.
Regulation of anthocyanin biosynthesis by PAP1 is controlled by the ratios of carbon/nitrogen used in the culture media. Martin et al. (2002) demonstrated that anthocyanin biosynthesis in A. thaliana seedlings is strongly induced by high ratios of sucrose/nitrogen (Martin et al. 2002). Recently, Gao et al. (2008) showed that the ratio of carbon/nitrogen (a balance or an imbalance) is critical in the regulation of anthocyanin biosynthesis in seedlings of both osu1 mutants and WT. In their study, they found that high ratios of carbon/nitrogen increases levels of anthocyanins in both osu1and WT seedlings and that OSU1 encodes a putative methyltransferase that represses the expression of the PAP1 gene under high carbon/nitrogen conditions (Gao et al. 2008). Another significant achievement was made by Rubin et al. (2009), who identified three nitrogen-induced transcription factors, LBD37, 38 and 39, which were shown to repress the expression of the PAP1 and PAP2 genes. Over expression of these genes decreased levels of anthocyanins that were induced by nitrogen limitation (Rubin et al. 2009). Both OSU1 and LBDs control anthocyanin biosynthesis upstream of PAP1 (Gao et al. 2008; Rubin et al. 2009). In our current study, we used 30 g/l (approximately 87.69 mM) of sucrose as the carbon source in three plant media. Three different combinations of NH4NO3 and KNO3 (medium-1: 20 mM NH4NO3 and 18.8 mM KNO3, medium-2: 10 mM NH4NO3 and 9.4 mM KNO3, and medium-3: 9.4 mM KNO3 and 0 mM NH4NO3) were used to compare their effects on cyanin biosynthesis in both pap1-D and WT plants. The concentrations of nitrogen in the three media were 58.8, 29.4, and 9.4 mM, respectively. In comparison with medium-1, the ratios of carbon/nitrogen were increased threefold and nearly sixfold in medium-2 and -3, respectively. Under each lighting condition (LL, ML, and HL), the increment of sucrose/nitrogen ratios in media led to obvious decreases in cyanin levels in rosette leaves of pap1-D plants (Fig. 4). These observations indicate that the PAP1 regulation on anthocyanin biosynthesis is controlled by the ratio of carbon/nitrogen in the medium. In rosette leaves of WT plants, however, the responses of anthocyanin biosynthesis to changes in the carbon/nitrogen ratio showed three different trends under the three lighting conditions (Fig. 4). The increase of the carbon/nitrogen ratio induced anthocyanin biosynthesis under LL, decreased levels of anthocyanins under ML, and increased then decreased levels of anthocyanins under HL (Fig. 4). We hypothesize that the mechanisms of these regulations are likely different in rosette leaves of pap1-D and WT plants. Ongoing studies are elucidating these mechanisms.
The regulation of cyanin biosynthesis by PAP1 is dependent upon several transcription factor-associated protein complexes. In our study, we observed that the total levels of cyanins in rosette leaves of pap1-D plants to some extent were independent of an absolute expression level of PAP1. This suggests that transcripts of other genes were likely involved in cyanin biosynthesis, thus controlled the total levels of cyanins in different growth conditions. Numerous studies have shown the biosynthesis of cyanins in vegetative tissues of A. thaliana is controlled by TTG1-dependent regulatory protein complexes (Cominelli et al. 2008; Feyissa et al. 2009; Gonzalez et al. 2008; Lillo et al. 2008; Ramsay and Glover 2005; Shan et al. 2009). TTG1 required for cyanin biosynthesis is actually expressed in all major tissues (Cominelli et al. 2008; Galway et al. 1994). The loss of functions of the TTG1 gene resulted in pleiotropic effects on development and secondary metabolism, including absence of leaf and stem trichomes; excessive roots hairs developing from the atrichoblast cells; loss of cell wall mucilage in the seed coat; no requirement for cold treatment and drying of seeds for germination; the lack of anthocyanins in hypocotyl, leaf and seeds; and no formation of procyanidins in seeds (Galway et al. 1994; Koornneef 1981; Walker et al. 1999). Several reports have shown that the cyanin biosynthesis of A. thaliana is likely differentially regulated by several TTG1-dependent protein complexes. The PAP1-associated MBW (MYB/PAP1- bHLH-WD40/TTG1) complex mainly regulates the biosynthesis of cyanins in rosette leaves and hypocotyls; For example, the complex of PAP1- GL3/EGL3-TTG1 was demonstrated to regulate cyanin biosynthesis particularly in vegetative tissues (Gonzalez et al. 2008; Ramsay and Glover 2005; Zhang et al. 2003). Transcriptional analysis showed an overlaying expression pattern for the PAP1, GL3 and EGL3 genes in the same tissues of seedlings (Gonzalez et al. 2008). In addition, in a given growth condition with nitrogen depletion, cyanin biosynthesis in seedlings was mainly regulated by the complex of PAP1-GL3-TTG1 (Feyissa et al. 2009). In certain specific conditions, however, the biosynthesis of cyanins in vegetative tissues may be independent of regulation by PAP1. Recently, MYB113 and MYB114 were demonstrated to regulate the biosynthesis of cyanins. The overexpression of either of these two genes increased cyanin production in seedlings (Gonzalez et al. 2008). The regulatory functions of these two genes are also dependent upon TTG1 (Gonzalez et al. 2008). PAP2 is highly homologous to PAP1 but has a different impact on pathway gene expression and cyanin biosynthesis in seedlings of A. thaliana (Cominelli et al. 2008). Unlike PAP1 transgenic seedlings but like wild-type ones, PAP2 transgenic seedlings produced very low levels of cyanins when transferred from dark to white or UV light (Cominelli et al. 2008). Furthermore, PAP2 transcription was reported to respond to nitrogen depletion much more strongly than that of PAP1 (Lea et al. 2007). Therefore, although more evidence is required, the PAP2/MYB113/114-EGL/GL3-TTG1 complexes are predicted to regulate cyanins in a manner independent of PAP1 in leaves of A. thaliana. In our study, the lack of a positive correlation between the expression levels of PAP1 and the production of cyanins (Fig. 4 and S-Fig. 1) as discussed in the previous paragraph likely supports this prediction. Although we have not investigated the gene expression levels for GL3, EGL3, and TTG1, we hypothesize that the expression levels of the three genes in different growth conditions likely control the final production of cyanins. In addition, although we have not analyzed the expression levels of PAP2, MYB113, MYB114, EGL3, and GL3, we also hypothesize that their expression levels likely lead to the lack of a positive correlation between the absolute expression level of PAP1 and the production of cyanins. Therefore, in the nine growth conditions, cyanin biosynthesis is likely regulated by different MYB complexes either alone or together, e.g. PAP1-GL3-TTG1 and/or PAP2/MYB113/MYB114-GL3/EGL3-TTG1.
Abbreviations
- pap1-D:
-
Production of anthocyanin pigmentation1-Dominant
- PAP1:
-
Production of anthocyanin pigmentation1
- HPLC-ESI-MS:
-
High performance-electrospray ionization-mass spectrometry
- HPLC-TOF-MS:
-
High performance-time of flight-mass spectrometry
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
References
Ang LH, Deng XW (1994) Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6:613–628
Bloor SJ, Abrahams S (2002) The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59:343–346
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2394
Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C (2008) Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 165:886–894
Dixon RA, Xie DY, Sharma SB (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165:9–28
Feyissa D, Løvdal T, Olsen K, Slimestad R, Lillo C (2009) The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta 230:747–754
Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The TTG1 gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Develop Biol 166:740–754
Gao P, Xin Z, Zheng Z-L (2008) The OSU1/QUA2/TSD2-encoded putative methyltransferase Is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. PLoS ONE 3:e1387
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Grotewold E (2006) The genetics and biochemistry of floral pigments. Ann Rev Plant Biol 57:761–780
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Kim J, Yi H, Choi G, Shin B, Song P-S, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15:2399–2407
Koornneef M (1981) The complex syndrome of ttg mutants. Arabidopsis Inf Serv 18:45–51
Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM (1992) Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4:1229–1236
Kubo H, Peeters AJM, Aarts MGM, Pereira A, Korneef M (1999) ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11:1217–1226
Lea U, Slimestad R, Smedvig P, Lillo C (2007) Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225:1245–1253
Lepiniec L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Ann Rev Plant Biol 57:405–430
Lillo C, Lea US, Ruoff P (2008) Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ 31:587–601
Lloyd AM, walbot V, Davis RW (1992) Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258:1773–1775
Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P, Parr A, Michael AJ, Saito K, Martin C (2007) Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J 50:678–695
Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128:472–481
Meyer P, Heidmann I, Forkmann G, Saedler H (1987) A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 330:677–678
Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud M-C (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. PNAS 102:11934–11939
Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, Scheible W-R (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant, Cell Environ 30:85–112
Muller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143:156–171
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Nakabayashi R, Kusano M, Kobayashi M, Tohge T, Yonekura-Sakakibara K, Kogure N, Yamazaki M, Kitajima M, Saito K, Takayama H (2009) Metabolomics-oriented isolation and structure elucidation of 37 compounds including two anthocyanins from Arabidopsis thaliana. Phytochemistry 70:1017–1029
Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell 95:657–667
Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C (2008) Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J Plant Physiol 165:1491–1499
Pourcel L, Irani NG, Lu Y, Riedl K, Schwartz S, Grotewold E (2009) The Formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol Plant: ssp071
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Rowan DD, Cao M, Kui L-W, Cooney JM, Jensen DJ, Austin PT, Hunt MB, Norling C, Hellens RP, Schaffer RJ, Allan AC (2009) Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol 182:102–115
Rubin G, Tohge T, Matsuda F, Saito K, Scheible W-R (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21:3567–3584
Shan X, Zhang Y, Peng W, Wang Z, Xie D (2009) Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot erp223
Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49:981–994
Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140:637–646
Song YH, Yoo CM, Hong AP, Kim SH, Jeong HJ, Shin SY, Kim HJ, Yun D-J, Lim CO, Bahk JD, Lee SY, Nagao RT, Key JL, Hong JC (2008) DNA-binding study identifies C-box and hybrid C/G-box or C/A-box motifs as high-affinity binding sites for STF1 and LONG HYPOCOTYL5 proteins. Plant Physiol 146:1862–1877
Springob K, Nakajima H, Yamazaki M, Saito K (2003) Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 20:288–303
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito K (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasdan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA 1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11:1349–1377
Winkel-Shirley B (2001) Flavonoid Biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Xie D-Y, Jackson LA, Cooper JD, Ferreira D, Paiva NL (2004) Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol 134:979–994
Xie D-Y, Sharma SB, Wright E, Wang Z-Y, Dixon RA (2006) Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J 45:895–907
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130:4859–4869
Zhou L-L, Zeng H-N, Shi M-Z, Xie D-Y (2008) Development of tobacco callus cultures over expressing Arabidopsis PAP1/MYB75 transcription factor and characterization of anthocyanin biosynthesis. Planta 229:37–51
Zuluaga DL, Gonzali S, Loreti E, Pucciariello C, Degl’Innocenti ED, Guidi L, Alpi A, Perata P (2008) Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol 35:606–618
Acknowledgments
This work was financially supported by North Carolina State University and USDA-NRI grant (USDA 2006-35318-17431). We thank Dr. LiLi Zhou for help running samples for LC–MS analysis. We thank Dr. Wendy Boss from our department for her critical reading of this manuscript and her many suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2010_1142_MOESM1_ESM.jpg
S-Fig. 1 Normalized values from semi-quantitative analysis of RT-PCR products show expression levels of 5 genes in rosette leaves of plants grown in different growth conditions. a: Normalized values show the impacts of three light intensities on transcript levels of five genes in rosette leaves of both WT and pap1-D plants grown on agar-solidified medium-2. Due to poor growth of WT plants in the condition of HLM-2, total RNA isolated from plants grown in the condition of HLM-3 were used as controls. HLM-2 (WT:M-3) means that total RNA for pap1-D plants was isolated from leaves on the condition of HLM-2 and total RNA for WT plants was isolated from leaves on the condition of HLM-3. b: Normalized values show the impacts of three media on transcript levels of 5 genes in rosette leaves of both WT and pap1-D plants grown in the ML condition. RT-PCR analysis for WT seedlings grown on medium-3 in the ML condition is not shown due to low yield and poor quality of total RNA from rosette leaves. LLM-2: low light: 50 μmol m−2 s−1 of light intensity and medium-2: ½ strength of NH4NO3 and KNO3 in MS medium MLM-2: middle light: 200 μmol m−2 s−1 of light intensity and medium-2 HLM-2: high light: 500 μmol m−2 s−1 of light intensity and medium-2 HLM-3: high light and medium-3: 0 NH4NO3 and ½ strength of KNO3 in MS medium MLM-1: middle light and medium-1: full strength of basal MS medium MLM-3: middle light and medium-3 (JPEG 942 kb)
425_2010_1142_MOESM2_ESM.jpg
S-Fig. 2 Mass spectra of 20 cyanins detected at 530 nm from rosette leaves of 20-day-old pap1-D plants grown in the condition of HLM-2 (high light: 500 μmol m−2 s−1 of light intensity and medium-2: ½ strength of both NH4NO3 and KNO3 in MS medium) (JPEG 2248 kb)
Rights and permissions
About this article
Cite this article
Shi, MZ., Xie, DY. Features of anthocyanin biosynthesis in pap1-D and wild-type Arabidopsis thaliana plants grown in different light intensity and culture media conditions. Planta 231, 1385–1400 (2010). https://doi.org/10.1007/s00425-010-1142-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1142-9