Abstract
Branched-chain amino acid transaminases (BCATs) play a crucial role in the metabolic pathway of leucine, isoleucine and valine by catalyzing the last step of synthesis and/or the initial step of degradation of these amino acids. In this study, we characterized a new BCAT from Nicotiana benthamiana (NbBCAT, GeneBank accession No. EU194916), the deduced amino acid sequence of which exhibits a very high percentage of identity to the homologous enzymes from Solanum tuberosum (StBCAT-2, 91.5%) and Arabidopsis thaliana (AtBCAT1-6, 56.4–68.6%). Complementation experiment using a Δbat1/Δbat2 double knockout yeast strain system demonstrated enzymatic activities for NbBCAT. Ectopically expressed NbBCAT::green fluorescence fusion protein was targeted predominantly to the chloroplasts in tobacco protoplasts. The highest levels of NbBCAT transcripts were found in open flowers as well as in young leaves. Virus-induced gene silencing of NbBCAT resulted in abnormal leaf development and loss of apical dominance. In NbBCAT-silenced plants, two KNOTTED1-type genes, NTH15 and NTH23, were upregulated. This was accompanied by various hormone changes, as a result of transcriptional regulation of gibberellin 20-oxidase (Ntc12) and adenosine phosphate isopentenyltransferase. The transcript levels of NbBCAT could also be repressed by hormone treatment. These results suggest that NbBCAT, an enzyme in the branched-chain amino acid metabolic pathway, may be involved in the regulation of endogenous hormones by its effect on KNOX genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Valine (Val), leucine (Leu) and isoleucine (Ile) contain certain short-branched hydrocarbon residues, which are responsible for their classification as branched-chain amino acids (BCAAs). They are essential amino acids in animals and plants. Animals cannot synthesize these amino acids and must obtain them through diet. In contrast, plants are able to synthesize them de novo, suggesting the existence of a unique metabolic pathway in plants (Singh and Shaner 1995; Singh 1999; Binder et al. 2007). The biosynthetic pathways of BCAAs in plants have been extensively investigated and a number of genes have been characterized, because numerous enzymes operating in this pathway are also targets of commercially important herbicides (Singh 1999). The only exception is the final biosynthetic step, which is catalyzed by a BCAA transaminase (BCAT). BCAT catalyzes the final transamination step in the pathway that produces the essential amino acids leucine, isoleucine and valine, and also the first step of degradation of these amino acids. BCAT activities have been detected in a wide range of eukaryotic organisms, including yeasts, plants, rats and humans (Singh 1999). Although experimental assays for the subcellular localization of the individual enzymes have only been performed in a few instances, it seems clear that these amino acids are synthesized in chloroplasts and catabolized in mitochondria (Aubert et al. 1996; Hagelstein et al. 1997; Anderson et al. 1998; Daschner et al. 1999, 2001; Fujiki et al. 2000, 2001).
There is a small family of BCAT genes in plants, which have been characterized in some species such as Solanum tuberosum (potato), Arabidopsis and Hordeum vulgare (barley) (Campbell et al. 2001; Diebold et al. 2002; Malatrasi et al. 2006). Two BCAT genes (StBCAT-1, StBCAT-2) isolated from S. tuberosum are differentially expressed. In contrast to StBCAT-2, StBCAT-1 is inducible by hormone treatment in leaf tissues (Campbell et al. 2001). In Arabidopsis, six transcribed members of the BCAT family are differentially localized. Transient expression of BCAT::green fluorescent protein (GFP) fusion in tobacco (Nicotiana tabacum) protoplasts revealed that the isoenzymes AtBCAT-2, -3 and -5 are imported into the chloroplasts, whereas AtBCAT-1 is directed into the mitochondria (Diebold et al. 2002). Interestingly, these isoenzymes also display different expression patterns. The mitochondrial isoenzyme (AtBCAT-1) is expressed in all tissues, with predominant transcription in seedlings and leaves. The plastid-located proteins (AtBCAT-2, -3 and -5) are expressed at rather low levels, with only AtBCAT-3 transcribed in all tissues. The most likely cytoplasm-located AtBCAT-4 and AtBCAT-6 are mainly expressed in tissues associated with transport functions and in meristematic tissues, respectively. Further investigation then demonstrated that AtBCAT-1 initiates the degradation of all BCAAs (Schuster and Binder 2005). Surprisingly, it was found that AtBCAT-4 participates in the chain elongation pathway of Met-derived glucosinolate biosynthesis, which has a close evolutionary relationship to the Leu biosynthesis pathway (Schuster et al. 2006). A newly identified BCAT gene from barley, HvBCAT-1, is inducible by drought stress (Malatrasi et al. 2006).
To better understand the BCAT gene family in higher plants, we characterized a new BCAT homolog in N. benthamiana (NbBCAT). Here, we report that NbBCAT is functional in yeast and is localized in chloroplasts. NbBCAT is transcribed predominantly in open flowers and young leaves. Suppressed expression of NbBCAT causes changes in various endogenous hormones and abnormal plant development that can be correlated with transcriptional induction of the KNOX genes, NTH15 and NTH23. The possible mechanisms for the association between NbBCAT and the KNOX genes are discussed.
Materials and methods
Plasmid construction
To clone the full length NbBCAT cDNA, RT-PCR was performed using 2 μg of total RNA isolated from N. benthamiana seedlings using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, with primers NbBCAT F1 and NbBCAT R1 (Table 1). The resulting RT-PCR fragments were cloned into the EcoRV site of pBluescriptIISK+ (Stratagene, La Jolla, CA, USA) to generate pBS-NbBCAT for direct sequencing.
To fuse NbBCAT with GFP for transient expression, we amplified the NbBCAT ORF with primers NbBCAT F5 and NbBCAT R5, which contained the NcoI site and lacked the stop codon. Subsequently, the PCR fragment was cloned into pRTL2-GFP using the NcoI site.
For TRV series constructs, various cDNA fragments of NbBCAT were PCR amplified and cloned into the pTRV2 vector (Liu et al. 2002) using EcoRI and BamHI sites. The PCR primers used were: NbBCAT F1 and NbBCAT R1 for fragment I; NbBCAT-II F2 and NbBCAT-II R2 for fragment II; NbBCAT F1 and NbBCAT-III R3 for fragment III; NbBCAT-IV F4 and NbBCAT R1 for fragment IV (Table 1).
All these constructs were verified by nucleotide sequencing.
Yeast (Saccharomyces cerevisiae) complementation analysis
For the complementation tests, the NbBCAT ORF fragment was sub-cloned from pBS-NbBCAT into pRS425GDP (Funk et al. 2002) to generate pRS425GDP-NbBCAT using the BamHI and SalI sites. The S. cerevisiae BAT1 gene was amplified with primer pairs BAT1 F and BAT1 R and cloned into pRS425GDP to generate pRS425GDP-ScBAT1 for using as a positive control. The vector without the insert was used as a negative control. About 3 μg of each DNA construct was delivered, via electroporation, into yeast strain Δbat1/Δbat2 (Kispal et al. 1996). The electroporated yeast was plated onto minimal medium (0.67% yeast nitrogen base without amino acids, 2% glucose, 20 mg l−1 adenine, His and Lys) for selection. The growth of yeast on the plate was examined at 30°C for 5 days (Diebold et al. 2002).
Transient expression of GFP fusions in N. benthamiana protoplasts
For preparation of N. benthamiana protoplasts, leaves from 3–4-week-old wild-type plants, which grew at 25°C in a growth chamber with a 16 h light/8 h dark cycle, were cut into 0.5–1 mm stripes. These stripes were then digested in an enzyme solution (1.5% cellulase R10, 0.4% macerozyme R10, 0.4 M mannitol, 20 mM KCl, 20 mM MES, pH 5.7, 10 mM CaCl2, 5 mM β-mercaptoethanol, 0.1% BSA) under darkness for 3 h at room temperature after vacuum infiltration for 30 min. The enzyme solution was filtered through a 35–75-μm nylon mesh and the collected protoplasts were washed once with cold WI solution (0.5 M mannitol, 4 mM MES, pH 5.7, 20 mM KCl) and resuspended at 2 × 105 cells ml−1 with WI solution. For transient gene expression, 300–400 μl protoplasts were mixed with 5 μg DNA plasmid and 50 μg ml−1 salmon sperm DNA in a 2-mm ice-cooled cuvette and electroporated at 150 V, 50 Ω and 900 μF (BTX ECM630, San Diego, CA, USA). After incubation in a culture medium (0.5 × MS salts and vitamins, 0.4 M mannitol, 3% sucrose, 5 mM MES pH 5.7, 50 μg ml−1 ampicillin) at 26°C under darkness for 16 h, the protoplasts were examined for GFP expression as described (Howard et al. 1992).
RNA isolation and RT-PCR analysis
Total RNA was extracted from N. benthamiana leaves using the TriReagent (Sigma-Aldrich, St Louis, MO, USA) and treated with RO1 RNase-free DNase (Promega, Madison, WI, USA), according to the manufacturer’s instructions. The first strand cDNA was synthesized using 2 μg of total template RNA, oligo d(T)18 primer (Promega, Madison, WI, USA) and superscript reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Semi-quantitative RT-PCR was performed as described previously (Lee et al. 2003). To analyze the endogenous NbBCAT mRNA levels in silenced and non-silenced N. benthamiana plants, we designed the RT-PCR primer pair Bcat-C (Table 1), which anneals outside the region targeted for silencing, to ensure that the bona fide endogenous gene was tested. All other RT-PCR primers are shown in Table 1. All RT-PCR results were verified at least three times.
Real-time PCR analysis
All real-time PCR runs were performed under identical conditions in DNA Engine Opticon 2 system (MJ Research) with 2 × SYBR Green master mix reagent (Toyobo Co., Ltd, Osaka, Japan). Quantifications of each cDNA sample were conducted with results from triplicate experiments, and consistent results from at least five separately prepared RNA samples were used for final analysis. The relative levels of the target gene expression were quantified by using the comparative Ct method (Roche Light-Cycler system). The primers used for PCR are listed in Table 1.
Virus-induced gene silencing assay
Nicotiana benthamiana plants were maintained at 25°C in a growth chamber with a 16 h light/8 h dark cycle. Plants of approximately four-leaf-stage were used for VIGS. VIGS assay was performed as described previously (Liu et al. 2002). Briefly, pTRV1 or pTRV2 and its derivatives were introduced into the Agrobacterium tumefaciens GV3101 strain by electroporation at 1,440 V, 125 Ω and 50 μF in BTX ECM630 (BTX, San Diego, CA, USA). Agrobacterium cultures at O.D. 600 = 0.8, containing pTRV1 and pTRV2 or its derivative plasmids, were mixed 1:1 and infiltrated into the lower leaves of four-leaf stage plants using a 1-ml needleless syringe. The phenotypes induced by VIGS were first observed at about 10 days post-infiltration.
Hormone treatments
Young leaves of 7-week-old N. benthamiana plants were placed in Falcon tubes filled with 10 μM indole-3-acetic acid (IAA), GA3 or zeatin riboside (ZR) for 12 h. Leaves incubated in double-distilled water were used as a control. The treated leaves were frozen in liquid nitrogen and stored at −70°C for RNA isolation.
Measurement of endogenous hormone levels
Leaves were homogenized in liquid nitrogen and then extracted with 4 ml of 80% (v/v) ice-cold, aqueous methanol containing butylated hydroxytoluene (1 mM). The samples were incubated at 4°C overnight and centrifuged at 10,000g for 10 min. Each of the resulting supernatants was divided into three equal samples and filtered through C18 Sep-Pak cartridges (Waters). The efflux from each sample was collected, dried by evaporation with N2, and GA1, IAA and ZR contents were determined with ELISA (He 1993; Yang et al. 2001).
Results
Cloning of the full-length NbBCAT cDNA
To obtain the cDNA sequence information of the N. benthamiana homolog of BCAT, we used the cDNA sequence of S. tuberosum BCAA aminotransferase (StBCAT-1) (GeneBank accession No. AF193845) as a query to search in the TIGR Plant Transcript Assembly BLAST Server (http://tigrblast.tigr.org/euk-blast/plantta_blast.cgi) (Childs et al. 2007). The N. benthamiana transcript assembly database was selected and program “blastn” was used with default parameters. We found a 1,891-bp transcript (TA8253_4100) that shares 92.49% identity with the cDNA sequence of StBCAT-1. To amplify the ORF of the N. benthamiana BCAT homolog, we used NbBCAT F1 and NbBCAT R1 primers (Table 1) designed according to this sequence and performed RT-PCR using template mRNA isolated from N. benthamiana seedlings. The full-length NbBCAT ORF is 1,257 bp in length and encodes a polypeptide consisting of 418 amino acids with a molecular mass of 45,279 Da (Fig. 1). The predicted NbBCAT protein has a very high degree of similarity in its amino acid sequence to BCAT proteins from S. tuberosum and Arabidopsis (Fig. 1).
Alignment of the deduced NbBCAT amino acid sequence with BCAT homologs from other organisms including StBCAT-1(AAF07191), StBCAT-2(AAF07192), AtBCAT-1 to AtBCAT-6 (Q93Y32, Q9M439, Q9M401, Q9LE06, Q9FYA6, Q9LPM9), human BCATm (mitochondrial, O15382) and BCATc (cytosolic, P54687), yeast BATm (mitochondrial, P38891) and BATc (cytosolic, P47176), E. coli iveE (BAB38127). Blue arrowheads pyridoxalphosphate-linking site, orange arrowheads 1 to 4 crucial function sites revealed by crystal structures in E. coli ILVE and human HsBCATm
NbBCAT has the highest amino acid sequence similarity (91.5%) with StBCAT-1 and StBCAT-2 from S. tuberosum. In contrast, it has weak amino acid sequence similarity to BCATs from human, yeast and E. coli. NbBCAT shares a number of conserved amino acids residues found in the BCATs from all the other species analyzed (Fig. 1). Lys202 in human HsBCATm and Lys159 in E. coli ilvE each represents the active site of the enzyme, to which pyridoxalphosphate is covalently linked (Diebold et al. 2002). This site is conserved in BCATs from all plants examined to date (Fig. 1, blue arrowheads). The sites for anchoring the PLP ring in the active site cavity (Y269 in NbBCAT, Y207 in HsBCATm, Y164 in EcILVE, and E299, E237 and E193) are also conserved in the plant BCAT sequences (Fig. 1, orange arrowheads 3 and 4). In addition, several other amino acids (Y133, Y70 and Y31 and R206, R143 and R97; Fig. 1 orange arrowheads 1 and 2) with crucial functions revealed by crystal structures of the respective E. coli ILVE and human HsBCATm (Okada et al. 1997; Yennawar et al. 2001) are also conserved in the NbBCAT sequences.
NbBCAT functions as a BCAT in yeast
The high amino acid sequence identity of the cloned NbBCAT with StBCAT-1 and StBCAT-2 strongly suggests that it functions as a BCAT. To test this experimentally, we performed experiments to determine whether NbBCAT could complement the auxotrophy for BCAAs in a Δbat1/Δbat2 double knockout yeast strain lacking the three BCAAs (Kispal et al. 1996; Diebold et al. 2002). The restored growth on minimal medium of Δbat1/Δbat2 double knockout yeast strain transformed with pRS425GPD-NbBCAT indicates that NbBCAT has the expected BCAT enzyme activity (Fig. 2). ScBAT-1 was used as a positive control for complementation.
Complementation assay of NbBCAT in the yeast double knockout mutant Δbat1/Δbat2. To examine the BCAT function in vivo, the ORF sequence of NbBCAT was cloned into pRS425GPD vectors and transformed into Δbat1/Δbat2. Restored growth on medium lacking Val, Leu and iso-Leu indicates the competent BCAT activity of NbBCAT. Complementation was also observed with ScBAT-1 as positive control. No growth is detected without transformation (Δbat1/Δbat2) or after transformation with the empty vector containing no insertion (pRS425GPD)
NbBCAT is localized to the chloroplasts
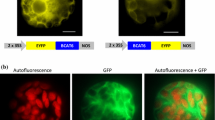
Diebold et al. (2002) reported that the Arabidopsis nuclear genome encodes four BCATs, with AtBCAT-2,-3,-5 imported into the chloroplasts and AtBCAT-1 imported into the mitochondria. NbBCAT exhibits the highest amino acid sequence similarity to AtBCAT-3 (68.5%) and AtBCAT-5 (64.1%), suggesting its possible localization to the chloroplasts. To test this, the NbBCAT ORF was cloned in frame with GFP at the C-terminus and transiently expressed in N. benthamiana protoplasts driven by an enhanced CaMV 35S promoter (Fig. 3a). The GFP fluorescence was localized exclusively to the chloroplasts, as shown by its perfect overlap with the red chlorophyll autofluorescence. In contrast, the control GFP alone was mainly detected in the cytoplasm (Fig. 3b). These data indicate that NtBCAT is a chloroplast-localized protein.
Subcellular localization of NbBCAT in N. benthamiana protoplasts. a Schematic diagram of constructs used for the transient transformation of N. benthamiana protoplasts. The ORF fragment of NbBCAT was driven by cauliflower mosaic virus 35S promoter, and a 5′-nontranslated region from the tobacco etch virus (TEV leader) was used to increase protein production. b Merged images of GFP green fluorescence and red chlorophyll autofluorescence demonstrate NbBCAT-GFP localization in the chloroplasts. Images were taken by the Axiovision software with a Zeiss LSM510 META confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany), using a 40× C-Apochromat water immersion objective lens (NA = 1.2). Excitation/emission settings were 488/505–530 nm for GFP and 488/650 nm for chlorophyll autofluorescence. Scale bars correspond to 10 μm
NbBCAT transcript levels in different plant organs
To assess the importance and possible role of NbBCAT in association with plant development, we examined its distribution in seedlings as well as in a range of organs of mature plants by semi-quantitative RT-PCR. EF-1α was used as an internal control and all amplifications were repeated at least three times. The NbBCAT mRNA was detected at different levels in the seedlings, roots, stems, young leaves, mature leaves and open flowers of N. benthamiana (Fig. 4). Its accumulation showed the highest level in open flowers and young leaves and relatively low levels in mature leaves, stems, roots and seedlings. This expression pattern was similar to that of AtBCAT-1 and AtBCAT-3 in Arabidopsis (Schuster and Binder 2005).
Semi-quantitative RT-PCR analysis of NbBCAT transcription level. The NbBCAT mRNA level in different organs of N. benthamiana was examined by semi-quantitative RT-PCR, and elongation factor 1-alpha (EF1α) was used as an internal control. OL mature leaves, YL young leaves, S stems, R roots, F open flowers, Se seedlings. The experiments were repeated with three independent samples and similar results were obtained
Suppressed expression of endogenous NbBCAT by VIGS results in abnormal leaf development and loss of apical dominance
In order to characterize the function of NbBCAT, we employed virus-induced gene silencing (VIGS). Briefly, viral vectors carrying a host-derived gene sequence can induce silencing of the corresponding endogenous gene in infected plants, which provides a simple and rapid approach to characterize gene functions (Ruiz et al. 1998; Baulcombe 1999; Liu et al. 2002). In this experiment, four different-sized fragments (I to IV) of NbBCAT were cloned into the pTRV2 vector, as shown in Fig. 5a. The leaves of young N. benthamiana plants (four-leaf stage) were infiltrated with Agrobacterium transformed with pTRV1, which was mixed with Agrobacterium transformed with four pTRV2 constructs containing different-sized fragments of NbBCAT, respectively. Due to the low transcription levels of NbBCAT, semi-quantitative RT-PCR was performed using the Bcat-C primers to examine the transcription levels of endogenous NbBCAT. Significantly reduced RT-PCR products were observed in NbBCAT-silenced plants compared to the TRV control plants, indicating that the endogenous NbBCAT mRNA level was greatly reduced in those plants, regardless of the specific NbBCAT fragments used for silencing. The transcripts of EF-1α, which served as an internal control, remained constant in all tested plants (Fig. 5b). Interestingly, the silenced plants with different NbBCAT fragments showed abnormal leaf development and loss of apical dominance (Fig. 6a I–IV, b III). The newly emerged leaves of all NbBCAT-silenced plants at 3 weeks post-infiltration were severely wrinkled and showed a palm-like shape with inconspicuous midrib, which indicates that the midrib and lateral veins of these leaves fail to develop normally. The protrudent regions of leaves also displayed darker color than the surrounding areas (Fig. 6a III). At 7 weeks post-infiltration, the NbBCAT-silenced plants were severely dwarfed and exhibited a loss of apical dominance (Fig. 6b III). Many leaves of these plants were initiated directly from the stems, and these leaves were very small and thick compared to those in the TRV control plants.
Suppression of endogenous NbBCAT mRNA by TRV vector containing different NbBCAT cDNA fragments-induced gene silencing. a Schematic representation of NbBCAT cDNA fragments from N. benthamiana. NbBCAT includes the open reading frame and portions of the 5′ and 3′ UTRs. Fragments I–IV were inserted into the pTRV2 vector in silencing experiments. No insertion vector was used as a control. Arrows show location and direction of primers Bcat-C used to measure transcript abundance. b The transcription levels of NbBCAT in five independent silenced plants with different insertions (TRV-NbBCT-I to TRV-NbBCAT-IV) and control plants were determined using RT-PCR with primers Bcat-C. For each TRV constructs, more than 15 plants were assayed individually. Here, we show one typical example of all results obtained. Products were visualized after 25 and 30 cycles for the NbBCAT and 25 cycles for EF1α
The typical phenotypes of TRV constructs induced NbBCAT-silencing plants. a VIGS phenotypes of the whole plants and leaves in NbBCAT-silenced (containing different insertions: I–IV) and TRV control plants at 3 weeks post-incubation. b VIGS phenotypes of III and TRV control plants at 7 weeks post-incubation
These phenotypes were observed reproducibly in all of the more than 100 plants treated with VIGS. In contrast, these abnormal phenotypes were never observed in the approximately 100 plants infiltrated with TRV control. These results demonstrated that NbBCAT expression is important for plant growth and development.
Hormone level was altered in NbBCAT-silenced plants and NbBCAT was regulated by hormones
Plant hormones such as gibberellin (GA), IAA and cytokinin (CK) play crucial roles in plant growth and development. To understand whether the abnormal phenotypes of NbBCAT-silenced plants are correlated with altered hormone levels, we assayed the levels of GA1, IAA and CK (ZR) in leaves of NbBCAT-silenced plants by ELISA. Since GA1 acts as the dominant GA compound in plants (Graebe 1987), we chose GA1 for analysis. As shown in Fig. 7a, the GA1 level in leaves of NbBCAT-silenced plants was decreased to approximately 56.8% of that from TRV control plants. Similarly, the IAA level in leaves of NbBCAT-silenced plants was decreased to approximately 37.6% of the TRV control plants. In contrast, the ZR level in leaves of NbBCAT-silenced plants was increased to approximately 1.6-fold of the TRV control plants. Thus, the suppressed expression of endogenous NbBCAT resulted in significant changes in the level of various plant hormones, suggesting a possible role of NbBCAT in hormone regulation in N. benthamiana.
Hormone changes in NbBCAT-silenced plants and the suppression of NbBCAT transcription by hormones. The NbBCAT-silenced plants, which were confirmed by RT-PCR analysis from Fig. 5 were used for hormone measurement. The upper leaves of NbBCAT-silenced (containing insertions fragment I–IV) plants were pooled for analysis. a GA1, IAA and ZR contents in TRV-NbBCAT and TRV plants. Each column represents the mean + SE of measurement from five independent plants. g.f.w gram fresh weight. b. The relative accumulation levels of NbBCAT by quantitative real-time RT-PCR. Data represent mean + SD based on results from three replicate experiments
To investigate whether NbBCAT is regulated by hormones as reported for StBCAT-1 (Campbell et al. 2001), we treated wild-type tobacco leaves with different hormones separately and determined the transcription levels of NbBCAT by quantitative real-time PCR. To our surprise, leaves treated with GA3, IAA and ZR all led to downregulation of the transcription levels of NbBCAT. As shown in Fig. 7b, the NbBCAT transcript levels were 20% in IAA-treated leaves, 50% in GA3-treated leaves and 80% in ZR-treated leaves, in comparison with those in H2O-treated control leaves. Thus, NbBCAT expression or transcript stability was negatively regulated by treatments with these hormones, albeit to different extent with different hormones.
Suppressed expression of NbBCAT by VIGS results in upregulation of Knotted-like homeobox (KNOX) transcription
Previous studies showed that transgenic tobacco plants overexpressing the KNOTTED1-type homeodomain gene NTH15 have severe morphological alterations: curved midrib, size-reduced wavy leaves, semi-dwarfing and loss of apical dominance (Tamaoki et al. 1997; Tanaka-Ueguchi et al. 1998; Sakamoto et al. 1999), which are very similar to those of the NbBCAT silenced plants described above.
These phenotypic similarities raise the possibility that the KNOX genes might be affected in NbBCAT-silenced plants. To test this suggestion, we performed quantitative real-time PCR to examine the transcript levels of the tobacco class I KNOX gene NTH15 and class II KNOX gene NTH23. As shown in Fig. 8, in NbBCAT-silenced plants the transcription levels of both NTH15 and NTH23 were significantly elevated as compared to those in TRV control plants (Fig. 8 a, b).
Transcription of KNOX genes and hormone synthesis-related genes. Total RNA was prepared from the pooled samples, which were used in hormone measurement experiment. Transcription levels of KNOX genes NTH15 (a) and NTH23 (b) and synthesis-related genes of gibberellin (Ntc12) (c) and cytokinin (Nbipt) (d) were determined by quantitative real-time RT-PCR. EF1α was used as an internal control. Data represent mean + SD based on results from five replicate experiments
Discussion
In the present study, we characterized a new branched-chain amino acid transaminase (BCAT) from N. benthamiana. Sequence alignments revealed that almost all of the amino acid residues identified in crucial positions by using the three-dimensional structures of homologous enzymes from human and E. coli (Okada et al. 1997; Yennawar et al. 2001) are also conserved in NbBCAT. These include residues that recognize BCAAs in the human and E. coli enzymes. For example, tyrosines in positions 70 and 207 of human mitochondrial BCAT are present in the respective positions in NbBCAT (positions 133 and 269) and E. coli ilvE (31 and 164) (Fig. 1). High amino acid sequence identities of plant BCATs and conservation of crucial residues in three-dimensional structures in most species suggest significant functions for BCATs in biochemical and physiological processes.
The complementation experiment using Δbat1/Δbat2 double knockout yeast strain system confirmed NbBCAT as a functional BCAT (Fig. 2). In mammals, there are mitochondrial and cytosolic isoforms of BCAT (Hutson et al. 1995; Bledsoe et al. 1997). Since mammals are not capable of synthesizing leucine, valine and isoleucine, it has been assumed BCAT is associated with catabolism. However, there is a small family of BCAT genes in plants, which contains enzymes involved in biosynthetic and catabolic pathways (Campbell et al. 2001; Diebold et al. 2002). The biosynthetic and catabolic pathways are localized in different subcellular compartments, with the former in chloroplasts and the latter probably in mitochondria (Aubert et al. 1996; Hagelstein et al. 1997; Anderson et al. 1998; Fujiki et al. 2000). To investigate the subcellular localization of NbBCAT, we fused GFP in the C-terminus of NbBCAT and transiently expressed it in N. benthamiana protoplasts. We found GFP fluorescence to be exclusively present in chloroplasts (Fig. 3b). The chloroplastic localization of NbBCAT is consistent with its high sequence similarity with AtBCAT-3 (68.5%), a chloroplast-localized enzyme (Diebold et al. 2002). This localization suggests NbBCAT to be likely involved in the BCAA biosynthetic pathway rather than the catabolic pathway.
Semi-quantitative RT-PCR showed accumulation of NbBCAT transcripts at different levels in different plant organs examined, with the highest levels in open flowers and young leaves (Fig. 4). This expression pattern is similar to that of AtBCAT-1, which is able to degrade all three BCAAs in mitochondria, and AtBCAT-3, which is considered to be able to synthesize all these amino acids in plastids in Arabidopsis (Schuster and Binder 2005). Thus, it is reasonable to speculate that the synthesis of BCAAs may occur in all of these plant organs with a preference in open flowers and young leaves, which can be explained by the high demand of these fast-growing tissues for new proteins and amino acids.
KNOX genes encode transcriptional factors that belong to the superclass homeodomain proteins and have been identified in all monocots and dicots examined to date. Subsets of these genes are essential for meristem maintenance and lateral organ formation in all higher plants (Burglin 1997; Sharma and Fletcher 2002). Ectopic expression of KNOX genes in various plants causes drastic alteration in leaf and flower morphogenesis in spontaneous mutants (Smith et al. 1992; Chen et al. 1997; Parnis et al. 1997) and dwarfism and loss of apical dominance in transgenic plants (Matsuoka et al. 1993; Sinha et al. 1993; Lincoln et al. 1994; Chuck et al. 1996; Chen et al. 1997; Parnis et al. 1997). Alterations in morphology caused by ectopic expression of KNOX genes were also accompanied by hormone level changes. The specific accumulation of cytokinins in leaves and altered leaf morphology were observed when KNAT1 was overexpressed in lettuce (Frugis et al. 2001). Elevated cytokinin levels were also observed in transgenic tobacco overexpressing NTH15 and OSH1, while the level of GAs decreased (Tamaoki et al. 1997; Kusaba et al. 1998).
Here, we showed that repressed expression of NbBCAT, a newly characterized BCAT, results in abnormal phenotypes in N. benthamiana: wrinkled and palm-like leaves with ill-defined midrib (Fig. 6a I–IV), dwarfism and loss of apical dominance (Fig. 6b III), which are very similar to the phenotypes of KNOX overexpressing transgenic tobacco plants (Tamaoki et al. 1997; Sakamoto et al. 1999). Similarly, changes in the hormone levels were also detected in these plants, that is, the GA1 and IAA levels were decreased by 43.2 and 62.4%, respectively, while ZR was increased by approximately 60% (Fig. 7a). Interestingly, the transcript levels of Class I KNOX genes, NTH15, and Class II KNOX genes, NTH23, were clearly elevated in NbBCAT-silenced plants (Fig. 8a, b).
Gibberellin (GA) 20-oxidase and adenosine phosphate isopentenyltransferase (IPT), respectively, catalyze the rate-limiting step of bioactive GA and CK biosynthesis. Previous studies revealed that KNOX proteins suppress the expression of gibberellin (GA) 20-oxidase genes in the dicot plants S. tuberosum, Arabidopsis and tobacco (Sakamoto et al. 2001; Hay et al. 2002; Chen et al. 2004) and induce the expression of adenosine phosphate isopentenyltransferase (IPT) in the monocot plant, rice (Sakamoto et al. 2006). To investigate whether upregulated expression of KNOX genes in leaves of the NbBCAT-silenced plants affects the expression of GA 20-oxidase (Ntc12) and Nbipt genes, the transcript levels of these genes in leaves of NbBCAT-silenced plants were examined by quantitative real-time PCR. Downregulation of Ntc12 and upregulation of Nbipt were observed in NbBCAT-silenced plants compared to the TRV control plants (Fig. 8c, d). Thus, suppression of NbBCAT activated the transcription of the KNOX genes in fully differentiated tissues, which consequently modulated the transcription of the downstream genes, GA 20-oxidase and Nbipt. As a result, hormone contents changed significantly and led to the formation of the abnormal leaf morphology and loss of apical dominance.
These data suggest that there might be a novel hormone regulation pathway existing in plants in which NbBCAT and KNOX genes are involved. It is very likely that NbBCAT regulates hormone levels by affecting KNOX genes. Alternatively, other NbBCAT-involved hormone regulation pathways may exist, for example, there may be a feedback loop between hormone and KNOX genes (Rupp et al. 1999; Scanlon 2003). Furthermore, NbBCAT was found to be regulated by plant hormones. The transcription level of NbBCAT was repressed differentially by plant hormones GA3, IAA and ZR (Fig. 7b). These data suggest that hormone contents have a feedback influence on NbBCAT expression.
Our results have uncovered evidence of a novel regulation pathway involving BCAT, KNOX genes and hormones in plants. However, to elucidate the actual role of BCAT in this pathway, overexpression in transgenic plants and knockout mutants will be required.
Abbreviations
- BCAA:
-
Branched-chain amino acids
- BCAT:
-
Branched-chain amino acid transaminase
- BSA:
-
Bovine serum albumin
- CK:
-
Cytokinin
- EF-1α:
-
Elongation factor 1-alpha
- GA:
-
Gibberellin
- GFP:
-
Green fluorescence protein
- IPT:
-
Isopentenyltransferase
- IAA:
-
Indole-3-acetic acid
- KNOX:
-
KNOTTED1-like HOMEOBOX
- MES:
-
2-(N-morpholine)-ethane sulfonic acid
- MS salt:
-
Murashige and Skoog basal salt
- RT-PCR:
-
Reverse transcription-PCR
- TRV:
-
Tobacco rattle virus
- VIGS:
-
Virus-induced gene silencing
References
Anderson MD, Che P, Song JP, Nikolau BJ, Wurtele ES (1998) 3-Methylcrotonyl coenzyme A carboxylase is a component of the mitochondrial leucine catabolic pathway in plants. Plant Physiol 118:1127–1138
Aubert S, Alban C, Bligny R, Douce R (1996) Induction of beta-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett 383:175–180
Baulcombe DC (1999) Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol 2:109–113
Binder S, Knill T, Schuster J (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129:68–78
Bledsoe RK, Dawson PA, Hutson SM (1997) Cloning of the rat and human mitochondrial branched chain aminotransferases (BCAT(m)). Biochim Biophys Acta 1339:9–13
Burglin TR (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25:4173–4180
Campbell MA, Patel JK, Meyers JL, Myrick LC, Gustin JL (2001) Genes encoding for branched-chain amino acid aminotransferase are differentially expressed in plants. Plant Physiol Biochem 39:855–860
Chen JJ, Janssen BJ, Williams A, Sinha N (1997) A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution. Plant Cell 9:1289–1304
Chen H, Banerjee AK, Hannapel DJ (2004) The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J 38:276–284
Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP (2007) The TIGR plant transcript assemblies database. Nucleic Acids Res 35:D846–D851
Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8:1277–1289
Daschner K, Thalheim C, Guha C, Brennicke A, Binder S (1999) In plants a putative isovaleryl-CoA-dehydrogenase is located in mitochondria. Plant Mol Biol 39:1275–1282
Daschner K, Couee I, Binder S (2001) The mitochondrial isovaleryl-coenzyme A dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol 126:601–612
Diebold R, Schuster J, Daschner K, Binder S (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129:540–550
Frugis G, Giannino D, Mele G, Nicolodi C, Chiappetta A, Bitonti MB, Innocenti AM, Dewitte W, Van Onckelen H, Mariotti D (2001) Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol 126:1370–1380
Fujiki Y, Sato T, Ito M, Watanabe A (2000) Isolation and characterization of cDNA, clones for the E1 beta and E2 subunits of the branched-chain alpha-ketoacid dehydrogenase complex in Arabidopsis. J Biol Chem 275:6007–6013
Fujiki Y, Ito M, Nishida I, Watanabe A (2001) Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett 499:161–165
Funk M, Niedenthal R, Mumberg D, Brinkmann K, Ronicke V, Henkel T (2002) Vector systems for heterologous expression of proteins in Saccharomyces cerevisiae. Methods Enzymol 350:248–257
Graebe JE (1987) Gibberellin biosynthesis and control. Annu Rev Plant Phys 38:419–465
Hagelstein P, Sieve B, Klein M, Jans H, Schultz G (1997) Leucine synthesis in chloroplasts: leucine/isoleucine aminotransferase and valine aminotransferase are different enzymes in spinach chloroplasts. J Plant Physiol 150:23–30
Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12:1557–1565
He Z (1993) A laboratory guide to chemical control technology on field crop. Beijing Agricultural University Press, Beijing
Howard EA, Zupan JR, Citovsky V, Zambryski PC (1992) The Vird2 protein of A. tumefaciens contains a C-terminal bipartite nuclear-localization signal—implications for nuclear uptake of DNA in plant-cells. Cell 68:109–118
Hutson SM, Bledsoe RK, Hall TR, Dawson PA (1995) Cloning and expression of the mammalian cytosolic branched chain aminotransferase isoenzyme. J Biol Chem 270:30344–30352
Kispal G, Steiner H, Court DA, Rolinski B, Lill R (1996) Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem 271:24458–24464
Kusaba S, Kano-Murakami Y, Matsuoka M, Tamaoki M, Sakamoto T, Yamaguchi I, Fukumoto M (1998) Alteration of hormone levels in transgenic tobacco plants overexpressing the rice homeobox gene OSH1. Plant Physiol 116:471–476
Lee SS, Cho HS, Yoon GM, Ahn JW, Kim HH, Pai HS (2003) Interaction of NtCDPK1 calcium-dependent protein kinase with NtRpn3 regulatory subunit of the 26S proteasome in Nicotiana tabacum. Plant J 33:825–840
Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6:1859–1876
Liu YL, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30:415–429
Malatrasi M, Corradi M, Svensson JT, Close TJ, Gulli M, Marmiroli N (2006) A branched-chain amino acid aminotransferase gene isolated from Hordeum vulgare is differentially regulated by drought stress. Theor Appl Genet 113:965–976
Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kanomurakami Y (1993) Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell 5:1039–1048
Okada K, Hirotsu K, Sato M, Hayashi H, Kagamiyama H (1997) Three-dimensional structure of Escherichia coli branched-chain amino acid aminotransferase at 2.5 angstrom resolution. J Biochem 121:637–641
Parnis A, Cohen O, Gutfinger T, Hareven D, Zamir D, Lifschitz E (1997) The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a knotted gene. Plant Cell 9:2143–2158
Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937–946
Rupp HM, Frank M, Werner T, Strnad M, Schmulling T (1999) Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J 18:557–563
Sakamoto T, Nishimura A, Tamaoki M, Kuba M, Tanaka H, Iwahori S, Matsuoka M (1999) The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell 11:1419–1431
Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Gene Dev 15:581–590
Sakamoto T, Sakakibara H, Kojima M, Yamamoto Y, Nagasaki H, Inukai Y, Sato Y, Matsuoka M (2006) Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol 142:54–62
Scanlon MJ (2003) The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 133:597–605
Schuster J, Binder S (2005) The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol Biol 57:241–254
Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S (2006) BRANCHED-CHAIN AMINOTRANSFERASE4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18:2664–2679
Sharma VK, Fletcher JC (2002) Maintenance of shoot and floral meristem cell proliferation and fate. Plant Physiol 129:31–39
Singh BK (1999) Biosynthesis of valine, leucine and isoleucine. In: Singh BK (ed) Plant amino acids: biochemistry and biotechnology. Marcel Dekker, New York, pp 227–247
Singh BK, Shaner DL (1995) Biosynthesis of branched-chain amino-acids—from test-tube to field. Plant Cell 7:935–944
Sinha NR, Williams RE, Hake S (1993) Overexpression of the maize homeo box gene, Knotted-1, causes a switch from determinate to indeterminate cell fates. Gene Dev 7:787–795
Smith LG, Greene B, Veit B, Hake S (1992) A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf-cells with altered fates. Development 116:21–30
Tamaoki M, Kusaba S, KanoMurakami Y, Matsuoka M (1997) Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol 38:917–927
Tanaka-Ueguchi M, Itoh H, Oyama N, Koshioka M, Matsuoka M (1998) Overexpression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J 15:391–400
Yang YM, Xu CN, Wang BM, Jia JZ (2001) Effects of plant growth regulators on secondary wall thickening of cotton fibres. Plant Growth Regul 35:233–237
Yennawar N, Dunbar J, Conway M, Hutson S, Farber G (2001) The structure of human mitochondrial branched-chain aminotransferase. Acta Crystallogr D Biol Crystallogr 57:506–515
Acknowledgments
We thank Dr Biao Ding for critical reading of the manuscript, Dr Yule Liu for providing the VIGS vectors and Prof. Dr Roland Lill for providing the Δbat1/Δbat2 double knockout yeast strain and yeast expressing vector pRS425GPD. This work was supported by grants from the National Science Foundation of China and 863 program (contract no. 2007AA10Z191, to Y. Li and 2006CB101903, to Chunhong Wei).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gao, F., Wang, C., Wei, C. et al. A branched-chain aminotransferase may regulate hormone levels by affecting KNOX genes in plants. Planta 230, 611–623 (2009). https://doi.org/10.1007/s00425-009-0973-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-009-0973-8