Abstract
The cellulose synthase-like (ZmCSL) gene family of maize was annotated and its expression studied in the maize mesocotyl. A total of 28 full-length CSL genes and another 13 partial sequences were annotated; four are predicted to be pseudogenes. Maize has all of the CSL subfamilies that are present in rice, but the CSLC subfamily is expanded from 6 in rice to 12 in maize, and the CSLH subfamily might be reduced from 3 to 1. Unlike rice, maize has a gene in the CSLG subfamily, based on its sequence similarity to two genes annotated as CSLG in poplar. Light regulation of glycan synthase enzyme activities and CSL gene expression were analyzed in the mesocotyl. A Golgi-localized glucan synthase activity is reduced by ~50% 12 h after exposure to light. β-1,4-Mannan synthase activity is reduced even more strongly (>85%), whereas β-1,4-xylan synthase, callose synthase, and latent IDPase activity respond only slightly, if at all, to light. At least 17 of the CSL genes (42%) are expressed in the mesocotyl, of which four are up-regulated at least twofold, seven are down-regulated at least twofold, and six are not affected by light. The results contribute to our understanding of the structure of the CSL gene family in an important food and biofuel plant, show that a large percentage of the CSL genes are expressed in the specialized tissues of the mesocotyl, and demonstrate that members of the CSL gene family are differentially subject to photobiological regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The plant cell wall is a complex network of several types of polymers including cellulose, hemicelluloses, pectin, proteins, and lignin (Cosgrove 2005). Hemicelluloses show a high degree of chemical diversity and specialization among cell types. The hemicelluloses of the cell walls of the commelinoid monocotyledons, which include the Poaceae, have a primary cell wall composition (called Type II) that is different from other monocotyledonous and dicotyledonous plants (Type I) (Carpita 1996). In particular, their primary cell walls contain mixed-linkage glucan (MLG), less pectin and xyloglucan, and higher levels of glucuronoarabinoxylan (GAX) (Smith and Harris 1999).
All plants have a superfamily of genes known as the cellulose synthase-like (CSL) genes, so denoted because of their amino acid sequence similarity to the CESA family encoding cellulose synthases (Saxena et al. 1995; Pear et al. 1996; Arioli et al. 1998; Hazen et al. 2002; Roberts and Bushoven 2007). The 29 members of the Arabidopsis (At) CSL gene family have been subdivided into six families called CSLA, CSLB, CSLC, CSLD, CSLE, and CSLG (Richmond and Somerville 2001). The 34 members of the rice (Oryza sativa) CSL gene family have been divided into the CSLA, CSLC, CSLD, CSLE, CSLF, and CSLH subfamilies. Rice does not have the CSLB and CSLG subfamilies, but it has two subfamilies (CSLF, CSLH) that are not found in dicotyledonous plants (Hazen et al. 2002).
Based on their similarity to the CESA family, it has been hypothesized that the CSL genes encode glycan synthases for the backbones of the hemicellulosic polymers of the cell wall. The functions of the CSL genes have been studied by forward and reverse genetic and biochemical approaches. In regard to the CSLA genes, mutants of AtCSLA7 are embryo-lethal and the AtCSLA9 mutant rat4 is resistant to transformation by Agrobacterium tumefaciens (Goubet et al. 2003; Zhu et al. 2003). When expressed in heterologous systems, several CSLA proteins encode β-1,4-(gluco)mannan synthases (Dhugga et al. 2004; Liepman et al. 2005, 2007; Suzuki et al. 2006). Of the CSLC family, AtCSLC4 and its ortholog from nasturtium (Tropaeolum majus) can synthesize β-(1,4)-linked glucan, the backbone of xyloglucan, when expressed in Pichia pastoris (Cocuron et al. 2007).
CSLD expression is positively correlated with cellulose biosynthesis in tip growing cells of tobacco and Arabidopsis, but no CSLD family members have been biochemically characterized (Doblin et al. 2001; Favery et al. 2001; Wang et al. 2001). AtCSLD3 mutants have reduced root hair elongation and decreased tensile strength of the cell wall (Favery et al. 2001; Wang et al. 2001). AtCSLD5 mutants have reduced root hair elongation and plant height (Bernal et al. 2007, 2008). A rice CSLD1 mutant shows a similar reduction in root hair elongation, suggesting a conserved function for the CSLD proteins in both monocotyledons and dicotyledons (Kim et al. 2007).
The CSLF family has to date been found only in grasses and cereals (Hazen et al. 2002; Farrokhi et al. 2006; http://www.brachypodium.org/). Expression of rice OsCSLF2 in Arabidopsis results in the appearance of immunologically detectable MLG in the cell walls, and therefore the CSLF subfamily probably encodes MLG synthases (Burton et al. 2006). This conclusion is consistent with the temporal and spatial patterns of expression of the CSLF genes in barley (Hordeum vulgare) (Burton et al. 2008). The functions of the other CSL subfamilies (CSLB, CSLE, CSLG, and CSLH) are not known. Many AtCSL mutants have no discernible phenotype, perhaps due to genetic redundancy (Richmond and Somerville 2001). However, consistent with their established or putative functions in hemicellulose biosynthesis, at least some of the CSL proteins are located in the Golgi, where hemicellulose synthases are known to be located (Ray et al. 1969; Walton and Ray 1982a; Dhugga et al. 2004; Dunkley et al. 2006; Bernal et al. 2007; Cocuron et al. 2007).
Plant growth and cell wall biosynthesis are tightly coupled processes. New cell wall synthesis is needed to prevent wall thinning during cell expansion, and maturation of many cell types involves deposition of a thick secondary wall (Cosgrove 2005). The photomorphogenetic and hormone regulation of tissue elongation has been intensively studied in the mesocotyl of cereals, particularly oats and maize. After germination, this organ elongates rapidly (over 5 cm/day), which serves to position the shoot meristem at the soil level (Tucker 1957). Growth of the cereal mesocotyl is strongly inhibited by exposure to very small light fluences (Blaauw et al. 1968). Light perception by phytochrome leads to decreased levels of auxin in the mesocotyl, which in turn leads to decreased growth (Iino and Carr 1982; Jones et al. 1991; Barker-Bridgers et al. 1998; Sawers et al. 2004).
After exposure to light, activity of a Golgi-localized glucan synthase activity in the mesocotyl is reduced by 50–60% (Walton and Ray 1982a). The decline in activity is mediated by auxin and is not due to a general effect on metabolic activity or on the Golgi because neither plasma membrane-localized callose (β-1,3-glucan) synthase nor Golgi-localized latent IDPase is affected by light (Walton and Ray 1982a, b). The relatively slow kinetics (half-life ~4 h) of the decline in glucan synthase activity, and the fact that auxin is known to regulate many genes at the transcriptional level (Pufky et al. 2003), are consistent with light-regulation of the glucan synthase activity at the level of transcription (Walton and Ray 1982a, b).
Because the kinetics of the decline in Golgi-localized glucan synthase after exposure to light suggested transcriptional control, which is mediated by auxin via phytochrome, and in view of the recent discovery that at least some CSLC genes encode Golgi-localized β-1,4-glucan synthases (Cocuron et al. 2007), we investigated the relationship between cell wall biosynthetic enzyme activities and ZmCSL gene expression after exposure to light. Before this was possible, however, it was first necessary to identify and annotate the maize CSL gene family.
Materials and methods
Annotation of the maize CSL family
The Iowa State Maize Assembled Genome Island 4 (MAGI4) database (http://magi.plantgenomics.iastate.edu/) (Fu et al. 2005) was searched with TBLASTN using OsCSLA1, OsCSLC1, OsCSLD1, OsCSLE1, OsCSLF1, and OsCSLH1 proteins as queries (Supplementary Table S1). Proteins encoded by the MAGI sequences were determined using the FGENESH monocotyledon ab initio gene prediction program (http://www.softberry.com) (Yao et al. 2005). The predicted protein sequences were aligned with the OsCSL protein sequences using ClustalW in MegAlign (DNAstar, Madison, WI). Gene models were refined, based on homology to the Oryza sativa (rice) CSL protein family (Hazen et al. 2002; http://waltonlab.prl.msu.edu/CSL_updates.htm) and all available EST sequences from NCBI (http://www.ncbi.nlm.nih.gov/, BLASTN, database EST_others, Zea mays) and MaizeSeq EST (http://www.maizeseq.org/, BLASTN) databases (Supplementary Table S2). The ESTs were assembled into contigs and aligned with the genomic contigs using Spidey (http://www.ncbi.nlm.nih.gov/spidey/).
MAGI sequences belonging to the same CSL subfamily were aligned (criteria: match size 12, mismatch percentage 80, minimum sequence length 100) using SeqMan (DNAstar). Sequences which aligned were assembled into contigs. These genomic contigs were used to search for additional genomic sequences in the MAGI4, TIGR (http://www.tigr.org/), and Plant Genome (http://www.plantgdb.org/) databases.
The poplar protein sequences were obtained from the Department of Energy Joint Genome Institute (Suzuki et al. 2006) (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). The Arabidopsis CSL, tobacco CSLG, guar mannan synthase (Dhugga et al. 2004) and loblolly pine CSLA (Liepman et al. 2007) sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov). The GenBank and JGI protein accession numbers for the Arabidopsis and poplar proteins can be found in Supplementary Tables S3 and S4. The GenBank accession number of the tobacco (Nicotiana tabacum) CSLG protein is DQ152918; for loblolly pine CSLA1 and A2, DQ641986 and DQ641987, respectively; and for guar mannan synthase the accession number is AAR23313.
Sorghum genomic sequences were obtained from the SAMI (Sorghum Assembled genoMic Islands) database (http://magi.plantgenomics.iastate.edu/). Wheat, sugarcane, barley, and sorghum ESTs were obtained from GenBank.
Proteins were aligned with ClustalX using the default settings (multiple alignment mode) (Thompson et al. 1997). Phylograms were constructed with the heuristic search method in PAUP, and a parsimony phylogram corresponding to the majority consensus of 1,000 bootstrap replicates was constructed (version 4.0b10, Sinauer Associates, Sunderland, MA). The phylogenetic trees were drawn with the Treeview program (Page 1996).
Growth of maize seedlings
Maize caryopses (cultivar FR1061 × FR9661, Midwest Seed Genetics, St. Carroll, IA) were imbibed at room temperature for 24 h in water in an Erlenmeyer flask shaken at 150 rpm. In the morning, the seeds were spread on trays of fine vermiculite soaked in water, covered with a thin layer of vermiculite, and covered with plastic. The trays were kept in complete darkness at 24°C, and after 48 h the plastic covers were removed. In the evening of day 3 (82 h from planting), one tray of seedlings was exposed for 10–15 min to white fluorescent light (97 μMol s−1 m−2), after which the tray was returned to total darkness; 16 h later the plants were harvested, and the second centimeter of the mesocotyl below the first node was collected.
RNA isolation
The tissue was frozen in liquid nitrogen and ground in a mortar and pestle, transferred to Eppendorf tubes (100 mg per tube), and mixed with 1 ml of TRIzole reagent (Invitrogen). All subsequent steps were performed at room temperature and all centrifugations at 4°C unless otherwise indicated. The mixture was incubated for 30 min and 0.2 ml of chloroform was added. The tubes were shaken for 15 s, incubated for 2 min, and centrifuged at 12,000×g for 15 min. The aqueous phase was transferred to a new tube and the RNA was precipitated with isopropyl alcohol. After collection by centrifugation at 12,000×g for 10 min, the RNA was washed with 75% (v/v) ethanol, precipitated by centrifugation at 7,500×g for 5 min, air-dried, and redissolved in 243 μl diethylpyrocarbonate (DEPC)-treated water. The RNA was treated with DNAse [27 μl 10x DNAse buffer (100 mM Tris pH 7.5, 25 mM MgCl2, 5 mM CaCl2) and 1 μl of RNase-free DNAse (Roche)] for 30 min, and then heated for 10 min at 85°C to inactivate the DNAse. The RNA concentration was calculated by OD260.
Quantitative real time (QRT)-PCR
RNA (1 μg) isolated from the second cm of the mesocotyl was reverse-transcribed into cDNA with the Superscript III kit (Invitrogen). The volume of the RNA solution was adjusted to 8 μl using DEPC-treated water. One microliter of 50 μM oligodT primer and 1 μl of 10 mM dNTP mixture were added followed by incubation at 65°C for 5 min. Two microliter of 10× reverse transcriptase buffer, 4 μl 25 mM MgCl2, 2 μl 0.1 mM DTT, 1 μl RNAse OUT and 1 μl of Superscript III reverse transcriptase were added followed by incubation at 50°C for 50 min. The reaction was terminated by heating at 85°C for 5 min.
To control for DNA contamination, most primer pairs were designed to span an intron, which allowed identification of inadvertent genomic DNA contamination by the size of the PCR products. All RNA samples were also treated with DNAse, and all experiments included a sample with RNA that had not been reverse-transcribed.
Primers for ZmCSL genes were designed based on the open reading frames using the PrimerSelect program (DNAstar, Madison, WI). Parameters for primer design were: primer length of 17–24 bp; product length of 400–600 bp; melting temperature of 39.1–70.2°C. The primers were tested on other ZmCSL genes using the PrimerSelect program in order to test their specificity. Only primers that did not give a predicted PCR product with the other ZmCSL genes were synthesized (Integrated DNA Technologies, Coralville, IA). The sizes of the amplified DNA fragments were determined by agarose gel electrophoresis. Primers amplifying the correct size fragments from genomic and cDNA were selected for quantitative real time PCR (QRT-PCR).
Sequences of the primers used for QRT-PCR can be found in Supplementary Table S5. Only primers with efficiency between 90 and 110% were used. The primer concentration in the PCR reactions was 300 nM. An equivalent of 5 ng of reverse-transcribed RNA and 7.5 μl of 2x SYBR Green I dye (Applied Biosystems) was added to each reaction. The PCR reactions were performed on the ABI PRISM 7700 Sequence Detection System (PE Biosystems) with the following cycling parameters: 2 min at 50°C for one cycle, 10 min at 95°C for one cycle, and 15 s at 95°C, and 1 min at 60°C for 40 cycles. The relative expression compared to EF1α was determined with the \( 2_{\text t}^{-\Updelta \Updelta {\text C}}\) method (Livak and Schmittgen 2001).
Isolation of membranes and enzyme assays
During its growth, including after inhibition of growth by light, the mesocotyl does not change more than 10% in diameter or weight per unit length (Walton and Ray 1982a). Therefore, fresh weight and numbers of mesocotyl segments are equivalent. An equal weight of mesocotyl segments isolated from either dark-grown or light-exposed maize seedlings were ground in buffer (50 mM Tris buffer, pH 8, 1 mM EDTA, 0.01 mM MgSO4, and 250 mM sucrose) at a concentration of 3 ml/g fresh weight in a mortar and pestle. All steps were performed at 4°C. After filtering through two layers of cheesecloth, the flow-through was centrifuged for 5 min at 10,000×g. The supernatant was recentrifuged for 20 min at 146,000×g in a Ti-50 rotor (Beckman). The pellet was resuspended with a glass homogenizer in grinding buffer at a final concentration of 1 ml per gm of original fresh weight.
Enzyme assays were performed in 15-ml Corex conical glass tubes. Each reaction contained 100 μl of resuspended microsomal membrane solution (25 μg protein). β-1,4-Glucan synthase assays contained 20 mM MgSO4 and 49 nM UDP-[3H]glucose (34 Ci/mmol, Sigma). The final total volume was 125 μl. Callose (β-1,3-glucan) synthase assays contained no added MgSO4, 49 nM UDP-[3H]glucose, and 1 mM nonradioactive UDP-glucose. Xylan synthase assays contained 2.0 mM MgSO4, 4.1 mM MnCl2 and 1.2 μM UDP-[14C]xylose (238.4 mCi/mmol, Perkin–Elmer). Mannan synthase assays contained 2.0 mM MgSO4, 4.1 mM MnCl2, and 1.4 nM GDP-[14C]mannose (260 mCi/mmol, Amersham). Assays for Gal-incorporating activity contained 1.2 mM MgSO4 and 1.37 μM UDP-[14C]galactose (367 mCi/mmol, Perkin–Elmer). Following incubation (1 h, 25°C), 4 ml 80% (v/v) ethanol was added and the reaction mixtures filtered through GF/A filters. The filters were washed twice with 70% (v/v) ethanol, dried, and counted in scintillation cocktail.
Latent inosine diphosphatase (IDPase) assays contained 0.4 ml working mixture and 50 μl microsomal membrane suspension. The working mixture was made by mixing 13.2 ml stock solution and 1.65 ml 3% (w/v) digitonin. The stock solution was made by mixing 30 ml of 80 mM Tris, pH 7.5, 200 μl 1 M MgCl2, 66 mg IDP, and 6.2 ml water. The assays were incubated for 45 min and stopped by adding 1 ml 14% (w/v) trichloroacetic acid followed by centrifugation for 20 min at 1,000×g. To measure released phosphate, 1 ml of the supernatant was added to 1 ml freshly prepared Taussky-Shorr reagent, which was prepared by mixing 5% (w/v) FeSO4, 1% (w/v) NH4MO4 and 1 N H2SO4 (Taussky and Shorr 1953). This mixture was vortexed and the OD710 measured after 10 min.
Glycan synthase product analysis
For analysis of in vitro reaction products, after addition of ethanol to a final concentration of 80% (v/v), the products were collected by centrifugation (14,000× g, 15 min, 4°C) and washed three times with 70% (v/v) ethanol (4°C). After drying under vacuum, the products were redissolved in 100 μl of the appropriate buffer plus 5 μl of the appropriate enzyme. After incubation, ethanol to a final concentration of 70% (v/v) was added, the samples were centrifuged (14,000×g, 10 min) and the supernatants analyzed by HPLC (see below).
The buffer for endo-β1,4-mannanase (from Bacillus sp., Megazyme E-BMABS [Wicklow, Ireland]) was 50 mM Tris-glycine, pH 8.8. The buffer for endo-β1,4-xylanase (from rumen microorganism, Megazyme E-XYRU6) was 50 mM sodium acetate buffer, pH 5.5. The buffer for endo-β1,3-glucanase (lamininarinase; from Trichoderma sp., Megazyme E-LAMSE) was 50 M acetate, pH 4.5. The buffer for endo-β1,4-glucanase (cellulase; from Trichoderma longibrachiatum; Megazyme E-CELTR) was 50 mM acetate, pH 5. The buffer for endo-β-1,4;1,3-glucanase (lichenase; Megazyme E-LICHN) was 50 mM sodium acetate, pH 6.0.
The HPAEC-PAD analysis (Dionex) was equipped with a 25 μl injection loop and an ED50 electrochemical pulsed amperometric detector (PAD). The radiolabeled digestion products were mixed with oligosaccharide standards (1–5 μg each) and analyzed using a CarboPac PA-1 column (4 × 250 mm) run at a flow rate of 1 ml/min; 1 or 2 min fractions were collected and neutralized with 1 N HCl before scintillation counting. The identity of the radiolabeled compounds was determined by co-elution with the standards.
Analysis of cell walls
For both total wall sugar monomer composition and MLG content, 10 second-centimeter mesocotyl segments of dark-grown or light-exposed seedlings were collected in Eppendorf tubes and frozen in liquid nitrogen. Samples were boiled for 5 min in 1 ml of water, ground in the tubes with a pestle, and centrifuged for 6 min at 14,000×g at 4°C. The supernatants were removed and the pellets washed three times with water, once with ethanol, and once with acetone. The pellets were dried under vacuum and weighed. Total cell wall monomer composition was determined by gas chromatography of the alditol acetate derivatives after acid hydrolysis (Hazen et al. 2003). For MLG analysis, one ml of 25 μM Bis–Tris (pH 6.5), 4 μl of 2% sodium azide, and 10 μl of lichenase (Megazyme E-LICHN) was added to the resuspended pellets and incubated overnight at 30°C. Ethanol was then added to a final concentration of 70% (v/v), the mixtures centrifuged for 10 min at 14,000×g, and the supernatants collected. The pellets were washed twice with 70% ethanol and the supernatants combined. The pooled supernatants were dried under vacuum and the pellets dissolved in 1 ml water. Twenty microliters of each sample were analyzed by high pressure anion exchange chromatography (HPAEC) (Dionex). The gradient conditions were 15 min from 0.5 M NaOH to 0.5 NaOH plus 0.25 M NaOAc. The chromatograph was equipped with a 25 μl injection loop and an ED50 electrochemical PAD. The column was a CarboPac PA-1 column (4 × 250 mm) operated at a flow rate of 1 ml/min.
Results
Annotation of the maize CSL gene family
In general, rice and maize have a comparable number of CSLA, CSLD, CSLE, and CSLF genes, based on the maize genome at the time this analysis was done (November 2007) (Hazen et al. 2002) (Supplementary Table S6). However, the ZmCSLC gene family is expanded from 6 in rice to 12 in maize, including 1 gene, ZmCSLC9 that is probably a pseudogene. ZmCSLA11, CSLE2, and CSLE4 are probably also pseudogenes, because all of these sequences contain stop codons interrupting their coding regions and there are no corresponding ESTs in the NCBI or MaizeSeq EST databases. Another difference between rice and maize is that whereas rice has three CSLH genes, maize appears to have only one. A third difference is that maize has a CSLG gene, in contrast to rice. Approximately eight full-length ZmCSL gene sequences are still lacking from the databases, further analysis of which must await availability of the whole maize genome (http://www.maizesequence.org) (Pennisi 2008).
A cereal-specific CSLA clade and ZmCSLG1
Figure 1 shows an unrooted parsimony phylogram of the deduced full length CSLA protein sequences of maize, rice, poplar (Populus trichocarpa), Arabidopsis, guar (Cyamopsis tetragonoloba), and loblolly pine (Pinus taeda) (Hazen et al. 2002; Suzuki et al. 2006; Dhugga et al. 2004). The tree indicates that there is a cereal-specific group of CSLA proteins that is statistically well-supported (bootstrap value 79%). The CSLA genes that have been experimentally demonstrated to encode (gluco)mannan synthases by heterologous expression are indicated in Fig. 1 (Dhugga et al. 2004; Liepman et al. 2005, 2007; Suzuki et al. 2006). Heterologous expression of OsCSLA7 and PtCSLA4 has been attempted, but OsCSLA7 was not expressed (Liepman et al. 2007) and PtCSLA4 was expressed but showed no activity (Suzuki et al. 2006). None of the CSLA proteins in the cereal-specific group have been shown experimentally to be mannan synthases.
Phylogenetic analysis of CSLA proteins of different plant species. The parsimony phylogram corresponds to the majority consensus of 1,000 bootstrap replicates of the CSLA proteins of Arabidopsis (At), guar (Ct), maize (Zm), pine (Pta), poplar (Pt), and rice (Os). The cereal-specific CSLA clade is indicated by blue background shading. CSLA proteins that have been shown to encode β-1,4-mannan synthases are shown in red. The maize CSLA proteins are shown in bold and underlined. Three predicted ZmCSLA proteins were excluded from the analysis because their full-length sequences are not available. Accession numbers for the Arabidopsis and poplar proteins are in Supplementary Tables S4 and S5
Figure 2 shows an unrooted phylogram of the Arabidopsis, maize, tobacco, poplar and rice CSLE and CSLG proteins, as well as an additional predicted protein from maize that we provisionally named CSLG1. The full length coding region of ZmCSLG1 could be confidently predicted from genomic sequences and overlapping ESTs (Supplementary Tables S1, S2). A related protein is not present in rice but ZmCSLG1 clusters most closely with a poplar-specific subset of the CSLG clade (Fig. 2). ZmCSLG1 clusters with two poplar proteins annotated as PtCSLG1 and PtCSLG3, but not with three other poplar proteins annotated as PtCSLG2, PtCSLG4, and PtCSLG5 (Suzuki et al. 2007) (Fig. 2).
Phylogenetic analysis of CSLE and CSLG proteins. The parsimony phylogram corresponds to the majority consensus of 1,000 bootstrap replicates of CSL proteins from Arabidopsis (At), maize (Zm), tobacco (Nt), poplar (Pt), and rice (Os). Accession numbers for the Arabidopsis and poplar proteins are in Supplementary Tables S4 and S5
The properties of ZmCSLG1 indicate that it is a true member of the CSL family because it has the D, D, D, and QXXRW motifs present in all of the members of the CESA superfamily (Saxena et al. 1995). ZmCSLG1 is predicted to have eight transmembrane domains (http://www.cbs.dtu.dk/services/TMHMM-2.0/). This is similar to the Arabidopsis CSLG proteins which have 6–8 transmembrane domains. The number of exons and the intron–exon structure is also conserved between ZmCSLG1 and AtCSLG1, AtCSLG2, and AtCSLG3. These data support the conclusion that ZmCSLG1 belongs to the CSLG family.
To determine if other cereals have genes related to ZmCSLG1, the available sorghum, rice, barley, sugarcane, and wheat EST and genome databases were searched using TBLASTN and BLASTN. Two ESTs were found in sorghum (Sorghum bicolor) (GenBank, est_others). No ESTs were found for rice and wheat (Triticum aestivum). Sugarcane (Saccharum officinarum) has one EST and barley (Hordeum vulgare) has three (Table 1). The high percentage of identity (Table 1) over their available ranges suggests that these are true relatives of ZmCSLG1. The predicted proteins encoded by these ESTs cluster most closely with ZmCSLG1 when aligned with all of the maize CSL proteins, and with the Arabidopsis, poplar, and tobacco CSLG proteins (data not shown). Therefore, it appears that the CSLG subfamily exists in maize, sugarcane, and barley, but not in rice or wheat.
Expression of the CSL gene family in the maize mesocotyl
Most (28 out of 41) of the maize CSL genes could be amplified from genomic DNA using gene-specific primers (Supplementary Tables S7, S8). The following ZmCSL genes could not be amplified from genomic DNA, although multiple primer pairs were tested for each: CSLC2, C9, C12, D4, F1, and F7. For CSLE2 and E4 insufficient genomic sequences were available to design primers. In the case of CSLC2 and C12, inability to amplify them might be related to the fact that their gene models are still incomplete.
From cDNA made from RNA extracted from the mesocotyls of maize grown in complete darkness, nine ZmCSLA genes, five ZmCSLC genes, one ZmCSLD gene, one ZmCSLE gene, two ZmCSLF genes, and none of the ZmCSLH or ZmCSLG genes could be amplified, a total of 18 CSL genes (44% of the total estimated number of CSL genes in maize).
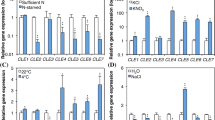
QRT-PCR was performed with cDNA made from RNA isolated from dark-grown mesocotyls and from mesocotyls exposed to 10 min white light 16 h before harvest. Figure 3a shows that the major CSLA expressed in the dark-grown maize mesocotyl is CSLA3 (57% of total CSLA transcript levels), followed by CSLA1. Transcript levels of two CSLA genes (CLSA3 and A7) are strongly affected by light (~7, ~13-fold, respectively). The transcript levels of CSLA4 and A10 are weakly (between two and threefold) affected by light and the transcript levels of CSLA1, A5, A6, A8 and A9 are affected less than twofold (Fig. 3a).
Quantitative real time (QRT)-PCR results for the maize CSL gene family in the maize mesocotyl in response to light. Black bars dark-grown maize seedlings. White bars seedlings exposed to light for 10 min 15 h before harvest. The numbers above the bars denote the fold increase or decrease in transcript levels caused by exposure to light. Note that the scale is logarithmic. a CSLA genes, b CSLC, CSLD, CSLE and CSLF genes. Data represent the mean (n = 3) ± 1 SD. Two-tailed t-test of significance: *** P < 0.001, ** 0.01 > P > 0.001, * 0.05 > P > 0.01. This experiment was repeated twice (two biological replicates using independently isolated RNA) with similar results. The data shown are from one experiment
Figure 3b shows the QRT-PCR results for the CSLC, CSLD, CSLE, and CSLF genes. In the dark-grown mesocotyl, none of the transcript levels are as high as CSLA3, but several are present at moderate levels. The most abundant CSLC is CSLC3 and its levels are moderately decreased by light (2.5-fold). The transcript levels of CSLC4 are not high in the dark-grown mesocotyl but are the most strongly reduced by light (~20-fold). Light causes a large increase in transcript levels of several CSL genes, especially CSLE1 (6.1-fold) and CSLF5 (9.4-fold). No conclusion could be made about the light-dependence of CSLC6 transcript levels because the efficiency of the primer pair was not in the acceptable range for QRT-PCR. Transcript levels of the one CSLD expressed in the maize mesocotyl, CSLD2, were not affected by light. Of the two CSLF genes expressed in the mesocotyl, CSLF5 shows a large increase in transcript levels whereas CSLF6 levels are low in the dark and moderately decreased further by exposure to light (Fig. 3b).
Light-regulation of enzymatic activities related to cell wall biosynthesis
Several studies have demonstrated a positive correlation between expression of specific CSL genes and temporal and spatial accumulation of the hemicellulosic polysaccharides synthesized by the products of those genes (Cocuron et al. 2007; Burton et al. 2008). There may therefore also be a positive correlation between CSL gene expression and measurable enzyme activities catalyzed by the encoded proteins. It was earlier shown that Golgi-localized β-1,4-glucan synthase is down-regulated by light in the maize mesocotyl (Walton and Ray 1982a). If this regulation occurs at the level of transcription of the encoding gene, then there should be a direct correlation between light regulation of its transcript levels and enzyme activity.
To test this hypothesis, we first studied the affect of light on additional glycan synthase activities. Microsomes were isolated from the rapidly elongating second cm of the mesocotyl below the node and incubated with radiolabelled UDP-Glc, GDP-Man, UDP-Gal, or UDP-Xyl (Fig. 4). The products were collected on glass fiber filters and hydrolyzed with specific enzymes. The identities of the hydrolysis products were determined by co-elution from HPAEC-PAD with standards. The products made from UDP-Glc at low (49 nM) and high (1 mM) concentrations are β-1,4-glucan and β-1,3-glucan, respectively (Fig. 4). The products made from UDP-Xyl and GDP-Man are β-1,4-xylan and β-1,4-mannan, respectively (Fig. 4). In the case of the product made from UDP-Gal (Fig. 5), the Gal is not converted to another sugar and the product is solubilized by a crude commercial pectinase (Sigma P2736), but the structure of the products have not been further characterized.
HPAEC-PAD analysis of in vitro products synthesized by microsomes from mesocotyls. In each case, standards (detected by PAD) were co-injected with digested enzyme products (detected by radioactivity). In each case, recovery of radioactivity was >90%. a–c Substrate was 49 nM [3H]UDP-Glc, and products were digested with endo-β-1,4-glucanase (cellulase). Less than 5% of this product was hydrolyzed with either endo-β-1,3-glucanase or lichenase. a standards, b microsomes from dark-grown mesocotyls, c microsomes from light-grown mesocotyls. d, e Substrate was 1 mM [3H]UDP-Glc, and products were digested with endo-β-1,3-glucanase (laminarinase). Products made by microsomes from dark-grown mesocotyls are shown. Less than 5% of this product was hydrolyzed with either cellulase or lichenase. f, g Substrate was 1.2 μM UDP-[14C]xylose, and products were digested with endo-β-1,4-xylanase. Products made by microsomes from dark-grown mesocotyls are shown. h, i Substrate was 1.4 μM UDP-[14C]mannose, and the products were digested with endo-β-1,4-mannanase. Products made by microsomes from dark-grown mesocotyls are shown. Standards: glucose (G1), xylobiose (X2), cellobiose (C2), xylotriose (X3), laminaribiose (L2), mannose (M1), mannobiose (M2), mannotriose (M3)
Enzyme activities in microsomes prepared from mesocotyls grown in total darkness (solid lines, solid symbols) or exposed to 15 min of light 16 h before harvest (dashed lines, open symbols). a β-1,4-glucan synthesis, b β-1,4-mannan synthesis, c UDP-Gal-incorporating activity, d β-1,4-xylan synthesis, e β-1,3-glucan synthesis, and f latent IDPase. Reactions were stopped by boiling and addition of ethanol after 0, 15, 30, 45, or 60 min. Data represent the mean ± range of two measurements of the same membrane sample. The entire experiment was repeated a total of twice with similar results. Values are expressed per mg membrane protein
To investigate the effects of light on glycan synthase activities, dark-grown seedlings were exposed to light for 10 min, returned to total darkness, and harvested 15 h later. Microsomes were isolated from the rapidly elongating second cm below the node. As reported earlier (Walton and Ray 1982a), light causes a decline in β-1,4-glucan synthesis by 43–53% (Fig. 5a). The synthesis of β-1,4-mannan and the ethanol-insoluble product formed from UDP-Gal are more dramatically affected by light (>80%) (Fig. 5b, c). Synthesis of β-1,4-xylan is only slightly affected by light (Fig. 5d). As also reported earlier, light causes no consistent major change in callose biosynthesis (Fig. 5e) or latent IDPase activity (Fig. 5f) (Walton and Ray 1982a; Mitsui et al. 1994).
The light-induced decrease in β-1,4-glucan synthase activity occurs with a half-life of ~4 h (Walton and Ray 1982a). The kinetics of the light-induced decline in β-1,4-mannan synthase and the unknown activity utilizing UDP-Gal (Fig. 5b, c) are similar to the decline in β-1,4-glucan synthase activity (data not shown).
Affect of light on cell wall sugar monomer and MLG content in the mesocotyl
Figure 6 shows that light treatment 15 h before harvest does not affect the content of Rha, Fuc, Ara, Xyl, Gal, or noncellulosic Glc in the mesocotyl by more than 15%. Because CSLF genes encode mixed-linked glucan (MLG) synthases and MLG is rapidly turned over in the coleoptile of etiolated maize seedlings (Burton et al. 2006; Gibeaut and Carpita 1991; Gibeaut et al. 2005), changes in CSLF gene expression might cause detectable changes in MLG content of the mesocotyl. Burton et al. (2008) found a reasonable correlation between expression of some HvCSLF genes and synthesis of MLG in barley endosperm and coleoptile.
Monosaccharide composition of cell walls of maize seedlings grown in complete darkness (shaded bars) or exposed to 15 min of light 16 h before harvest (open bars). Data represent the mean of duplicate measurements of two biological replicates ±1 SD. The experiment was repeated three times with similar results
Expression of only two CSLF genes (ZmCSLF5 and ZmCSLF6) could be detected in the maize mesocotyl. The dominant one, CSLF5, is strongly upregulated by light (Fig. 3). However, Fig. 7 shows that neither the total MLG content nor the concentration of any of the diagnostic MLG oligomers changes in the second cm of the maize mesocotyl after light treatment.
Mixed-linkage glucan (MLG) content of cell walls of maize mesocotyls from seedlings grown in complete darkness (shaded bars) or exposed to 15 min of light 16 h before harvest (open bars). Data represent the mean of two biological replicates (n = 6) ± 1 SD. The experiment was repeated three times with similar results
Discussion
Analysis of the CSL gene family of maize indicates strong overall similarity but also several intriguing differences from rice, the only cereal completely analyzed to date (Hazen et al. 2002). Like rice, maize lacks the CSLB subfamily found in dicotyledonous plants, but does have the two subfamilies (CSLF and CSLH) that have not been found in any dicotyledons. Of particular note, maize apparently has a member of the CSLG subfamily, which we provisionally call CLSG1. It is most closely related to a subgroup of the CSLG proteins from poplar. CSLG genes appear to be present in some, but not all, other cereals. A tree of the CSL superfamily published recently by Farrokhi et al. (2006) indicates a cereal-specific CSL clade, called CSLJ, that contains proteins from maize, barley, wheat, and sorghum, but not rice. Our CSLG1 may be the same as CSLJ, but this cannot be resolved until the CSLJ sequences are published. Further resolution of the existence and phylogenetic position of ZmCSLG1, including whether other cereals have true members of the CSLG subfamily, will need to await more full-length sequences from additional plants.
The maize mesocotyl is an anatomically simple, nonphotosynthetic organ (Tucker 1957). Nevertheless, it expresses 18 of the 39 ZmCSL genes. Nine of them are ZmCSLA, five are CSLC, one is CSLD, one is CSLE, and two are CSLF. Based on maize EST databases, all of these CSL genes are also expressed in other maize tissues (Supplementary Table S2). Therefore, there is no evidence for any mesocotyl-specific CSL genes. An explanation for the large number of CSL genes expressed in a single organ is that CSL genes are specialized for particular cell or tissue types. Some CESA genes show such a pattern, e.g., ZmCESA-3 and ZmCESA-8 are expressed in vascular tissue throughout the maize seedling (Holland et al. 2000).
A question addressed by our experiments is whether expression of any CSL genes is photomorphogenetically co-regulated with known enzyme activities. Some CSLA proteins have been shown to encode β-1,4-mannan synthases (Dhugga et al. 2004; Liepman et al. 2005). We found that mannan synthase activity is strongly down regulated by light (Fig. 5), and mRNA levels of the most highly expressed CSLA (ZmCSLA3) are strongly reduced by light (Fig. 3). ZmCSLA3 is most closely related to OsCSLA1 (Fig. 1), which encodes a mannan synthase (Liepman et al. 2007). The mRNA levels of the third most abundant CSL (ZmCSLA7) are also strongly down-regulated by light (Fig. 3). Taken together, our results are consistent with at least some of the maize CSLA genes encoding β-1,4-mannan synthases. It remains an open question, however, whether all CSLA proteins in all plants have this activity. It has previously been pointed out that although CSLA genes are the largest and most highly expressed family of CSL genes, mannan is a minor component of most cell walls, including those of maize (Liepman et al. 2005, 2007; Suzuki et al. 2006; Carpita et al. 2001). Of particular interest in this regard is the cereal-specific clade of CSLA proteins, none of which have yet been shown to encode mannan synthases (Fig. 1). It is therefore possible that these proteins synthesize cell wall polymers other than (gluco)mannan. This could be one explanation for the discrepancy between the large number of CLSA genes and their strong expression, and mannose scarcity.
In regard to the CSLC family, at least two members (AtCSLC4 and TmCSLC) are localized in the Golgi and catalyze synthesis of the β-1,4-linked glucan backbone of xyloglucan (Cocuron et al. 2007). Although sometimes considered to be a minor component of cereal cell walls, in fact xyloglucan levels in cereals can approach those in dicotyledons (Hayashi 1989). Consistent with xyloglucan being an important hemicellulose in cereals, rice has a large family of xyloglucan endotransglucosylase/hydrolases (XTH) (Yokoyama et al. 2004). Based on the aforementioned results, members of the ZmCSLC family are candidates to encode the light-regulated Golgi-localized β-1,4-glucan synthase (Walton and Ray 1982a, b) (Figs. 3, 5). One explanation of the fact that light causes only a ~50% decrease in β-1,4-glucan synthase activity, compared to a greater than 80% decrease in two other enzyme activities in the same microsomal preparations (Fig. 5), is that mesocotyls contain two or more β-1,4-glucan synthase activities, and not all of them are subject to light regulation. Of the four ZmCSLC genes that are expressed in the mesocotyl, CSLC3 and CSLC4 are candidates to encode the light-regulated β-1-4-glucan (Fig. 3). Expression of CSLC7 was affected by light less than twofold, but CSLC1 expression was enhanced almost fourfold. Therefore, insofar as CSLC gene encode β-1,4-glucan synthases in maize, the situation is complex and there is not a simple relationship between transcript abundance, response to light, and enzyme activity.
Expression of only one CSLD gene (ZmCSLD2) was detected in the maize mesocotyl, and its level of expression is not regulated by light. Therefore, ZmCSLD2 is not a good candidate to encode the β-1,4-glucan synthase, the mannan synthase, or the Gal-incorporating activity. Of the CSLE genes, only CSLE1 is expressed in the mesocotyl, and its transcript levels increase sixfold after light exposure (Fig. 3). Therefore, this gene is also not a good candidate to encode any of the assayed activities (Fig. 5).
Of the seven maize CSLF genes, only CSLF5 and CSLF6 have been shown by our experiments to be expressed in the maize mesocotyl. Light induces a large increase in CSLF5 transcript levels, which pattern is not consistent with CSLF5 encoding proteins with any of the assayed enzymatic activities (Fig. 5) nor with the lack of change in MLG levels after light treatment (Fig. 7). However, our results cannot exclude the possibility that any light-induced changes in de novo MLG synthesis might be undetectable because of background levels present in the mesocotyl before exposure to light, or because of counteracting light-regulation of MLG levels by a MLG-specific glycosyl hydrolase (Inouhe and Nevins 1991).
Only one ZmCSLH was found in maize, compared to three in rice. However, it is possible that more ZmCSLH family members will be found once the complete genome of maize is analyzed. Expression of ZmCSLH1 could not be detected in the maize mesocotyl, and therefore it is probably not involved in the synthesis of MLG nor does it encode any of the assayed enzyme activities in this tissue.
Of the other enzyme activities assayed (Fig. 5), callose synthase is known to be encoded by the GSL family of proteins, which are located in the plasma membrane (Jacobs et al. 2003; Li et al. 2003). Our results predict that transcript levels of the responsible gene are not strongly regulated by light or auxin in the maize mesocotyl. Likewise, in vitro xylan synthesis is not strongly affected by light (Fig. 5). If the xylan synthase is encoded by a member of the CSL superfamily, it is not one of the strongly light-regulated genes.
Although enzyme activities either stayed approximately the same or declined sharply after light (Fig. 5), transcript levels of some CSL genes increased after exposure to light (CSLC1, CSLE1, and CSLF5). It is not known what activities are encoded by these up-regulated CSL genes, but since secondary wall synthesis takes place when growth of the mesocotyl ceases, these CSL genes might encode enzymes involved in secondary wall biosynthesis (Goodwin 1942). A gene for polyamine oxidase, involved in secondary wall lignin biosynthesis, is up-regulated by light in the mesocotyl (Piazza et al. 2002; Cona et al. 2003).
In conclusion, the results in this paper extend our knowledge of the phylogenetic distribution of the large and important CSL superfamily. A large percentage of the maize CSL genes are expressed in the mesocotyl, and transcript levels of some are regulated by light. The proximal cause of the light-inhibition of mesocotyl growth is a decrease in active auxin levels, and therefore the light-regulation of CSL transcript levels is probably regulated by auxin. Because auxin is known to be a critical regulator of growth, and because of the essential coupling between plant cell growth and cell wall biosynthesis, it is reasonable to expect that CSL gene expression changes in parallel with changes in growth rate of the mesocotyl.
Abbreviations
- CSL:
-
Cellulose synthase-like
- MLG:
-
Mixed-linked glucan
- HPAEC:
-
High performance anion exchange chromatography
- PAD:
-
Pulsed amperometric detector
References
Arioli T, Peng L, Betzner SA, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazenski J, Birch R, Cork A, Glover J, Redmond J, Williamson RE (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279:717–720
Barker-Bridgers M, Ribnicky DM, Cohen JD, Jones AM (1998) Red-light-regulated growth. Changes in the abundance of indole acetic acid in the maize (Zea mays L.) mesocotyl. Planta 204:207–211
Bernal AJ, Jensen JK, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, de Lotto R, Pauly M, Scheller H, Willats W (2007) Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J 52:791–802
Bernal AJ, Yoo C-M, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sorensen I, Blancaflor EB, Scheller HV, Willats WGT (2008) Functional analysis of the cellulose synthase like genes CSLD1, CSLD2 and CSLD4 in tip growing Arabidopsis cells. Plant Physiol 148:1238–1253
Blaauw OH, Blaauw-Jansen G, Van Leeuwen WJ (1968) An irreversible red light-induced growth response in Avena. Planta 82:87–104
Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1, 3;1, 4)-β-d-glucans. Science 311:1940–1942
Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCSslF gene family in barley. Plant Physiol 146:1821–1833
Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47:445–476
Carpita NC, Defernez M, Findlay K, Wells B, Shoue DA, Catchpole G, Wilson RH, McCann MC (2001) Cell wall architecture of the elongating maize coleoptile. Plant Physiol 127:551–565
Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson C (2007) A gene from the cellulose synthase-like C family encodes a β-1, 4 glucan synthase. Proc Natl Acad Sci USA 104:8550–8555
Cona A, Cenci F, Cervelli M, Federico R, Mariottini P, Moreno S, Angelini R (2003) Polyamine oxidase, a hydrogen peroxide-producing enzyme, is up-regulated by light and down-regulated by auxin in the outer tissues of the maize mesocotyl. Plant Physiol 131:803–813
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell biol 6:850–861
Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, Anderson P (2004) Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303:363–366
Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125:2040–2052
Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, Watson RB, Dupree P, Lilley KS (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103:6518–6523
Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB (2006) Plant cell wall biosynthesis:genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotech J 4:145–167
Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15:79–89
Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, Aluru S, Schnable PS (2005) Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc Natl Acad Sci USA 102:12282–12287
Gibeaut DM, Carpita NC (1991) Tracing cell wall biogenesis in intact cells and plants. Selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol 97:551–561
Gibeaut DM, Pauly M, Bacic A, Fincher GB (2005) Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221:729–738
Goodwin RH (1942) On the development of xylary elements in the first internode of Avena in dark and light. Am J Bot 29:818–828
Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P (2003) AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131:547–557
Hayashi T (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol 40:139–168
Hazen SP, Scott-Craig JS, Walton JD (2002) Cellulose synthase-like genes of rice. Plant Physiol 128:336–340. Available at http://waltonlab.prl.msu.edu/CSL_updates.htm
Hazen SP, Hawley RM, Davis GL, Henrissat B, Walton JD (2003) Quantitative trait loci and comparative genomics of cereal cell wall composition. Plant Physiol 132:263–271
Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123:1313–1324
Iino M, Carr DJ (1982) Sources of free IAA in the mesocotyl of etiolated maize seedlings. Plant Physiol 69:1109–1112
Inouhe M, Nevins DJ (1991) Auxin-enhanced glucan autohydrolysis in maize coleoptile cell walls. Plant Physiol 96:285–290
Jacobs A, Lipka V, Burton R, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher G (2003) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15:2503–2513
Jones AM, Cochran DS, Lamerson PM, Evans ML, Cohen JD (1991) Red light-regulated growth. Changes in the abundance of indoleacetic acid and a 22-kilodalton auxin-binding protein in the maize mesocotyl. Plant Physiol 97:352–358
Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han C (2007) OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol 143:1220–1230
Li J, Burton RA, Harvey AJ, Hrmova M, Wardak AZ, Stone BA, Fincher GB (2003) Biochemical evidence linking a putative callose synthase gene with (1 → 3)-β-d-glucan biosynthesis in barley. Plant Mol Biol 53:213–225
Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102:2221–2226
Liepman AH, Nairn CJ, Willats WGT, Sørensen I, Roberts AW, Keegstra K (2007) Functional genomic analysis supports conservation of function among cellulose synthase-like A gene family members and suggests diverse roles of mannans in plants. Plant Physiol 143:1881–1893
Livak KJ, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Mitsui T, Honma M, Kondo T, Hashimoto N, Kimura S, Igaue I (1994) Structure and function of the Golgi complex in rice cells. II. Purification and characterization of Golgi membrane-bound nucleoside diphosphatase. Plant Physiol 106:119–125
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93:12637–12642
Pennisi E (2008) Corn genomics pops wide open. Science 319:1333
Piazza P, Jengkins ProcissiA, GI TonelliC (2002) Members of the c1/pl1 regulatory gene family mediate the response of maize aleurone and mesocotyl to different light qualitities and cytokinins. Plant Physiol 128:1077–1086
Pufky J, Qiu Y, Rao MV, Hurban P, Jones AM (2003) The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct Integr Genomics 3:135–143
Ray PM, Shininger TL, Ray MM (1969) Isolation of β-glucan synthetase particles from plant cells and identification with Golgi membranes. Proc Natl Acad Sci USA 64:605–612
Richmond TA, Somerville CR (2001) Integrative approaches to determining Csl function. Plant Mol Biol 47:131–143
Roberts AW, Bushoven JT (2007) The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63:207–219
Sawers RJ, Linley PJ, Gutierrez-Marcos JF, Delli-Bovi T, Farmer PR, Kohchi T, Terry MJ, Brutnell TP (2004) The Elm1 (ZmHy2) gene of maize encodes a phytochromobilin synthase. Plant Physiol 136:2771–2781
Saxena IM, Brown RM Jr, Fevre M, Geremia RA, Henrissat B (1995) Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol 177:1419–1424
Smith BG, Harris PJ (1999) The polysaccharide composition of Poales cell walls: Poaceae cell walls are not unique. Biochem System Ecol 27:33–53
Suzuki S, Li L, Sun YH, Chiang VL (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142:1233–1245
Taussky HH, Shorr E (1953) A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem 202:675–685
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Tucker SC (1957) Ontogeny of the etiolated seedling mesocotyl of Zea mays. Bot Gaz 118:160–174
Walton JD, Ray PM (1982a) Inhibition by light of growth and Golgi-localized glucan synthase in the maize mesocotyl. Planta 156:302–308
Walton JD, Ray PM (1982b) Auxin controls Golgi-localized glucan synthase activity in the maize mesocotyl. Planta 156:309–313
Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M (2001) AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol 126:575–586
Yao H, Guo L, Fu Y, Borsuk LA, Wen TJ, Skibbe DS, Cui X, Scheffler BE, Cao J, Emrich SJ, Ashlock DA, Schnable PS (2005) Evaluation of five ab initio gene prediction programs for the discovery of maize genes. Plant Mol Biol 57:445–460
Yokoyama R, Rose JK, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 134:1088–1099
Zhu Y, Nam J, Carpita NC, Matthysse AG, Gelvin SB (2003) Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol 133:1000–1010
Acknowledgments
This work was supported by grants from the National Science Foundation (DBI 0211797) and the US Department of Energy, Division of Energy Biosciences (DE-FG02-91ER20021). We thank members of Ken Keegstra and Curt Wilkerson’s laboratories for many helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
van Erp, H., Walton, J.D. Regulation of the cellulose synthase-like gene family by light in the maize mesocotyl. Planta 229, 885–897 (2009). https://doi.org/10.1007/s00425-008-0881-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0881-3