Abstract
FLOWERING LOCUS T (FT) like genes are crucial regulators (both positive and negative) of flowering in angiosperms. We identified two FT homologs in Chenopodium rubrum, a short-day species used as a model plant for the studies of photoperiodic flower induction. We found that CrFTL1 gene was highly inducible by a 12-h dark period, which in turn induced flowering. On the other hand, photoperiodic treatments that did not induce flowering (short dark periods, or a permissive darkness interrupted by a night break) caused only a slight increase in CrFTL1 mRNA level. We demonstrated diurnal oscillation of CrFTL1 expression with peaks in the middle of a light period. The oscillation persisted under constant darkness. Unlike FT homologs in rice and Pharbitis, the CrFTL1 expression under constant darkness was very low. The CrFTL2 gene showed constitutive expression. We suggest that the CrFTL1 gene may play a role as a floral regulator, but the function of CrFTL2 remains unknown.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transition from vegetative growth to flowering is a crucial commitment in plant development. The proper timing of flowering ensures successful reproduction and the persistence of populations and species. Multiple signaling pathways receptive to the environmental cues are responsible for the accurate regulation of flower induction (reviewed by Lee et al. 2006).
FLOWERING LOCUS T (FT) located in the photoperiod-dependent pathway acts as a floral promoter (Kardailsky et al. 1999) and also integrates signals from other pathways (Lee et al. 2006). The FT gene codes for a small protein similar to mammalian PEBP (phosphatidyl ethanolamine-binding protein, Schoentgen et al. 1987). In Arabidopsis thaliana, FT interacts with the transcription factor FD which binds to the promoters of florally expressed genes, such as APETALA1 (AP1) (Wigge et al. 2005). Recent experiments suggest that FT protein may be a florigen (Zeevaart 1976), a substance that moves from leaves to the apex and induces flowering (Jaeger and Wigge 2007; Tamaki et al. 2007).
The FT gene belongs to a small gene family of floral regulators, which in A. thaliana involves five other members. The closest relative of FT in A. thaliana is TWIN SISTER OF FT (TSF). It promotes flowering in a redundant manner with FT (Yamaguchi et al. 2005), but differs in how it responds to environmental conditions. TSF shows higher expression at low temperature (16°C) than FT (Blazquez et al. 2003) and also plays a role in flowering under short-day conditions (Yamaguchi et al. 2005). TERMINAL FLOWER 1 (TFL1) (Bradley et al. 1997) is an inhibitor of flowering despite exhibiting a high sequence homology with FT. The antagonistic activity of TFL1 is mediated by aspartic acid (position 144) and histidine (position 88) which forms a ligand-binding site (Ahn et al. 2006). These residues are conserved in all the proteins of the FT/TFL1 family which inhibit flowering, including a third member of this family, the A. thaliana CENTRORADIALIS homologue (ATC) (Mimida et al. 2001). MOTHER OF FT AND TFL1 (MFT) is a weak floral inducer (Kobayashi et al. 1999; Yoo et al. 2004), but its loss-of-function mutation had no influence on flowering under long-day conditions in A. thaliana (Yoo et al. 2004). Very little information is known about the function of the final member of this gene family BROTHER OF FT AND TFL1 (BFT) (Kobayashi et al. 1999).
FT and TFL1 are conserved in sequence and function across angiosperms (Carmel-Goren et al. 2003; Hecht et al. 2005; Kojima et al. 2002; Hsu et al. 2006). The degree of conservation and the functional role of other FT/TFL1 family members are less well known. As plants have variable life history strategies (perennial, annual) and growth forms (trees, shrubs, herbs), different FT/TFL1 family members may have evolved to take on different roles and functional requirements specific to particular life forms. One mechanism that generates several FT duplicates is polyploidization. It is common in higher plants and is considered to be a main force in the evolution of novel gene functions (Ohno 1970). Each polyploidization event results in the doubling (or other higher multiples) of the entire FT/TFL1 gene family. Because these regulator genes play critical roles in plant development, tight control of the evolutionary fate of new gene copies is necessary to ensure polyploid survival (Otto and Yong 2002). Duplicated copies have three potential fates: (1) they can retain the original function, (2) they may escape from selective constraints, accumulate mutations and finally lose coding capacity, or (3) their evolutionary trajectories may be redirected to establish different functions (Lynch and Force 2000).
We chose the genus Chenopodium, which consists of species with various ploidy levels (Rahiminejad and Gornall 2004), to search for putative FT homologs with altered function that might have arisen following polyploidization. Although the genus Chenopodium, especially the tetraploid species Chenopodium rubrum (short-day ecotype 374), has been used extensively in studies of flowering (Cumming et al. 1965; Cumming 1967; Cumming and Seabrook 1985; Ullmann et al. 1985), a very little is known about the molecular background of floral regulatory pathways in this species. Recently, a partial sequence of CrFL, a FLORICAULA/LEAFY ortholog was found in C. rubrum by Veit et al. (2004). The function of LFY orthologs in many plant species is to promote floral development (Wada et al. 2002). No member of the FT/TFL1 gene family has been yet identified in C. rubrum.
Ecotype 374 of C. rubrum shows an interesting photoperiodic regulation of flowering. Flowering can be induced by a single 12-h dark period in plants as young as 4–5 days, shortly after the opening of cotyledons. However, this permissive developmental period is followed by a period of reduced photoperiodic sensitivity during which two to three 12-h nights are required for full flower induction (Seidlová and Krekule 1973; Ullmann et al. 1985). The response to a single period of 12-h darkness is restored in plants older than about 14 days. The number of short-day cycles required for flowering depends on leaf formation on the shoot apex (Ullmann et al. 1985; Blažková et al. 2000). Identification of key regulatory genes and analysis of their expression in various developmental stages and under various photoperiodic regimes are necessary to understand this unique flowering response.
In this study, we describe two counterparts of FT genes in C. rubrum, CrFTL1 and CrFTL2. We analyzed the expression of both the genes under various photoperiodic regimes, including continuous light and prolonged darkness (72 h). We also applied a short exposure to light in the middle of night, called a night break, which inhibits flowering in short-day plants (Thomas and Vince-Prue 1997). While the CrFTL1 gene was strongly upregulated under the experimental conditions that induced flowering, the CrFTL2 gene expression was invariable.
Materials and methods
Plant material
In all experiments, plants of Chenopodium rubrum L., ecotype 374 (seed from the Institute of Experimental Botany AS CR v.v.i, originally donated by B.G. Cumming University of New Brunswick, Canada) were cultivated in growth chambers at 20°C and in continuous light (cool-white fluorescent, 100 μmol m−2 s−1), except when exposed to photoperiodic treatments as indicated below.
Seedlings of C. rubrum used in flower induction experiments were planted into 96-well flat-bottom ELISA plates (one seedling per well). The wells were filled with perlite (particle size 0.4–0.8 mm) and had a 1-mm hole at the bottom to provide nutrient solution. The plates were floated in half-strength Hoagland solution which was replaced every 2 days. At the age of 5 days, when 40–60% of seedlings had their cotyledons fully opened, average-sized plants with fully opened cotyledons were selected for the experiment and all other plants were discarded. Six hours after this selection, plants were exposed to various photoperiodic treatments as described in the “Results”. Controls were kept in constant light.
For night-break treatments, plants were exposed to 10 μmol m−2 s−1 of red light for 15 min (cool-white fluorescent light filtered through one layer of red plastic filter and two layers of yellow filter, manufactured by Rand Strand Electric, London, UK; this combination had no measurable transmission below 580 nm). The exposure started 6 h after the beginning of a dark period.
Sampling for expression during darkness was performed under a dim green fluorescent light to avoid eliciting photoperiodic signaling responses from the plants. To characterize flower development, 10–20 plants from each variant were left intact and grown in continuous light until the age of 14 days. Flowering stages were determined under a stereomicroscope.
In a separate set of experiments, plants of C. rubrum were grown in 8 × 8 cm pots filled with perlite and supplied with half-strength Hoagland solution. At the age of 15 days, plants were induced to flower by exposure to a 12-h light/12-h darkness photoperiodic regime for 1–3 days (i.e., they experienced 1, 2, or 3 12-h nights); control individuals were kept in continuous light. Plants were then returned to constant light. Flowering of plants was assessed 8 days later by dissecting their shoot apices under a stereomicroscope.
Cloning of CrFTL1 and CrFTL2 cDNAs
Total RNA was extracted with the RNeasy Plant Mini kit (Qiagen, Hilden, Germany) from the apex and upper leaves of 3-week-old plants of C. rubrum, which were induced to flower by a single 12-h darkness. RNA was treated with DNA-free (Ambion, Austin, TX, USA) to eliminate the traces of genomic DNA. cDNA was prepared from the total RNA by means of a SMART PCR cDNA Synthesis kit (Clontech, BD Biosciences, Palo Alto, CA, USA) according to manufacturer instructions. This approach leads to enrichment of full length cDNA. This cDNA was cloned into a T/A vector using pGEM-T Easy kit (Promega, Madison, WI, USA). The cDNA library was used as a template in a modified RACE procedure based on seminested PCR (Huang and Chen 2006).
A pair of primers, CrFT345For and CrFT501Rev (Table 1), was designed using a conserved region from well-studied and confirmed FT genes in dicots. C. rubrum cDNA was amplified in a T Gradient cycler (Biometra, Goettingen, Germany) using these primers and Taq polymerase (Promega) under the following conditions: initial denaturation 2 min at 94°C; 36 cycles consisting of 40 s at 93°C, 45 s at 54°C and 2 min at 72°C; final extension 10 min at 72°C. The PCR fragment was sequenced and new specific CrFT primers were designed using this sequence as a template (Table 1). RACE was performed with the cDNA library as a template and primers targeted to the sequence of the pGEM-T Easy vector (GGT AAC GCC AGG GT and TGT TAT CCG CTC ACA ATT CC) and specific CrFT reverse primers (to search for 5′ end) or forward primers (to search for the 3′ end). PCR conditions were: 2 min at 94°C; 35 cycles 40 s at 93°C, 45 s at 60°C and 2 min at 72°C; final extension 10 min at 72°C. If no band was generated, the product of the first round of PCR (10× to 100× diluted) was reamplified with the primer targeted to the vector sequence and the gene-specific primer (Table 1) closer to the end of the known FT sequence (seminested PCR). The PCR products were cloned using the pGEM-T Easy (Promega) and transformed into E. coli XL-1 Blue. To verify the sequence of full length CrFTL cDNAs, new primers in untranslated regions (UTRs) were designed (Table 1), and the cDNA library was amplified using Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland). All sequencing was performed on an ABI 3100 capillary sequencing machine (Applied Biosystems, Foster City, CA, USA).
Southern hybridization
A sorbitol extraction method (Štorchová et al. 2000) was applied to extract DNA from about 5 g of fresh leaf tissue. Three micrograms of DNA were digested with HindIII (Fermentas UAB, Vilnius, Lithuania), electrophoresed overnight on a 0.7% agarose gel and transferred to a positively charged membrane (Hybond N+, Amersham, Little Chalfont, UK) by capillary blotting. A PCR fragment of CrFTL1 gene was amplified using the primers CrFT39For and CrFTL1Rev (Table 1) and labeled with digoxigenin (DIG) using PCR labeling kit (Roche Applied Science, Germany) according to the manufacturer. The genomic blots were hybridized in an EasyHyb buffer (Roche, Applied Science, Mannheim, Germany) with the DIG probe at 42°C overnight, washed twice at high stringency (0.1× SSC, 65°C), and detected using CDPStar (Roche) as a substrate. Exposure time of 4 h was sufficient to visualize single genes on Hyperfilm (Amersham).
Gene expression estimation by means of qRT PCR
One microgram of total RNA and 500 ng of oligodT25V primers (Metabion, Planegg, Germany) were heated for 5 min at 70°C, chilled on ice and mixed with reverse transcription (RT) buffer, 0.5 μl of Protector RNase Inhibitor (Roche), 1.2 μl of 10 mM dNTPs and 100 U of M-MLV Reverse Transcriptase RNaseH Minus, Point Mutation (Promega), final volume 25 μl. The first strand cDNA was synthesized at 42°C for 60 min.
First strand cDNA was diluted 20× and qPCR was performed using FastStart DNA MasterPLUS SYBR Green I Kit (Roche) in a final volume of 10 μl with 300 nM of each of the HPLC purified primers (Metabion). Glass (Roche) or plastic capillaries (Genaxxon, Biberach, Baden-Wurttemberg, Germany) were applied. The LightCycler 1.2 (Roche) was programmed as follows: after 10 min of initial denaturation at 95°C, 45 cycles of 10 s at 95°C for melting, 4 s at 58°C (actin, CrFTL1) or 60°C (CrFTL2, CrFL) for annealing and 7 s at 72°C for extension were performed. cDNA derived from calibrator RNA was included in each LightCycler run to correct for run-to-run differences. qPCR with all the samples was repeated using a new real-time device LC480 (Roche) under exactly the same cycling conditions as with the LightCycler 1.2. for actin and CrFTL2; or under slightly changed cycling program [10 s at 95°C for melting, 8 s at 58°C (CrFTL1) or 60°C (CrFL) for annealing and 15 s at 72°C for extension]. PCR efficiencies were estimated from calibration curves generated from serial dilution of cDNAs. The calibrated normalized ratios of the relative amount of the target and reference gene were calculated as follows:
where ET, ER are the efficiency for target or reference gene qRT PCR assay; CpT, CpR a crossing point for target or reference genes; S sample; C calibrator.
Expression levels in C. rubrum were normalized against actin (GenBank acc. number AM263428; Table 1).
Sequence and phylogenetic analysis
PCR products were cloned using the pGEM-T Easy kit (Promega) and sequenced. Contig assembly was accomplished using Vector NTI Suite 9 (InforMax, Invitrogen, Paisley, UK). A multiple alignment was performed using AlignX in Vector NTI followed by manual correction.
The phylogenetic relationships among sequences were estimated using the maximum parsimony method (MP) in PAUP* 4 b10 (Swofford 2003). We calculated phylogenies based on the amino acid sequences of 16 genes, including CrFTL1 and CrFTL2, which are reported here. We incorporated a simple codon-based model of evolution that weights transitions between amino acid residues by the minimum numbers of nucleotide changes required to evolve from one to another (Felsenstein 2004). This same methodology is employed by PROTPARS described at http://bioportal.cgb.indiana.edu/docs/tools/phylip/protpars.html. The weight matrix was kindly supplied to the authors by D. Swofford (Florida State University, Tallahassee, USA) and will be provided by request. Consensus bootstrap trees and bootstrap values were calculated using 1,000 pseudo-replicates from random seeds. Heuristic search strategies were employed to reduce tree search times.
Sequence data from this study have been deposited in GenBank data library under the accession numbers EU128013 (CrFTL1, complete cds); EF445636 (CrFTL2, complete cds); EF422350 (CrFTL1, intron); EF422358 (CrFTL2, intron). Sequences of FT like proteins were taken from GenBank and their accession numbers are indicated under the respective figures.
Results
Cloning and characterization of the cDNAs of CrFTL1 and CrFTL2 from C. rubrum
Two cDNA clones were isolated from the cDNA library prepared from the apex and upper (folded) leaves of a 3-week-old plant of C. rubrum ecotype 374 that was induced to flower by 12 h of darkness. Complete coding sequence and 3′ and 5′ UTRs of CrFTL2 were obtained by modified 3′ and 5′ RACE with single primers CrFT345For and CrFT142Rev (Table 1). RACE with nested primers (CrFT39For, CrFT78For for 3′ end and CrFT116Rev, CrFT83Rev for 5′ end; Table 1) was used to find the 3′ and 5′ ends of CrFTL1.
Figure 1 shows an alignment of the predicted complete amino acid sequences of CrFTL1 and CrFTL2 with representative members of FT/TFL1 family from angiosperms. CrFTL2 has 52% identity (62% similarity) with A. thaliana FT and differs in several conserved regions from all other family members (positions 1–18, 104–107 in the alignment; Fig. 1). It also has a premature stop codon located in position 179 and lacks the last 9–12 amino acids, which are present in other FT/TFL1 proteins. Moreover, isoleucine occupies position 144 in CrFTL2. This position has importance as a potential ligand-binding pocket (Ahn et al. 2006); glutamine is present at this position in all known FT orthologs, and aspartic acid in TFL1 orthologs. These features of amino acid sequence suggest that the function of CrFTL2 could be different from that of FT or TFL1. By contrast, 128 amino acid residues of CrFTL1 correspond to A. thaliana FT (73% identity) including the functionally important glutamine in position 144 and tyrosine in 93.
The alignment of amino acid sequences of CrFTL1 and CrFTL2 with other FT/TFL1 family members from angiosperms. Dashes denote gaps. Slashes denote exon boundaries, capital letters above the alignment indicate functionally important conserved amino acids (Tyr85 and Gln140 of AtFT). Protein sequences taken from GenBank include the following: SlSP3D, AAO31792; and SlS5PG, AAO31793 (from Lycopersicon esculentum); AmCEN, AAB36112 (from Antirrhinum majus); AtMFT, AAD37380; AtTSF, Q9S7R5; AtFT, NP_176726; AtTFL1, NP_196004 and AtBFT, Q9F1T4 (from Arabidopsis thaliana); MdTFL1, BAD06418 from Malus × domestica; OsHd3a, BAB61030 (from Oryza sativa)
To evaluate the relationship of CrFTL1 and CrFTL2 with other plant FT/TFL1 proteins more closely, we performed a phylogenetic analysis (Fig. 2). The bootstrap consensus tree was not well resolved, but did support a hypothesis that CrFTL1 and CrFTL2 are more closely related to other FT homologs than to Brother of FT (BFT) or Mother of FT (MFT). This also indicates that both CrFTL1 and CrFTL2 are members of the FT/TFL1 family.
The Maximum parsimony bootstrap consensus tree based on amino acid sequences of angiosperm FT proteins. Transitions between amino acid residues were weighed by the minimum numbers of nucleotide changes required to evolve from one to another. AtBFT was used as the outgroup. The bootstrap support values calculated from 1,000 pseudoreplicates are indicated near the nodes. Protein sequences taken from GenBank are the following: SlSP3D, AAO31792; SlSP2G, AAO31791; and SlS5PG, AAO31793 (from Lycopersicon esculentum); AtMFT, AAD37380; AtTSF, Q9S7R5; AtFT, NP_176726; and AtBFT, Q9F1T4 (from Arabidopsis thaliana); MdFT, BAD08340 (from Malus × domestica); CsFT, BAA77836 (from Citrus sinensis), AAR04684; PnFT4a, BAD08337; PnFTL4, BAD22677; PnFT2a, BAD01576; and PnFT1b, bAD01561 (from Populus nigra); OsHd3a, BAB61030 (from Oryza sativa)
We also analyzed the genomic structure of the CrFTL1 and CrFTL2 genes and compared it with FT gene of A. thaliana. The position and number of introns were identical, but their lengths differed (Fig. 3).
To estimate the number of FT like genes in the C. rubrum genome we performed Southern hybridization with a probe derived from the coding region of the CrFTL1 gene. Two bands were visible on the autoradiogram (Fig. 4a). As the target region of the CrFTL1 gene recognized by the probe did not contain HindIII restriction site, the bands corresponded to CrFTL1 and another FT like gene. This gene could not be CrFTL2, as the CrFTL1 probe was highly specific and gave very weak signal with the CrFTL2 gene (Fig. 4b). Therefore, one gene, closely related to CrFTL1 exists in C. rubrum. Restriction analysis data of the genomic regions are not presented.
Southern hybridization with DIG labeled probe derived from the CrFTL1 gene. a Two bands were revealed in total DNA of C. rubrum, digested with HindIII. b The plasmids with cloned CrFTL1 or CrFTL2 genes were diluted as indicated and applied in double dots on the membrane and hybridized. The CrFTL2 gene gave about 1,000× weaker signal than the CrFTL1 gene
Diurnal oscillations of CrFTL1 mRNA levels in the seedlings of C. rubrum
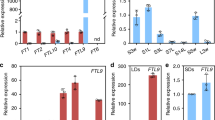
At the age of 5 days, C. rubrum seedlings with fully opened cotyledons were exposed to one of the two treatments: (1) continuous light to a 12-h dark/12-h light photoperiodic regime or (2) a single12-h darkness and then returned to continuous light. Control seedlings were kept under continuous light. mRNA levels for CrFTL1, CrFTL2, and CrFL were estimated in whole shoots by qRT PCR using actin as a reference gene. In the continuous 12-h dark/12-h light regime CrFTL1 expression displayed a diurnal rhythm with a peak in the middle of the light period (Fig. 5a). When only one 12-h dark period was applied, the robust diurnal rhythmicity disappeared after returning the plants to continuous light; an initial high peak of a CrFTL1 mRNA was followed by a slight increase 30–36 h after the first maximum. This observation documents strong damping of the CrFTL1 expression rhythm in the absence of darkness. Constantly low levels of CrFTL1 mRNA were found in continuous light. These observations emphasize an essential role of a dark period for the induction of CrFTL1 transcription.
a The CrFTL1 expression in 5-day-old C. rubrum seedlings displays a diurnal rhythm at 12-h dark/12-h light photoperiod. If permanent light follows the first dark period, the oscillation nearly disappears. b The CrFTL2 expression is not influenced by dark or light periods. c CrFL expression starts to increase about 30 h after the treatment by at least one 12-h dark period. The aerial parts of seedlings for RNA extraction were sampled every 3 h for 72 h. White and black bars below the graph represent light and dark periods, respectively. Mean values ± SD were calculated from four independent measurements of two seedlings
Because the maximum CrFTL1 expression occurred in the middle of the light period, we investigated the transcription pattern during permanent darkness for comparison. The 5-day-old, light grown seedlings were transferred to darkness for 72 h. Unlike arrhythmic expression exhibited under continuous light, the CrFTL1 transcript abundance showed clear rhythmicity with the first peak around 12 h after the transfer into darkness (Fig. 6). This corresponded to the length of a dark period, which induced maximal flowering. The second peak occurred at 45 h and was slightly lower, suggesting the period of the CrFTL1 rhythmic expression was about 33 h. Because the first peak could be affected by the light to dark transition, the time span between the second and third peaks (15 h) would be a better estimate of the period. However, the position of the third maximum was not exactly to determine as the third peak was burdened with high standard error. Generally, the amplitudes of the CrFTL1 expression in dark were very low, comparable with the levels of expression of this gene in permanent light. Thus, transfer of C. rubrum seedlings from light to dark, induced rhythmic CrFTL1 expression, but with very low amplitudes.
The CrFTL1 transcript abundance in C. rubrum seedlings follows a rhythmic pattern under constant darkness with a period of about 33 h. The CrFTL1 expression under constant light is arrhythmic. The CrFTL1 transcript abundance under constant darkness and constant light is comparable. Mean values ± SD were calculated from four independent measurements of two seedlings
In contrast to CrFTL1, expression of CrFTL2 (Fig. 5b) was almost identical in seedlings growing under permanent light and seedlings treated by darkness. CrFTL2 expression was about 20–50 times higher than the expression of CrFTL1 under non-permissive (permanent light) conditions.
We extracted RNA from the above-ground tissues of 5-day-old C. rubrum seedlings, which contained both cotyledons and shoot apices. It enabled us to measure the CrFTL1 and CrFL expression in the same specimen of total RNA, even if the two genes are expressed in different organs (cotyledons or apices). CrFL mRNA levels were constantly low in continuous light. However, they started to increase on the third day in the seedlings treated by at least one dark period (Fig. 5c).
At the age of 14 days, flowering was assessed in 17–20 plants from each treatment. One 12-h dark period was sufficient to induce flowering in 94% of plants, three dark periods induced flowering in all examined plants. No flowering was found in plants grown in continuous light. Hence CrFL transcript levels rose only in those treatments that induced flowering (Fig. 5c). Plants treated by the 72-h dark period showed no flowering.
Expression of CrFTL1 depends on the length of dark period in the seedlings of C. rubrum
Five-day-old seedlings of C. rubrum ecotype 374 can be induced to flower by a single dark period; its optimal length is about 12 h, shorter dark periods are only partially inductive. To investigate whether CrFTL1 expression correlates with the strength of photoperiodic flower induction, we measured CrFTL1 transcript levels in shoots after a single 12, 6 or 4-h period of darkness and in continuous light. CrFTL1 expression rose about 20-fold after a single 12-h dark period, while a 6-h dark period resulted in only 3-fold increase (Fig. 7). No change in constantly low CrFTL1 expression was observed if a 4-h period of darkness or permanent light were applied (Fig. 7). The increase in CrFTL1 expression indeed correlated with the efficiency of flower induction, estimated at the age of 14 days in ten individuals. After a single 12-h dark period 90% of plants flowered, whereas only 20% were induced by a 6-h dark period and no flowering was observed in those exposed to a 4-h darkness or permanent light. Thus, the estimated CrFTL1 mRNA levels increased with the length of darkness.
The induction of CrFTL1 expression correlates with the length of a period of darkness. Seedlings were grown at light for 5 days and then transferred to darkness at time 0, except for the plants treated by 6-h darkness, which were transferred at −2 h. Control plants were grown under continuous light. The aerial parts of seedlings for RNA extraction were sampled every 4 h for 24 h. Black and white bars below the graph represent dark and light periods, respectively. Mean values ± SD were calculated from eight independent measurements of four seedlings
As in the previous experiment, CrFTL2 expression was almost identical in seedlings growing under permanent light and seedlings treated by different periods of darkness (data not shown).
The effects of night break on CrFTL1 and CrFTL2 expression
A short exposure to light in the middle of night, called a night break, inhibits flowering in short-day plants, including C. rubrum (Cumming et al. 1965). To test the effects of night break on CrFTL1 and CrFTL2 expression, C. rubrum seedlings were grown under continuous light for 5 days, transferred to darkness for 12 or 18 h, and then back to permanent light. One set of plants was exposed to a break of red light for 15 min in the middle of 12-h dark period—6 h after the transfer to darkness. Another set of plants was treated by a night break at the same time and left in dark for additional 12 h—they were subjected to dark for 18 h on the whole. Control seedlings were exposed to 12-h darkness without a night break. Flowering response was analyzed in 20 individuals from each set. In control plants 80% were induced to flower, whereas no plants flowered in the variants exposed to the night break.
The CrFTL1 mRNA abundance was examined every 4 h for 36 h, starting at the time of shift to darkness. CrFTL1 expression in control seedlings peaked about 6 h after the return to light, reaching about 50-fold level than in darkness (Fig. 8). By contrast, plants exposed to night break showed just a low increase (5-fold) in CrFTL1 transcript abundance (Fig. 8), about 6–8 h after the return to permanent light. The peak of CrFTL1 expression in the seedlings treated with 18-h dark period (6-h dark, night break, 12-h dark) occurred about 8 h later than in the seedlings treated with 12-h dark. Therefore, the CrFTL1 gene was upregulated after the return from dark to light, in both control and night-break treated plants. However, the transcript level was 20-fold higher in controls than in plants exposed to a night break. As in the previous experiments, high CrFTL1 transcript abundance was associated with those experimental conditions which also led to flower induction.
The effect of a night break on the CrFTL1 transcript level in C. rubrum seedlings. Seedlings were grown at permanent light for 5 days and then transferred to darkness at time 0. Night break was applied at 6 h after the transfer to darkness. The aerial parts of seedlings for RNA extraction were sampled every 4 h for 28 h. White and black bars below the graph represent light and dark periods, respectively. Mean values ± SD were calculated from four independent measurements of two seedlings
In contrast to CrFTL1, the expression of the CrFTL2 gene was invariable and not influenced by a nigh break.
Expression of CrFTL1 and CrFTL2 in the plants of C. rubrum with true leaves after photoperiodic induction
As the sensitivity to photoperiodic flower induction depends on age in C. rubrum (Seidlová and Krekule 1973; Ullmann et al. 1985), we investigated the expression of CrFTL1 and CrFTL2 in 15-day-old plants. They were induced to flower by one, two or three periods of 12-h darkness. At the time of induction they had one pair of fully expanded true leaves and still green cotyledons. Expression levels of CrFTL1 in the leaves were estimated immediately after the end of the first and second dark period and 0, 3 and 6 h after the third dark period. The transcript level of CrFTL1 in leaves examined after the first dark period increased slightly (7×; Fig. 9a). The increase was higher (about 15×) than in the control after the second dark period and further increased with time since the end of the third dark period. Six hours after the third dark period it was approximately 50× higher than in the control with uninduced leaves. The efficiency of flower induction was estimated 8 days after the application of dark periods in 20 plants. 100% of plants treated by three or two dark periods developed flower buds, but only 58% of plants were induced to flower by a single period of darkness. No flowering was observed in plants grown at permanent light.
Relative expression levels of CrFTL1 (a) and CrFTL2 (b) in the leaves of 15-day-old C. rubrum. Plants previously grown in constant light were exposed to a 12-h dark/12-h light photoperiodic regime and sampled immediately after the end of the first (1 × D) and second (2 × D) dark period or 0, 3 and 6 h after the third (3 × D) dark period. Light gray bars refer to controls grown in continuous light and black bars refer to induction treatments. Mean values ± SD were calculated from four independent measurements of two plants
CrFTL2 expression showed no significant variation (Fig. 9b).
Discussion
Although the short-day species C. rubrum has been used to investigate flowering for many years (Seidlová and Krekule 1973; Ullmann et al. 1985), regulator genes governing this process have only recently been discovered when Veit et al. (2004) described a LEAFY/FLORICAULA homolog. In this study, we focused on the genes located upstream of LEAFY in the floral signaling pathway and identified two C. rubrum homologs of FT – CrFTL1 and CrFTL2.
CrFTL1 and CrFTL2 show similarity to FT of A. thaliana (Figs. 1, 2), but differ from one another in DNA (and amino acid) sequence and expression pattern. Whereas CrFTL1 transcription is highly induced by a 12-h period of darkness, expression of CrFTL2 is not affected by this treatment at all. CrFTL2 was continuously maintained at about 50 times the level of the background expression of CrFTL1 in non-induced plants.
The nucleotide substitutions in the CrFTL2 result in the replacement of some conservative amino acids (Fig. 1) that are necessary for the correct functioning of FT genes across angiosperms (Ahn et al. 2006). We also note that compared to other genes in the FT/FTL1 family, CrFTL2 has a premature stop codon at the 3′ end and a divergent sequence at the 5′ end of its coding region. These observations might lead to the conclusion that CrFTL2 is a non-functional pseudogene. The preserved open reading frame, correct splicing of the third intron and particularly the high levels of transcription suggest, however, that CrFTL2 does have a biological function, albeit one that is unknown.
The CrFTL1 transcript level displayed diurnal rhythmicity under the 12-h dark/12-h light regime with a peak in the middle of a light period. In A. thaliana, the diurnal expression pattern of FT was characterized by a maximum at the end of a long day (Suarez-Lopez et al. 2001; Yamaguchi et al. 2005). Transcription of Hd3a, an FT ortholog in a short-day Oryza sativa peaks around dawn with a gradual decrease during the 9-h short day (Kojima et al. 2002). Putative Pharbitis (Ipomoea nil) FT orthologs PnFT1 and PnFT2 showed a diurnal rhythm of expression under short day with a peak after dawn (Hayama et al. 2007). The phase of rhythmic expression of FT orthologs is therefore most similar between rice and Pharbitis, two short-day plants studied in the greatest detail. The CrFTL1 mRNA reaches its maximum at midday, which is different from rice, Pharbitis, and A. thaliana.
The CrFTL1 expression dampened to a low level when the first dark period was followed by permanent light. Alternation of light and dark periods was therefore necessary to maintain diurnal oscillation of CrFTL1 expression, similarly to other short-day plants—rice and Pharbitis (Kojima et al. 2002; Hayama et al. 2007).
Under constant darkness, the CrFTL1 expression followed a rhythmic pattern with very low amplitude. This behavior was in deep contrast with recent observations in Pharbitis and rice. Pharbitis FT homologs cycled with higher amplitude under constant darkness than under constant light (Hayama et al. 2007), whereas Hd3a expression in rice peaked in the middle of the night (Ishikawa et al. 2005). Our finding demonstrated that the transfer of seedlings from continuous light to constant darkness caused a change from an arrhythmic pattern of CrFTL1 expression to a rhythmic pattern, but was not sufficient to activate this gene to the level necessary for flower induction. To achieve this level, transfer of plants from darkness to light was necessary (Fig. 6). Taken together, our experiments suggest that upregulation of the CrFTL1 gene in C. rubrum requires (1) a permissive period of darkness and (2) light at the end of dark period.
Similar to the other well-studied short-day plants (Ishikawa et al. 2005; Hayama et al. 2007), flowering of C. rubrum was inhibited by night break, a pulse of red light given at midnight. Night break also inhibited CrFTL1 expression. Interestingly, slight gene activation was observed when the plants treated by night break in the course of 12-h dark period were returned to light. Similar slight upregulation after the transfer to light occurred in plants subjected to an 18-h dark period interrupted with a night break. Thus, the CrFTL1 gene activation was driven by the transfer to light, not by time since the transfer to dark or since a night break. This finding is in agreement with the supposed activation effect of light on the CrFTL1 expression. If a night break was applied, activation effect of a dark period was canceled and light induced just a very weak upregulation of the CrFTL1 expression.
In all the experiments, a high level of CrFTL1 mRNA was always associated with flower induction, whereas a low level of its expression was found when plants did not flower. Based on these transcription results, we conclude, that the CrFTL1 gene may function as one of the floral inducers in C. rubrum. A close relative of the CrFTL1 gene, different from CrFTL2, was revealed by Southern hybridization in the C. rubrum genome (Fig. 4) and multiple floral inducers have been identified in other model species. Despite detailed knowledge of rice and A. thaliana, the roles of floral inducers such as TSF and FTL1 that are related to FT and Hd3a, respectively, are not fully clarified (Kobayashi et al.1999; Doi et al. 2004).
Hayama et al. (2007) concluded that PnFT expression in Pharbitis is regulated by different mechanism than Hd3a expression in rice, apart from the phenotypic similarity of flowering promotion between both short-day species. They assumed that short-day plants adopted various modes of control of flower induction. The results of our study, particularly very low expression of the CrFTL1gene under constant darkness in C. rubrum seedlings supports this hypothesis.
CrFL, the LEAFY/FLORICAULA homolog in C. rubrum is responsible for the transition from vegetative to reproductive meristem (Veit et al. 2004). The CrFL expression started to increase about 30 h after the end of the first dark period. This is several hours after the floral stimulus from cotyledons (florigen) was found to reach the shoot apex (King 1972). Activation of the FT transcription is mediated by CONSTANS (CO) in Arabidopsis (Putterill et al. 1995; Samach et al. 2000), whereas its rice homolog Hd1 (Yano et al. 2000) is proposed to have a dual effect (inhibition or activation) on Hd3a expression (Kojima et al. 2002; Hayama et al. 2003). To elucidate the regulation of CrFTL1 expression, we have recently identified CO homologs in C. rubrum and we are analyzing their expression patterns (Storchova et al. data not shown).
Our gene expression data suggest that CrFTL1 may be a floral regulator. However, more experiments are necessary to bring conclusive evidence. The transformation of A. thaliana (wt and FT loss-of-function mutants) with CrFTL1 and CrFTL2 genes under the control of 35S promoter is currently under way in our laboratory with the aim of analyzing the function of both the genes in a heterologous plant system.
Although focused on C. rubrum, we anticipate that our study will facilitate the identification of flowering related genes in related species such as C. quinoa, an important grain crop in South America (Partap et al. 1998). Understanding developmental processes in this species, particularly regulation of flowering, is of great importance for its future broader use in agriculture.
Abbreviations
- AP1:
-
APETALA1
- ATC:
-
Arabidopsis thaliana CENTRORADIALIS homolog
- BFT:
-
BROTHER OF FT AND TFL1
- CEN:
-
CENTRORADIALIS
- CrFL:
-
FLORICAULA/LEAFY
- FT:
-
FLOWERING LOCUS T
- FTL:
-
FLOWERING LOCUS T- like
- Hd1:
-
Heading date 1
- Hd3a:
-
Heading date 3a
- MFT:
-
MOTHER OF FT AND TFL1
- SP:
-
SELF PRUNING
- TFL:
-
TERMINAL FLOWER
- TSF:
-
TWIN SISTER OF FT
References
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25:605–614
Blažková A, Vondráková Z, Krekule J (2000) The shoot apex as a marker of the responsivity to photoperiodic treatment inducing flowering of Chenopodium rubrum L. Biol Plant 43:31–34
Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33:168–171
Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen C (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275:80–83
Carmel-Goren L, Liu YS, Lifschitz E, Zamir D (2003) The SELF-PRUNING gene family in tomato. Plant Mol Biol 52:1215–1222
Cumming BG (1967) Early-flowering plants. In: Wilt FH, Wessels NK (eds) Methods in developmental biology. Crowell, New York, pp 277–299
Cumming BG, Seabrook JEA (1985) Chenopodium. In: Halevy AH (ed) CRC handbook of flowering, vol II. CRC Press, Boca Raton, pp 96–228
Cumming BG, Hendricks SB, Borthwick HA (1965) Rhythmic flowering responses and phytochrome changes in a selection of Chenopodium rubrum. Can J Bot 43:825–853
Doi K, Izawa T, Fuse T, Yamaguchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT like gene expression independently of Hd1. Gene Dev 18:926–936
Felsenstein J (2004) Inferring phylogenies. Sinauer Associates, Inc, Sunderland Mass
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422:719–722
Hayama R, Agashe B, Luley E, King R, Coupland G (2007) A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19:2988–3000
Hecht V, Foucher F, Ferrandiz C, Macknight R, Navarro C, Morin J, Vardy ME, Ellis N, Beltran JP, Rameau C, Weller JL (2005) Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol 137:1420–1434
Hsu CY, Liu YX, Luthe DS, Yuceer C (2006) Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18:1846–1861
Huang JC, Chen F (2006) Simultaneous amplification of 5′ and 3′ cDNA ends based on template-switching effect and inverse PCR. BioTechniques 40:187–189
Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, Takano M, Shimamoto K (2005) Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17:3326–3336
Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17:1050–1054
Kardailsky I, Shukla V, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286:1962–1965
King RW (1972) Timing in Chenopodium rubrum of export of the floral stimulus from the cotyledons and its action at the shoot apex. Can J Bot 50:697–702
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–1962
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short day conditions. Plant Cell Physiol 43:1096–1105
Lee JH, Hong SM, Yoo SJ, Park OK, Lee JS, Ahn JH (2006) Integration of floral inductive signals by flowering locus T and supressor of overexpression of Constans 1. Physiol Plantarum 126:475–483
Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154:459–473
Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W (2001) Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6:327–336
Ohno S (1970) Evolution by gene duplication. Springer-Verlag, Berlin
Otto SP, Yong P (2002) The evolution of gene duplicates. Adv Genet 46:451–483
Partap T, Joshi BD, Galway NW (1998) Chenopods. Chenopodium spp. Promoting the conservation and use of underutilized and neglected crops, vol 22. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Rahiminejad MR, Gornall RJ (2004) Flavonoid evidence for allopolyploidy in the Chenopodium album aggregate (Amaranthaceae). Plant Syst Evol 246:77–87
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Schoentgen F, Saccoccio F, Jolles J, Bernier I, Jolles P (1987) Complete amino-acid-sequence of a basic 21 kDa protein from bovine brain cytosol. Euro J Biochem 166:333–338
Seidlová F, Krekule J (1973) The negative response of photoperiodic floral induction in Chenopodium rubrum L. to preceding growth. Ann Bot 37:605–614
Štorchová H, Hrdličková R, Chrtek J Jr, Tetera M, Fitze D, Fehrer J (2000) An improved method of DNA isolation from plants collected in the field and conserved in saturated NaCl/CTAB solution. Taxon 49:79–84
Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036
Ullmann J, Seidlová F, Krekule J, Pavlová L (1985) Chenopodium rubrum as a model plant for testing the flowering effects of PGRs. Biol Plant 27:367–372
Veit J, Wagner E, Albrechtová JTP (2004) Isolation of a FLORICAULA/LEAFY putative orthologue from Chenopodium rubrum and its expression during photoperiodic flower induction. Plant Physiol Biochem 42:573–578
Wada M, Cao Q, Nobuhiro K, Soejima J, Masuda T (2002) Apple has two orthologs of FLORICAULA/LEAFY involved in flowering. Plant Mol Biol 49:567–577
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohman JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46:1175–1189
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484
Yoo SY, Kardailsky I, Lee JS, Weigel D, Ahn JH (2004) Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol Cells 17:95–101
Zeevaart JAD (1976) Physiology of flower formation. Annu Rev Plant Physiol Plant Mol Biol 27:321–348
Acknowledgments
We appreciate stimulating discussion and helpful comments by Frideta Seidlová and Ivana Macháčková of the Institute of Experimental Botany, Prague; Gary J. Houliston of the University of Alaska, Fairbanks, USA; Edgar Wagner of the University of Freiburg, Germany; and two anonymous reviewers. We are thankful to Justyna Veit for designing some primer sequences and helping with qRT PCR optimization. The excellent technical assistance of Martina Dostálová was highly appreciated. Financial support was provided through the grants GACR 522/05/0300 to H.S. and Z-5038910 to IEB AS CR, v.v.i, Prague.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cháb, D., Kolář, J., Olson, M.S. et al. Two FLOWERING LOCUS T (FT) homologs in Chenopodium rubrum differ in expression patterns. Planta 228, 929–940 (2008). https://doi.org/10.1007/s00425-008-0792-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0792-3