Abstract
Oligonucleotide microarrays corresponding to over 16,000 genes were used to analyze changes in transcript accumulation in root tips of the Al-sensitive Medicago truncatula cultivar Jemalong genotype A17 in response to Al treatment. Out of 2,782 genes with significant changes in transcript accumulation, 324 genes were up-regulated and 267 genes were down-regulated at least twofold by Al. Up-regulated genes were enriched in transcripts involved in cell-wall modification and abiotic and biotic stress responses while down-regulated genes were enriched in transcripts involved in primary metabolism, secondary metabolism, protein synthesis and processing, and the cell cycle. Known markers of Al-induced gene expression including genes associated with oxidative stress and cell wall stiffening were differentially regulated in this study. Transcript profiling identified novel genes associated with processes involved in Al toxicity including cell wall modification, cell cycle arrest and ethylene production. Novel genes potentially associated with Al resistance and tolerance in M. truncatula including organic acid transporters, cell wall loosening enzymes, Ca2+ homeostasis maintaining genes, and Al-binding were also identified. In addition, expression analysis of nine genes in the mature regions of the root revealed that Al-induced gene expression in these regions may play a role in Al tolerance. Finally, interfering RNA-induced silencing of two Al-induced genes, pectin acetylesterase and annexin, in A17 hairy roots slightly increased the sensitivity of A17 to Al suggesting that these genes may play a role in Al resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum (Al) is one of the most important factors limiting crop production on acid soils worldwide (Foy et al. 1978). Under acidic conditions (pH < 5.0) phytotoxic species of Al, such as Al3+, are released into the soil solution at levels that affect normal plant growth (Kinraide 1991). The primary symptom of Al toxicity is a rapid reduction in root growth and lateral root formation resulting in roots with a stunted appearance. The rapidity of this response indicates that Al quickly disrupts root cell expansion and elongation, prior to inhibiting cell division (Kochian et al. 2005).
Al rapidly affects a number of cellular processes in the root apex, which is considered to be the primary target of Al stress (Ryan et al. 1993). Al binds rapidly in the apoplast with as much as 30–90% of the total absorbed Al in the root tissue being localized to the extracellular compartments (Zheng and Yang 2005). Within the cell wall (CW), Al binds to negatively charged sites in the pectin matrix (Schildknecht and Vidal 2002) leading to the displacement of Ca2+ and thereby reducing CW extensibility and inhibiting root growth (Ma et al. 2004). Al has a strong affinity for the negatively charged plasma membrane (PM) and causes catastrophic depolarization of the PM (Kochian et al. 2005), with resulting disruption of the H+ gradient and ion fluxes across the membrane (Ahn et al. 2002), particularly Ca2+ fluxes (reviewed in Rengel and Zheng 2003). Furthermore, Al-induced reactive oxygen species (ROS) accumulation causes peroxidative damage to membrane lipids (Yamamoto et al. 2001). Al has also been shown to rapidly accumulate in the symplasm (Lazof et al. 1994) and in the nuclei of root tip cells within 30 min of Al treatment (Silva et al. 2000). Binding of Al to the nuclei may inhibit mitotic activity via alterations in DNA composition (Matsumoto et al. 1976), chromatin structure (Matsumoto 1988), template activity (Matsumoto and Morimura 1980), or by affecting the mechanisms controlling the organization and polymerization of microtubules (Frantzois et al. 2000). Collectively, these findings demonstrate that Al has deleterious effects on various cellular components and some of these Al-dependent responses are the effects of Al toxicity rather than the cause.
Occurrence of genotypes with contrasting Al resistance in several plant species has led to the identification of Al resistance mechanisms. The most widely studied mechanism of Al resistance involves the Al-induced release of organic acids (e.g. malate, citrate, and oxalate) from root tips, which form stable complexes with Al rendering it non-toxic. Some plant species secrete organic acids into the root rhizosphere preventing Al accumulation within root tips (Al resistance) while others utilize an organic acids-mediated mechanism to detoxify Al internally (Al tolerance) (Kochian et al. 2005). Recently, a number of Al-activated organic acid transporters that exude either malate (e.g. AtALMT1) or citrate (e.g. FRD3) have been identified in several plant species (Delhaize et al. 2007). In addition, Larsen et al. (2005, 2007) identified two Arabidopsis ABC transporters, ALS1 and ALS3, which may act in mobilizing and sequestering Al within the plant to confer Al tolerance.
Despite the considerable progress made in deciphering the physiological and genetic basis of Al toxicity and resistance over the last decade, our understanding of the molecular mechanisms underlying these processes is limited. Several studies have identified genes that are up-regulated under Al-stress conditions using a variety of methods. However, most of these genes were also induced under other stress conditions including other metal toxicities, calcium deprivation, wounding, and attack by plant pathogens (Snowden et al. 1995; Hamel et al. 1998; Richards et al. 1998). Microarrays have become an important technology for the global analysis of gene expression but have yet to be used widely to better understand Al toxicity effects. Microarray results can facilitate the understanding of pathways of expression and regulation of plant genes under Al stress and provide insights into the genetic control of Al resistance in plants.
A majority of the research in the area of Al toxicity and resistance has focused on monocot crop species such as wheat, maize and rice and dicot species such as Arabidopsis, while limited research has been conducted in legumes. Genetic and genomic studies of Al resistance in agriculturally important legumes such as soybean and alfalfa are complicated by the complex large genomes of these species. M. truncatula is a useful system to study Al toxicity in legumes due to its simple diploid genetics, synteny with agriculturally important legumes, and abundant genetic and genomic resources. In this study, we used oligonucleotide microarrays to evaluate changes in gene expression in response to Al stress in M. truncatula. Our goal was to identify a suite of genes that are regulated by Al stress and use the results obtained to better understand the molecular mechanisms underlying Al toxicity, resistance and tolerance. Approximately 10% of the genes represented on the array were regulated by Al in root tips. The functional relevance of Al-specific induction or repression of selected genes is discussed. Also, two genes were examined by interfering RNA (RNAi)-induced gene silencing to evaluate their roles in Al stress response.
Materials and methods
Plant culture
Seeds of M. truncatula genotype A17 were scarified using concentrated sulfuric acid and surface sterilized with 5% (v/v) household commercial bleach for 3 min. The seeds were placed at 4°C in sterile water for 2 days and germinated overnight on 1% agar plates in the dark at room temperature. Seedlings with roots of about 1 cm in length were sown through the mesh bottoms of polypropylene cups. The cups were placed in precut holes of a plastic insert placed over a plastic tub that held 7.3 L of aerated control nutrient solution. The solution contained the following nutrients: 1.2 mM KNO3, 0.8 mM Ca(NO3)2, 0.1 mM NH4H2PO4, 0.2 mM MgSO4, 10 μM FeNaEDTA, 50 μM KCl, 12.5 μM H3BO3, 1 μM MnSO4, 1 μM ZnSO4, 0.5 μM CuSO4, 0.1 μM Na2MoO4, and 0.1 μM NiCl2 (Pence et al. 2000). The pH of the control nutrient solution was adjusted to 4.5 using 1 N HCl. Seedlings were grown for 72 h in a growth chamber (light/dark, 14/10 h) under a light intensity of 440 μmol photons m−2 s−1. Al treatment was initiated after 72 h by replacing the control growth solution with identical solutions that contained 0 (control), 10, 25, 50 and 100 μM Al, respectively, added as AlK(SO4)3. The pH of the control and Al treatment solutions was adjusted to pH 4.5 using 1 N HCl and 1 N KOH, respectively. Due to the addition of base, the Al concentrations in the solution culture corresponded to free Al3+ activities of {0}, {0.9}, {2.3}, {5.1}, and {11.9} (braces indicate Al3+ activity), respectively, as estimated with the speciation software program VISUAL MINTEQ v 2.4b (Gustaffson JP, KTH, Department of Land and Water Resources Eng., Stockholm, Sweden; Allison et al. 1991). Root growth measurements were made at 0 and 48 h following Al exposure. Relative root growth was calculated as: RRG (%) = (root growth in Al solution/root growth in control solution) × 100.
For tissue collection for microarrays, plants were grown in control solution culture for 72 h then transferred to nutrient solutions containing either 0 μM (control) or 25 μM Al. The pH of the control and Al treatment solutions was adjusted to 4.5 using 1 N HCl and 1 N KOH, respectively. Root tips (0.5 cm) from 0 μM control and 25 μM Al treated seedlings were harvested after 12 h and immediately frozen in liquid nitrogen. Samples were collected from three independent biological replicates and stored immediately at −80°C. To demonstrate that the seedlings in the three replicates were exposed to similar levels of Al stress, root growth measurements of 12 sample roots were made at 0 and 12 h following Al exposure.
RNA preparation, cDNA synthesis, and microarray hybridization
Total RNA from control and Al treated root tips was extracted using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. First-strand cDNA synthesis was carried out as described by Lohar et al. (2006) using the 3DNA Array 50 Expression Array Detection Kit for cDNA Microarrays (Genisphere, Hatfield, PA).
The M. truncatula AROS (Array-Ready Oligo Set) Version 1.0 array (Qiagen) containing 70-mer probes representing 16,086 M. truncatula genes was used for microarray experiments. The probes were printed on Telechem Super Amine slides (Sunnyvale, CA) with spot size approximately 100–110 μm in diameter. Slides were processed prior to use by rehydrating over a 50–55°C water bath for 5–10 s and snap-drying on a 65°C heating block for 5 s, approximately three to four times. Probes were cross linked by exposing the spots to 65 mJ in a UV crosslinker. Slides were washed in 1% SDS at room temperature for 5 min, dipped in 100% ethanol for 30 s with gentle agitation, centrifuged at 1,500 rpm for 2–5 min and stored in a light proof box under cool and dry conditions before use. Microarrays were hybridized to cDNAs from both Al- treated and control roots, with cDNAs from the two different treatments labeled with different dyes. Each hybridization was repeated at least six times to account for technical variability, with triplicates of each dye combination to control for dye effects. A modified one-step hybridization reaction was performed as described in the 3DNA Array 50 kit (Genisphere). Slide scanning and data analysis was performed as described by Lohar et al. (2006).
Plasmid construction and plant transformation using Agrobacterium rhizogenes
To create the construct silencing pectin acetylesterase (PAE) a 573 bp fragment was amplified from the 5′ end of the cDNA clone BG646197. The construct silencing annexin (ANN) was made using a 652 bp fragment amplified from the 5′ end of CA922466. Fragments were introduced into pHellsgate 8 (Helliwell et al. 2002), creating the PAEi and ANNi constructs. The pHellsgate 8 vector with a LacZ fragment was used as the control. The resulting recombinant constructs were introduced into Agrobacterium rhizogenes strain ARqua1 and used for transformation of A17 as described previously (Boisson-Dernier et al. 2001).
Root growth of RNAi plants on Al-containing medium
The medium for Al-plate assays contained 0.5 mM CaCl2 in 1% agarose. The pH was adjusted to 4.5 using 5 mM HCl. For the 10 μM Al medium, Al was added as AlK(SO4)3 after autoclaving, and poured into 25.4 × 25.4 cm2 plates (Genetix, Boston, MA). The Al3+ activity was {5.2} as estimated with the speciation software program VISUAL MINTEQ v 2.4b (Gustaffson JP, KTH, Department of Land and Water Resources Eng., Stockholm, Sweden; Allison et al. 1991).
Three-week-old A. rhizogenes-transformed A17 seedlings were transferred either to control (0 μM Al) or 10 μM Al plates and root growth measurements were made at 0 and 5 days after Al treatment.
RNA gel blot analysis
Total RNA was separated on a 1.2% agarose gel with 1.8% formaldehyde and transferred to Hybond N+ (Amersham, Pittsburgh, PA) overnight in 10X SSC (1.5 M NaCl, 0.15 M sodium citrate, pH 7.0). 32P-labeled cDNA probes were synthesized from cDNA clones [BG582785 (TC100646), BF520966 (TC102452), AW981429 (TC107354), AW684706 (TC98950), BG452060 (TC103000), AW686146 (TC111698), BQ155427 (TC94419), AW776496 (TC100583), BQ156790 (TC102756)] using the DecaPrime II kit (Ambion, Austin, TX), and blots were hybridized in 0.25 M Na2HPO4, 7% SDS, 1 mM EDTA, pH 7.4 and washed under highly stringent conditions of 40 mM Na2HPO4, 1% SDS, 1 mM EDTA.
Quantitative real-time PCR (q-PCR)
Root tip RNA samples from the three biological replicates that were used for microarray experiments were used for quantitative real-time PCR (q-PCR) assays. cDNA synthesis and PCR conditions are as described previously (Tesfaye et al. 2006). Primers were designed using the Primer Express Software (v 2.0, Applied Biosystems, Foster City, CA) and are listed in Supplemental Table S1. For each treatment, three q-PCR reactions were run from a cDNA synthesis and the mean values presented. Amplification of 18S ribosomal RNA was used as the endogenous control. The delta delta Ct (threshold cycle) method was used to calculate relative fold changes between Al-treated and control (-Al) cDNA. The specificity of the product was confirmed by a single peak in a dissociation curve at the end of the PCR reaction.
To determine transcript levels of ANN and PAE in transgenic A17 roots, total RNA was isolated from individual hairy roots and RNA isolation, cDNA synthesis and q-PCR were performed as described above.
Results and discussion
Sensitivity of M. truncatula to Al treatment
The effect of Al treatment on root growth of M. truncatula genotype A17 (henceforth referred to as A17) was evaluated using Al dose-response treatments in solution culture. Root growth of A17 was inhibited by approximately 40, 50 and 80% at 10, 25 and 50 μM added Al concentrations, respectively (Fig. 1a). Complete inhibition of root growth was observed by 48 h at 100 μM Al. Based on these results, an Al concentration of 25 μM was selected to profile global changes in gene expression in A17 in response to Al stress. A reduction in root growth by approximately 50% was observed in the 12 h Al treatments in the three biological replicates (Fig. 1b) for the microarrays, demonstrating that the three independent replicates were exposed to similar levels of Al stress.
Effect of Al-dose on root growth in Medicago truncatula A17. a Relative root growth ±SE of 4-day-old A17 seedlings grown at 0, 10, 25, 50 and 100 μM Al for 48 h. b Root growth (mm) ± SE of 4-day-old A17 seedlings grown in 0 μM Al (control) and 25 μM Al for 12 h from three independent biological replicate microarray experiments
Microarray analysis reveals hundreds of genes that are significantly regulated in response to Al toxicity
To obtain a global picture of gene transcript accumulation during Al stress, cDNAs synthesized from control and Al-treated A17 root tip RNA were used to hybridize to oligonucleotide microarrays. Out of the 16,086 spots on the array, approximately 33% produced signals on the microarray that could be used for further analysis. For the vast majority of transcripts, expression appeared unchanged with Al treatment (Fig. 2a). We used the controlled False Discovery Rate (FDR) method as implemented in the software Significance Analysis of Microarrays (Tusher et al. 2001) to identify genes with reproducible changes in transcript accumulation (FDR < 1%). As a result, 2,742 genes were identified as statistically significant. Approximately 80% of these significant genes showed under two-fold change in transcript accumulation in the Al treatment (Fig. 2b). Normalized and raw data for all genes is available from the Gene Expression Omnibus (Accession No. GSE7066). A fold change cut-off value of 2.0 identified 607 genes with Al-responsive transcript accumulation. Of these, 336 genes were up-regulated and 271 genes were down-regulated (Fig. 2a, b). Selected Al-responsive genes are presented in Supplemental Tables S2 and S3. A limited summary of this information is presented in Table 1.
Detection of genes with altered transcript accumulation in Al-treated A17 root tips by differential microarray analysis. a Scatter plot of normalized signal intensities for approximately 5,500 genes on the microarray. Log2 intensities for each spot on the microarray are plotted with signals from control (-Al) and Al-treated root tips on the x and y axes, respectively. The diagonal lines represent 2.0-fold change cutoffs. b Distribution of genes in various fold-change categories based on ratio of transcript abundance in Al-treated root tips compared to control (-Al) root tips
Functional classification of genes significantly up-and down-regulated in response to Al toxicity
All significant genes with at least a twofold change in transcript abundance were grouped into functional categories based on gene ontology information (Fig. 3). We evaluated the Z score, a standardized difference between observed and expected values, to determine whether up- or down-regulated genes in a functional group were found in numbers greater than would be expected by chance. A Z score of ≥2.0 was considered to be significant. Approximately 40% of the genes were of unknown function in both up- and down-regulated gene classes (Fig. 3a, b). Genes involved in abiotic and biotic stress response were represented to a greater extent in the Al-induced class as indicated by a significant Z score (3.5). Al-induced damage to the cell wall and membrane might induce secondary stresses including oxidative stress, which could result in an increase in abiotic and biotic stress response genes. In addition, genes belonging to the CW modification and cell growth group were overrepresented in the Al-induced class (Z score 2.9). Since Al can easily and rapidly interact with the CW and cause its rigidification, it is conceivable that plant cells might respond by modifying their CW components.
Functional distributions of Al-responsive genes showing at least twofold change in transcript abundance. a Functional distribution of genes with fold-change values of ≥2.0. b Functional distribution of genes with fold-change values of ≤0.5. All genes were assigned functions according to gene ontology information and grouped into broader classes as defined by Journet et al. (2002), with modifications. Z scores for each class are given in parentheses. A Z score of ≥2.0 indicates that the gene group is represented significantly above what would be expected by chance among up-regulated or down-regulated genes in Al-treated roots
In contrast, genes associated with primary metabolism, protein synthesis and processing, and secondary metabolism and hormone metabolism were overrepresented in the Al-repressed class (Z scores 2.2, 2.2 and 3.6, respectively). Although, release of organic acids, which are primary metabolites, is one of the major mechanisms of Al resistance in many plant species, several studies have shown that there is no apparent correlation between Al-induced expression of any of the organic acid biosynthetic genes and increased activity or exudation of organic acids (Mariano et al. 2005). Also, under stressful conditions, down-regulation of certain metabolic processes may be necessary to allow cells to redirect resources towards adaptation mechanisms. This suggests that down-regulation of primary metabolism might be necessary to cope with stressful conditions, and that activation of an organic acid transporter and not the synthesis of the organic acid itself might be required for Al resistance. It has also been suggested that adaptive reprogramming may require a higher number of down-regulated genes than up-regulated genes (Arnholdt-Schmitt 2004) and that inhibition of protein synthesis is a metabolic response of plants under stress. Finally, genes belonging to the cell division cycle, chromatin and DNA metabolism group were overrepresented in the Al-repressed class (Z score 3.5) potentially reflecting the effect of Al toxicity on cell division in root tips of A17.
Validation of microarray results
To validate the microarray data, the transcript abundance of selected genes was monitored by q-PCR and RNA blotting. Quantitative PCR data confirmed that expression of the ten genes analyzed was Al-responsive with the same direction of fold change of transcript abundance in all three biological replicates (Table 2). The absolute values of fold change were not identical to those from the array and in most cases the ratios based on q-PCR were higher than those obtained from microarray hybridizations, reflecting the difference in sensitivity of the q-PCR and microarray techniques (Manthey et al. 2004). RNA blot analysis conducted for nine out of the ten genes tested by q-PCR showed similar trends of transcript abundance as observed from the microarray (Fig. 4).
RNA gel blot analyses of selected Al-responsive genes. a Ca2+/H+ exchanger (TC100646). b Putative ABC transporter (TC102452). c Pectin acetylesterase precursor (TC100583). d Putative MATE efflux family protein (TC98950). e Periaxin-like protein (TC103000). f COBRA-like GPI anchored protein (TC111698). g Unknown (TC102756). h MtN9 protein (TC107353). I Unknown (TC94419). j An ethidium bromide stain of the gel is shown to confirm equal RNA loading
Known markers of Al-induced gene expression were differentially regulated in M. truncatula
A number of genes previously shown to be up-regulated in response to Al in other plant species were also up-regulated in this study. ADR6 (Sali5-4a) (Ragland and Soliman 1997) and wali-7 (Richards et al. 1998), two Al-induced genes of unknown function previously identified in soybean and wheat, respectively, were up-regulated in A17 root tips in response to Al treatment (Table 1).
Genes encoding peroxidases and germin-like proteins, which are potential reactive oxygen species (ROS) generators, and a number of antioxidant genes including blue copper binding proteins, glutathione reductases, and glutathione S-transferases were highly up-regulated (2.0- to 13-fold) (Table 1, Supplemental Table S2). These genes have been previously shown to be up-regulated in response to Al stress in Arabidopsis (Richards et al. 1998) and wheat roots (Delisle et al. 2001). Al stress induced ROS production in plant roots can cause peroxidative damage to membrane lipids (Yamamoto et al. 2001; Boscolo et al. 2003) and a positive correlation between Al-induced peroxidase activity and root growth inhibition was found in Al-sensitive barley roots (Tamas et al. 2003). Furthermore, expression of antioxidant genes has been observed to be higher in Al-tolerant lines (Devi et al. 2003; Darko et al. 2004), suggesting that the high antioxidant status of root cells may play a role in Al tolerance.
A number of pathogenesis-related (PR) genes including a PR-4 like protein, thaumatin-like proteins, β-1,3-glucanase, and chitinase were up-regulated in response to Al treatment (Table 1, Supplemental Table S2). The up-regulation of β-1,3-glucanases in response to Al has been previously reported in wheat (Cruz-Ortega et al. 1997) and rice (Mao et al. 2004). Oxidative stress, and in some cases lipid peroxides, have been shown to induce PR protein gene expression (Klessig et al. 2000; Hong and Hwang 2005), therefore suggesting that Al induces oxidative stress and peroxidative damage to lipid membranes.
Genes encoding 4-coumarate:CoA ligase and caffeic-acid O-methyltransferase, which catalyze key steps in the lignin biosynthetic pathway, were up-regulated in response to Al stress in A17 root tips (Table 1, Supplemental Table S2). Mao et al. (2004) reported that four genes encoding key enzymes in the lignin biosynthetic pathway were up-regulated by Al stress more rapidly and in greater amounts in an Al-sensitive rice cultivar. Sasaki et al. (1996) demonstrated that the extent of root growth inhibition in wheat in response to Al was closely correlated with the extent of lignin deposition, and proposed that Al-sensitive varieties would accumulate more lignin in the roots than Al-tolerant varieties. It is conceivable that Al-induced increase in activity of peroxidases may trigger lignin production resulting in CW stiffening and reduced root growth in A17.
Cell wall modifying genes including pectin esterases and pectin esterase precursors were up-regulated in response to Al stress in A17 (Table 1, Supplemental Table S2). Previously, Schmohl et al. (2000) demonstrated that transgenic potato plants over-expressing pectin methyl esterase (PME) proved to be more Al-sensitive than the wild type potato cultivar. It has been proposed that the interaction of Al with pectin alters CW structural and mechanical properties, leading to a stiffening of the CW, which inhibits cell elongation. Pectin esterases (PEs) such as PME and pectin acetylesterases (PAE) can deesterify pectin and potentially regulate CW degradation by altering the accessibility of pectin to the action of CW hydrolyzing enzymes (Wen et al. 1999; Vercauteren et al. 2002). Therefore, it is conceivable that this enzymatic function could promote CW loosening in response to Al-induced stiffening of the CW. However, free carboxylic groups produced as a result of pectin deesterification by PEs may serve as additional binding sites for Al in the apoplast (Eticha et al. 2005) making this response detrimental to the plant root under Al toxic conditions.
Identification of novel genes associated with Al toxicity
Several genes not previously identified as Al-inducible or known to be associated with Al toxicity were identified in this study. A gene encoding a carbohydrate oxidase, which is an ROS generator and genes encoding a quinone oxidoreductase and a thioredoxin-like protein, which are ROS scavenging proteins (Kuge and Jones 1994), were up-regulated in this study, suggesting that these genes may also play a role in the oxidative burst response following Al stress (Table 1). In addition, up-regulation of genes encoding probable mannitol dehydrogenase and proline dehydrogenase enzymes, which are involved in degrading osmolytes with antioxidant properties (Jennings et al. 1998; Chen and Dickmann 2005), may also promote oxidative damage in root tip cells.
Although Al toxicity can inhibit cell division, little is known about the transcriptional changes involved in this response. Silva et al. (2000) detected Al within the nucleus of Al-sensitive soybean roots within 30 min of Al exposure. It has been suggested that the presence of Al at the surface of the nucleus can potentially lead to microtubule binding at the membrane surface during the G2 phase of the cell cycle, which could lead to an arrest of the cell cycle (Kochian et al. 2005). It is possible that down-regulation of various cell cycle genes might result in an arrest in the cell cycle. In this study, a number of cell cycle genes including a cyclin-dependant kinase (cdc2MsF), cytokinesis-specific syntaxin KNOLLE, B-type cell cycle switch protein (ccs52B), and a number of histone genes were down-regulated in response to Al stress (Table 1), suggesting that repression of these genes might result in inhibition of cell division. Alternatively, Al-induced inhibition of cell division might result in a down-regulation of these genes. Furthermore, a putative serine/threonine protein kinase potentially involved in down-regulating histone transcription was also down-regulated. Thus, key cell cycle players that may be down-regulated either in response to Al-mediated inhibition of cell division or be the cause for inhibition of cell division were identified.
A gene involved in ethylene biosynthesis, ACC oxidase, was significantly down-regulated in response to Al treatment (Table 1, Supplemental Table S3). Massot et al. (2002) reported that Al rapidly stimulates ethylene synthesis and inhibits root elongation in bean. Sun et al. (2007) suggested that ethylene production is associated with inhibition of root elongation in Lotus japonicus and M. truncatula and the ACC oxidase gene was induced by 2 h of Al treatment. In our study, inhibition in root growth of A17 had already occurred by 12 h, suggesting that this gene might be involved in Al toxicity and that the down-regulation of this gene might be an essential adaptive response to prevent further inhibition in root growth.
Identification of novel genes potentially associated with Al resistance and tolerance
Exudation of organic acids, including malate and citrate, in response to Al stress is one of the major resistance mechanisms reported in several plant species. Recently, a number of malate and citrate transporters, which belong to the Al-activated malate transporter (ALMT) and multi-drug and toxin efflux (MATE) protein families, respectively, have been identified in certain Al-resistant plant species (Delhaize et al. 2007). To date, the role of organic acid exudation in Al resistance in M. truncatula is not certain, although Al-induced secretion of malate and citrate was found to occur in preliminary experiments with three-week-old M. truncatula plants propagated from stem cuttings (Chandran, unpublished). Interestingly, the gene with the highest fold-change value in our microarray experiment (approximately 53-fold induced) encodes a putative multidrug and toxin efflux (MATE) protein (Table 1). The M. truncatula MATE gene shows 77% similarity to the FDR3 (FERRIC REDUCTASE DEFECTIVE 3) antiporter gene from Arabidopsis, the SbMATE gene from sorghum and the HvAACT1 gene from barley. The MATE proteins comprise a large family and were shown to function as drug/cation antiporters that remove toxic compounds and secondary metabolites from the cytosol by exporting them out of the cell or sequestering them to the vacuole (Delhaize et al. 2007). FRD3 was recently identified as a citrate efflux transporter and Arabidopsis plants ectopically expressing FRD3-GFP had significantly higher amounts of citrate in their root exudates and greater resistance to Al compared to untransformed control plants (Durrett et al. 2007). Furthermore, both SbMATE and HvAACT1 have also shown to be Al-activated citrate efflux transporters (Furukawa et al. 2007; Magalhaes et al. 2007). Since the M. truncatula MATE gene is highly induced by Al and shows high homology to citrate efflux transporters from other plant species, it is very likely that it is a citrate efflux transporter. However, since A17 is Al-sensitive, it is not clear whether the up-regulation of this gene in response to Al stress corresponds with increased protein activity and therefore greater efflux of citrate. One hypothetical model for the Al-activated efflux of organic anions by members of the MATE family of proteins is that Al first induces the expression of these proteins through a signal transduction pathway possibly involving a specific receptor or non-specific stress responses (Delhaize et al. 2007). Al then activates organic acid efflux by interacting with the newly synthesized proteins in the plasma membrane. In A17, it is possible that although Al can induce expression of the MATE gene, subsequent activation of organic acid efflux might be delayed or inhibited. We also did not observe up-regulation of genes encoding organic acid biosynthetic enzymes in our study, which is not surprising since several studies have shown that there is no apparent correlation between Al-induced expression of organic acid biosynthetic enzymes and increased activity and exudation of organic acids (Mariano et al. 2005). Future work will involve determining whether the M. truncatula MATE gene is a citrate efflux transporter and whether its over-expression will result in enhanced Al resistance.
A putative ABC transporter gene was also up-regulated by 3.3-fold in response to Al treatment (Table 1). The ABC transporters constitute a large family and have been shown to detoxify organic and inorganic substances by sequestering these substances into vacuoles. Al-inducible ABC transporters have been identified in wheat (TaMDR1) (Sasaki et al. 2002) and Arabidopsis (ALS3) (Larsen et al. 2005) but these proteins showed no significant similarity to the M. truncatula ABC transporter. However, ALS1, an Arabidopsis ABC transporter, which is not Al-inducible showed 56% similarity to the M. truncatula ABC transporter. ALS1 is almost exclusively localized to the vacuolar membrane and has been suggested to play a role in intracellular movement of some substrate, possibly chelated Al, as part of a mechanism of Al sequestration (Larsen et al. 2007). On the other hand, ALS3 has been suggested to be involved in mediating Al uptake and sequestration in leaf tissue in order to remove Al from sensitive growing tissues such as the root tip. The Al-induced expression of the M. truncatula ABC transporter in both the root tip and the mature regions of the root (Fig. 4b) suggests that this transporter may be involved in sequestering Al into the vacuole either in the root tip or in more mature regions of the plant and thus play a role in Al tolerance.
Programmed cell death (PCD) can occur as a result of an oxidative burst following exposure to abiotic stresses. ROS production via Al toxicity was shown to induce cell-death in wheat (Delisle et al. 2001), barley (Simonovicova et al. 2004), and maize root tip cells (Vazquez 2002), and this process was proposed to remove cells that accumulate Al and therefore serve as a mechanism of Al resistance. It is highly plausible that Al stress induces a similar oxidative burst in M. truncatula root tips, since genes involved in ROS generation and scavenging were significantly up-regulated. Furthermore, this oxidative burst could lead to cell death in M. truncatula root tips. In this study, genes encoding cysteine protease, cysteine protease precursor and cysteine protease inhibitors were up-regulated in response to Al by 2.2- to 3.4-fold (Table 1). Cysteine proteases and cysteine protease inhibitors have previously been implicated in the regulation of oxidative stress-mediated PCD (Solomon et al. 1999). In addition, a number of senescence related genes were also up-regulated in response to Al stress and since senescence involves PCD it is likely that cells programmed to undergo cell death may display enhanced expression of these genes. An ACD11 gene, which has been shown to suppress PCD in Arabidopsis (Brodersen et al. 2002), was down-regulated in A17 suggesting activation of a cell death-like response. In contrast, a gene encoding a BAG6 like protein, which has been shown to induce cell death in response to stress in Arabidopsis (Kang et al. 2006), was down-regulated in response to Al treatment (Table 1), suggesting a tight regulation of the Al-induced cell death response, probably associated with timing or location of the response. A number of ROS generating genes including peroxidase and peroxidase precursors, a germin-like protein, an amine oxidase, and a lipoxygenase and antioxidant genes including glutathione peroxidase, PR-proteins, and production of isoflavonoids were down-regulated (Supplemental Table S3). It is conceivable that the production and scavenging of ROS may be a tightly regulated process, with the location of ROS production and/or accumulation important in mediating the Al-induced cell death-like response.
Disruption of cytoplasmic Ca2+ homeostasis has been suggested as a primary trigger of Al toxicity. While some studies have demonstrated that the disruption of Ca2+ homeostasis is achieved through an Al-mediated decrease in Ca2+ influx via blockade of Ca2+ channels, others have shown that an increase in intracellular Ca2+ concentration triggered by Al was responsible for this effect (reviewed in Rengel and Zhang 2003). Despite these differences, it is apparent that Al alters or disrupts Ca2+ homeostasis, which can potentially disrupt numerous biochemical and physiological processes, including those involved in root growth. In this study, genes encoding a Ca2+/H+ exchanger and a putative plasma membrane type Ca2+-ATPase were up-regulated in response to Al treatment (Table 1, Supplemental Table S2). It has been hypothesized that root cells might use these proteins to reduce intracellular Ca2+ levels following an Al-induced spike in Ca2+ levels. Therefore, identification of these genes in our study suggests the involvement of these specific proteins in maintaining Ca2+ homeostasis following Al exposure.
The cytoskeleton is considered to be a potential target for Al toxicity because of its role in both cell expansion and cell division (Kochian et al. 2005). However, the genes potentially involved in this process have yet to be identified. It has been shown that Al can depolymerize cortical microtubules within 1 h of Al exposure (Sivaguru et al. 2003). However, during long-term Al exposure (3–24 h) microtubule reorientation and stabilization occurs in the inner and outer cortical cells, respectively (Blancaflor et al. 1998). Al-induced microtubule reorientation may cause an uneven and radial expansion of cells of the inner cortex causing root swelling and mechanical stress on the epidermis resulting in the appearance of cracks at the root surface (Ciamporova 2002). The arrangement of newly synthesized cellulose microfibrils (CMFs) is thought to play a prominent role in determining the rate and direction of cell expansion and cortical microtubules are usually implicated in the alignment of CMFs (Sugimoto et al. 2000). Interestingly, a COBRA-like gene (COB) encoding a putative glycosylphosphatidylinositol-anchored protein that potentially modulates cellulose deposition and orients cell expansion in roots was induced by approximately 40-fold in response to Al treatment in A17 root tips (Table 1). COBRA-like gene RNA levels are dramatically up-regulated in cells entering the zone of rapid longitudinal expansion (Schindelman et al. 2001) and root cells in the elongation zone of an Arabidopsis cob mutant expand radially to a greater extent compared to wild type. Thus, it is conceivable that the up-regulation of COB transcripts might be necessary to reorient cell expansion in the longitudinal direction in response to the Al-induced radial expansion in root cortical cells, thereby promoting root growth. This gene can serve as a candidate gene for over-expression in legume crops since it is might be required for continued root growth under Al toxic conditions.
Root growth inhibition upon exposure to Al is the earliest symptom of Al toxicity and occurs as a result of a rapid inhibition of cell elongation (Kochian et al. 2005). A number of genes encoding CW-modifying enzymes including four xyloglucan endotransglycosylase genes with roles in the CW loosening process necessary for cell elongation were up-regulated in response to Al treatment. In addition, a gene encoding a periaxin-like protein displayed 17-fold induction in response to Al treatment. Periaxin-like proteins accumulate in high amounts in the CW after cessation of elongation and play a role in determining physical characteristics of the plant CW (Cassab 1998; Bucher et al. 2002). This gene has been previously shown to be up-regulated in response to drought stress (Chen et al. 2008) with no known function. Up-regulation of these genes suggests that A17 roots respond to Al-induced CW stiffening by enhancing the expression of CW loosening enzymes in a perhaps futile attempt at stimulating CW expansion.
A putative metal-binding isoprenylated protein was up-regulated by Al stress by approximately ninefold (Table 1). A class of proteins known as farnesylated proteins, which are capable of being isoprenylated and binding transition metal ions, has been identified in plants (Dykema et al. 1999). A comparison of the putative M. truncatula isoprenylated protein sequence with known farnesylated proteins revealed the presence of a conserved metal-binding site (CEGC) and a consensus isoprenylation site (CSIM). Although farnesylated proteins have been shown to bind Cu2+, Ni2+, and Zn2+, the metal-binding capacity of the M. truncatula protein is not known. The involvement of this protein in Al binding requires further investigation.
Al-induced gene expression is not always restricted to the root-tip
To determine whether Al-induced changes in gene expression were restricted to the root tip, transcript abundance of nine genes was analyzed in A17 root tips and mature regions of the root (region above 5 mm from the root tip). Unlike the root tips, the mature regions of the root are generally not as sensitive to Al (Ryan et al. 1993); however, relative expression of Al-induced genes in different root segments has not been described. Gene transcripts encoding a putative ABC transport protein (TC102452) (Fig. 4b) and an unknown protein (TC102756) (Fig. 4g), accumulated to similar levels after Al treatment in both root tips and mature roots, while transcripts of genes encoding a Ca2+/H+ exchanger (TC100646) (Fig. 4a), a putative MATE protein (TC98950) (Fig. 4d), a periaxin-like protein (TC103000) (Fig. 4e) and a COBRA-like GPI-anchored protein (TC111698) (Fig. 4f) accumulated at lower levels in mature roots as compared to root tips. However, the relative transcript abundance (Al stressed/control roots) of the putative ABC transporter appeared to be higher in the root tip than the mature root. This ABC transporter is similar to ALS1, which is highly expressed in the root tip and in vasculature throughout the plant (Larsen et al. 2007). The transcript level of a PAE precursor gene (TC100583) (Fig. 4c) and a gene encoding an MtN9 protein (TC107353) (Fig. 4h) appeared to be up- and down-regulated, respectively by Al to a much greater extent in root tips than in mature roots. Finally, transcripts of an unknown protein (TC94419) (Fig. 4i) appeared to be down-regulated by Al in both tissue types. With the exception of the PAE and MtN9 genes, the majority of the genes tested were expressed in both root-tips and mature roots, suggesting a role for these genes in Al response in both root segments. Sivaguru and Horst (1998) showed that lower amounts of Al accumulated in the more-basal root segments (5–10 mm from the root tip) than the root apex. However, lower amounts of Al in the mature regions of the root may still trigger gene expression changes similar to those observed in the root apex, albeit to lower magnitudes. Moreover, the expression of some of these genes in the mature regions of the root, such as the ALS1-like ABC transporter, may be required for transporting chelated Al away from the Al-sensitive root apices.
Evaluating the roles of pectin acetylesterase and annexin genes in Al response
In response to Al stress, a gene for a putative pectin acetylesterase (TC100583) was up-regulated by approximately threefold (Table 2). We tested whether a reduction in PAE gene expression, which would potentially decrease Al binding sites in the CW, would increase Al resistance in transgenic A17 roots. Quantitative PCR analysis of RNA isolated from transformed roots containing an RNAi-inducing construct (PAEi) designed to inhibit the expression of PAE (TC100583) revealed a decrease in transcript abundance by 6- to 175-fold compared with that of control roots (Fig. 5a). We selected 10 μM as the concentration for the Al treatment in the plate assay since preliminary Al dose-response curves showed that 10 μM Al resulted in approximately 50% inhibition in root growth (data not shown). Since the agarose media used for the plate assays contained only CaCl2, the availability of Al to the plant roots would be greater in the plate assay compared to the nutrient solution. This would result in greater root growth inhibition at lower Al concentrations in the plate assay.
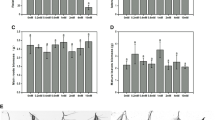
Transcript abundance and root growth in ANN-silenced (ANNi) and PAE-silenced (PAEi) roots. Error bars represent ± SE of measurements from 25 roots. a q-PCR analysis of ANN and PAE transcripts in transgenic roots. b Five-day root growth (mm) of LacZ vector-transformed control, ANNi, and PAEi plants on 0 μM and 10 μM Al. An asterisk above the bar indicates a significant difference (P < 0.05) between the root growth of the transgenic and control plants on 0 μM Al
The LacZ vector-transformed roots showed approximately 60% reduction in root growth in the presence of 10 μM Al, which corresponded with root growth inhibition observed between 25–50 μM Al concentrations in nutrient solutions. In 0 μM Al, growth of roots with the PAEi construct was significantly reduced (P ≤ 0.03) to 63% as compared to the LacZ vector-transformed control roots (Fig. 5b). Since deesterification of pectin by pectinesterases plays an important role in determining the extent to which pectin is accessible to degradation by CW hydrolyzing enzymes, it is possible that the down-regulation of PAE transcripts prevents the deacetylation of pectin and activation of CW hydrolyzing enzymes involved in cell expansion, thereby restricting root growth. Wen et al. (1999) showed that the partial inhibition of pectin methyl esterase by antisense RNA reduced root elongation in transgenic pea hairy roots. In 10 μM Al, growth of PAEi-transformed roots was reduced by 40% compared to no Al controls (Fig. 5b). Therefore, it appears that Al can further reduce root growth in PAEi plants, suggesting that a down-regulation of the PAE transcript may not necessarily affect Al-binding to the CW. The reduced CW permeability of Al bound-CW may limit the movement of wall-loosening enzymes and the access to their substrates, which could consequently result in reduced cell wall expansion (Eticha et al. 2005). PAE might be partly required for CW loosening following Al-induced cell elongation inhibition. Therefore, PAE might play a role in conferring a slight increase in Al resistance in M. truncatula; however the exact mode of action is to be determined.
An annexin-like gene (TC95776) was up-regulated by approximately sevenfold in response to Al treatment (Table 2). Annexins are a family of calcium-dependent membrane-binding proteins, which have been shown to possess a diverse array of functions including golgi-mediated secretion of newly synthesized plasma membrane and wall materials, nucleotide phosphodiesterase activity, peroxidase activity, vacuole biogenesis and cell expansion, DNA replication, and Ca channel activity (Delmer and Potikha 1997; Clark et al. 2001). Since a majority of these functions have been shown to be disrupted by Al toxicity, the annexin gene may be an important candidate for Al response studies. The protein sequence of the M. truncatula annexin gene showed 87% similarity to a previously identified M. truncatula annexin, MtAnn1 (AJ441322, De Carvalho-Niebel et al. 1998), which was shown to be transcriptionally activated in root tissues in response to rhizobial nodulation factors (De Carvalho-Niebel et al. 2002).
Transcript abundance in roots transformed with the construct ANNi, designed to silence the annexin gene (TC95776), was reduced 2- to 11-fold compared to vector-transformed roots (Fig. 5a). In 0 μM Al plates, growth of ANNi-transformed roots was significantly reduced (P ≤ 0.0007) by approximately 54% as compared to LacZ vector-transformed control roots (Fig. 5b). Previously, an Arabidopsis annexin T-DNA knockout mutant was identified, which showed inhibited root growth compared to wild-type seedlings (Clark et al. 2005). In 10 μM Al, growth of ANNi-transformed roots was reduced by approximately 6% compared to no Al controls (Fig. 5b). Since growth of ANNi-transformed roots is compromised in 0 μM Al, it is possible that Al cannot further inhibit root growth. Due to the potential role of annexin in multiple cellular processes that were previously shown to be affected by Al, it is possible that down-regulating annexin in ANNi roots mimics the Al toxicity effect consequently minimizing any further impact of Al on its growth. Thus, the M. truncatula annexin may play an important but indirect role in promoting root growth during Al stress conditions; however its exact function is yet to be determined.
Concluding remarks
In legumes, a molecular evaluation of Al toxicity and resistance mechanisms has not been fully explored. The primary goal of this study was to use large-scale transcript profiling to examine cellular processes affected by Al stress in root tips of an Al-sensitive genotype of M. truncatula. A number of genes previously shown to be induced in response to Al-stress, including oxidative stress induced genes, pathogenesis-related genes and cell-wall modifying genes were up-regulated in M. truncatula. Novel CW modifying genes potentially associated with Al toxicity were induced by Al, attributing a major role for the apoplasm in sensing and responding to the Al stress signal. In addition, cell cycle genes were down-regulated suggesting that transcriptional regulation of these genes might contribute to Al-induced arrest in cell division. One of the major Al resistance mechanisms includes Al exclusion and/or internal detoxification using organic acid chelators. To date, no strong evidence exists for Al-induced expression of any of the enzymes catalyzing organic acid synthesis and metabolism. In this study, we identified the first Al-inducible putative organic acid transporter gene in M. truncatula. Future work will involve determining whether this MATE gene is a citrate efflux transporter. However, since A17 is Al sensitive, it appears that the induction in expression of the organic acid transporter gene is not sufficient for Al resistance in M. truncatula, and that activation of the transporter protein might also be required. RNAi-induced gene silencing of two Al-induced genes demonstrated the importance of these genes in root growth and their potential role in Al resistance response. Further investigations of these genes and the putative MATE transporter may facilitate the understanding of the mechanistic functioning of Al toxicity and/or resistance processes and will provide researchers with new molecular resources to develop more Al-tolerant crops.
Abbreviations
- Al:
-
Aluminum
- ANN:
-
Annexin
- AROS:
-
Array-ready oligo set
- CMF:
-
Cellulose microfibrils
- COB:
-
COBRA-like gene
- CW:
-
Cell wall
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- MATE:
-
Multidrug and toxin efflux
- PAE:
-
Pectin acetylesterase
- PCD:
-
Programmed cell death
- PE:
-
Pectinesterase
- PM:
-
Plasma membrane
- PME:
-
Pectin methylesterase
- PR:
-
Pathogenesis-related
- q-PCR:
-
Quantitative real-time PCR
- RNAi:
-
Interfering RNA
- ROS:
-
Reactive oxygen species
- TC:
-
Tentative consensus
References
Ahn SJ, Sivaguru M, Chung GC, Rengel Z, Matsumoto H (2002) Aluminium-induced growth inhibition is associated with impaired efflux and influx of H+ across the plasma membrane in root apices of squash (Cucurbita pepo). J Exp Bot 53:1959–1966
Allison JD, Brown DS, Novo-Gradac KJ (1991) MINTEQA2/PRODEFA2, a geochemical assessment model for environmental systems, v 3.0 User’s manual; U.S. EPA; Athens, GA
Arnholdt-Schmitt B (2004) Stress-induced cell reprogramming. A role for global genome regulation? Plant Physiol 136:2579–2586
Blancaflor EB, Jones DL, Gilroy S (1998) Alterations in the cytoskeleton accompany aluminum-induced growth inhibition and morphological changes in the primary roots of maize. Plant Physiol 118:159–172
Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14:695–700
Boscolo PR, Menossi M, Jorge RA (2003) Aluminum-induced oxidative stress in maize. Phytochemistry 62:181–189
Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jorgensen LB, Brown RE, Mundy J (2002) Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 16:490–502
Bucher M, Brunner S, Zimmermann P, Zardi GI, Amrhein N, Willmitzer L, Riesmeier JW (2002) The expression of an extensin-like protein correlates with cellular tip growth in tomato. Plant Physiol 128:911–923
Cassab GI (1998) Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 49:281–309
Chen C, Dickman MB (2005) Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc Natl Acad Sci USA 102:3459–3464
Chen D, Liang M-X, Dewald D, Weimer B, Peel MD, Bugbee B, Michaelson J, Davis E, Yajun W (2008) Identification of dehydration responsive genes from two non-nodulated alfalfa cultivars using Medicago truncatula microarrays. Acta Physiol Plant 30:183–199
Ciamporova M (2002) Morphological and structural responses of plant roots to aluminum at organ, tissue and cellular levels. Biol Plant 45:161–171
Clark G, Cantero-Garcia A, Butterfield T, Dauwalder M, Roux SJ (2005) Secretion as a key component of gravitropic growth: implications for annexin involvement in differential growth. Gravit Space Biol Bull 18:113–114
Clark GB, Sessions A, Eastburn DJ, Roux SJ (2001) Differential expression of members of the annexin multigene family in Arabidopsis. Plant Physiol 126:1072–1084
Cruz-Ortega R, Cushman JC, Ownby JD (1997) cDNA clones encoding 1, 3-β-glucanase and a fimbrin-like cytoskeletal protein are induced by Al toxicity in wheat roots. Plant Physiol 114:1453–1460
Darko E, Ambrus H, Stefanovits-Banyai E, Fodor J, Bakos F, Barnabas B (2004) Aluminum toxicity, Al tolerance and oxidative stress in an Al-sensitive wheat genotype and in Al-tolerant lines developed by an in vitro microspore selection. Plant Sci 166:583–591
De Carvalho-Niebel F, Lescure N, Cullimore JV, Gamas P (1998) The Medicago truncatula MtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Mol Plant Microbe Interact 11:504–513
De Carvalho-Niebel F, Timmers AC, Chabaud M, Defaux-Petras A, Barker DG (2002) The Nod factor-elicited annexin MtAnn1 is preferentially localized at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. Plant J 32:343–352
Delhaize E, Gruber BD, Ryan PR (2007) The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett 12:2255–2262
Delisle G, Champoux M, Houde M (2001) Characterization of oxalate oxidase and cell death in Al-sensitive and tolerant wheat roots. Plant Cell Physiol 42:324–333
Delmer DP, Potikha TS (1997) Structures and functions of annexins in plants. Cell Mol Life Sci 53:546–553
Devi SR, Yamamoto Y, Matsumoto H (2003) An intracellular mechanism of aluminum tolerance associated with high antioxidant status in cultured tobacco cells. J Inorg Biochem 97:59–68
Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144:197–205
Dykema PE, Sipes PR, Marie A, Biermann BJ, Crowell DN, Randall SK (1999) A new class of proteins capable of binding transition metals. Plant Mol Biol 41:139–150
Eticha D, Stass A, Horst WJ (2005) Cell-wall pectin and its degree of methylation in the maize root-apex: significance for genotypic differences in aluminum resistance. Plant Cell Environ 28:1410–1420
Foy CD, Chaney RL, White MC (1978) The physiology of metal toxicity in plants. Annu Rev Plant Physiol 29:511–566
Frantzois G, Galatis B, Apostolakos P (2000) Aluminium effects on microtubule organization in dividing root-tip cells of Triticum turgidum. I. Mitotic cells. New Phytol 145:211–224
Furukawa N, Yamaji H, Wang N, Mitani Y, Murata K, Sato M, Katsuhara K, Takeda, Ma JF (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48:1081–1091
Hamel F, Breton C, Houde M (1998) Isolation and characterization of wheat aluminum-regulated genes: possible involvement of aluminum as a pathogenesis response elicitor. Planta 205:531–538
Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29:1217–1225
Hong JK, Hwang BK (2005) Induction of enhanced disease resistance and oxidative stress tolerance by over expression of pepper basic PR-1 gene in Arabidopsis. Physiol Plant 124:267–277
Jennings DB, Ehrenshaft M, Pharr DM, Williamson JD (1998) Roles for mannitol and mannitol dehydrogenase in active oxygen-mediated plant defense. Proc Natl Acad Sci USA 95:15129–15133
Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, Godiard L, Micheli F, Kahn D, Gianinazzi-Pearson V, Gamas P (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30:5579–5592
Kang CH, Jung WY, Kang YH, Kim JY, Kim DG, Jeong JC, Baek DW, Jin JB, Lee JY, Kim MO, Chung WS, Mengiste T, Koiwa H, Kwak SS, Bahk JD, Lee SY, Nam JS, Yun DJ, Cho MJ (2006) AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ 13:84–95
Kinraide TB (1991) Identity of the rhizotoxic aluminum species. Plant Soil 134:167–178
Klessig DF, Durner J, Noad R, Navarre DA, Wendehenne D, Kumar D, Zhou JM, Shah J, Zhang S, Kachroo P, Trifa Y, Pontier D, Lam E, Silva H (2000) Nitric oxide and salicylic cid signaling in plant defense. Proc Natl Acad Sci USA 97:8849–8855
Kochian LV, Piñeros MA, Hoekenga OA (2005) The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274:175–195
Kuge S, Jones N (1994) YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J 13:655–664
Larsen PB, Geisler MJ, Jones CA, Williams KM, Cancel JD (2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J 41:353–363
Larsen PB, Cancel J, Rounds M, Ochoa V (2007) Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225:1447–1458
Lazof DB, Goldsmith JG, Rufty TW, Linton RW (1994) Rapid uptake of aluminum into cells of intact soybean root tips: A microanalytical study using secondary ion mass spectrometry. Plant Physiol 106:1107–1114
Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KA, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140:221–234
Ma JF, Shen R, Nagao S, Tanimoto E (2004) Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol 45:583–589
Magalhaes JV, Liu J, Guimarães CT, Lana UGP, Alves VMC, Wang Y-H, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, Klein PE, Carneiro NP, Coelho CM, Trick HN, Kochian LV (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39:1156–1161
Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Puhler A, Perlick AM, Kuster H (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant Microbe Interact 17:1063–1077
Mao C, Yi K, Yang L, Zheng B, Wu Y, Liu F, Wu P (2004) Identification of aluminum-regulated genes by cDNA-AFLP in rice (Oryza sativa L.): aluminum-regulated genes for the metabolism of cell wall components. J Exp Bot 55:137–143
Mariano ED, Jorge RA, Keltjens WG, Menossi M (2005) Metabolism and root exudation of organic acid anions under aluminium stress. Braz J Plant Physiol 17:157–172
Massot N, Nicander B, Barceló J, Poschenrieder Ch, Tillberg E (2002) A rapid increase in cytokinin levels and enhanced ethylene evolution precede Al3+-induced inhibition of root growth in bean seedlings (Phaseolus vulgaris L.). Plant Growth Regul 37:1573–1587
Matsumoto H (1988) Changes of the structure of pea chromatin by aluminum. Plant Cell Physiol 29:281–287
Matsumoto H, Hirasawa E, Torikai H, Takahashi E (1976) Localization of absorbed aluminum in pea root and its binding to nucleic acids. Plant Cell Physiol 17:127–137
Matsumoto H, Morimura S (1980) Repressed template activity of chromatin of pea roots treated by aluminum. Plant Cell Physiol 21:951–959
Pence NS, Larsen PB, Ebbs SD, Letham DL, Lasat MM, Garvin DF, Eide D, Kochian LV (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97:4956–4960
Rengel Z, Zhang WH (2003) Role of dynamics of intracellular calcium in aluminium-toxicity syndrome. New Phytol 159:295–314
Ragland M, Soliman KM (1997) Sali-4a (Accession No. U64866) and Sali3-2 (Accession No. U89693). Two genes induced by aluminum in soybean roots. Plant Physiol 114:395
Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC (1998) Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol 116:409–418
Ryan PR, Ditomaso JM, Kochian LV (1993) Aluminum toxicity in roots: An investigation of spatial sensitivity and the role of the root cap. J Exp Bot 44:437–446
Sasaki M, Yamamoto Y, Matsumoto H (1996) Lignin deposition induced by aluminum in wheat (Triticum aestivum) roots. Physiol Plant 96:193–198
Sasaki T, Ezaki B, Matsumoto H (2002) A gene encoding multidrug resistance (MDR)-like protein is induced by aluminum and inhibitors of calcium flux in wheat. Plant Cell Physiol 43:177–185
Schildknecht PHPA, Vidal BC (2002) A role for the cell wall in Al3+ resistance and toxicity: crystallinity and availability of negative charges. Int Arch Biosci 2000:1087–1095
Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15:1115–1127
Schmohl N, Pilling J, Fisahn J, Horst WJ (2000) Pectin methylesterase modulates aluminum sensitvity in Zea mays and Solanum tuberosum. Physiol Plant 109:419–427
Silva IR, Smyth TJ, Moxley DF, Carter TE, Allen NS, Rufty TW (2000) Aluminum accumulation at nuclei of cells in the root tip. Fluorescence detection using lumogallion and confocal laser scanning microscopy. Plant Physiol 123:543–552
Simonovicova M, Huttova J, Mistrik I, Siroka B, Tamas L (2004) Root growth inhibition by aluminum is probably caused by cell death due to peroxidase-mediated hydrogen peroxide production. Protoplasma 224:91–98
Sivaguru M, Horst WJ (1998) The distal part of the transition zone is the most aluminum sensitive apical root zone of maize. Plant Physiol 116:155–163
Sivaguru M, Pike S, Gassman W, Baskin TI (2003) Aluminum rapidly depolymerizes cortical microtubules and depolarizes the plasma membrane: evidence that these responses are mediated by a glutamate receptor. Plant Cell Physiol 44:667–675
Snowden KC, Richards KD, Gardner RC (1995) Aluminum-induced genes. Induction by toxic metals, low calcium, and wounding and pattern of expression in root tips. Plant Physiol 107:341–348
Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11:431–444
Sugimoto K, Williamson RE, Wasteneys GO (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124:1493–1506
Sun P, Tian QY, Zhao MG, Dai XY, Huang JH, Li LH, Zhang WH (2007) Aluminum-induced ethylene production is associated with inhibition of root elongation in Lotus japonicus L. Plant Cell Physiol 48:1229–1235
Tamas L, Huttova J, Mistrik I (2003) Inhibition of Al-induced root elongation and enhancement of Al-induced peroxidase activity in Al -sensitive and Al-resistant barley cultivars are positively correlated. Plant Soil 250:193–200
Tesfaye M, Samac DA, Vance CP (2006) Insights into symbiotic nitrogen fixation in Medicago truncatula. Mol Plant Microbe Interact 19:330–341
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121
Vazquez MD (2002) Aluminum exclusion mechanism in root tips of maize (Zea mays L.): Lysigeny of aluminum hyperaccumulator cells. Plant Biol 4:234–249
Vercauteren I, de Almeida Engler J, De Groodt R, Gheysen G (2002) An Arabidopsis thaliana pectin acetylesterase gene is upregulated in nematode feeding sites induced by root-knot and cyst nematodes. Mol Plant Microbe Interact 15:404–407
Wen F, Zhu Y, Hawes MC (1999) Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11:1129–1140
Yamamoto Y, Kobayashi Y, Matsumoto H (2001) Lipid peroxidation is an early symptom triggered by aluminum, but not the primary cause of elongation inhibition in pea roots. Plant Physiol 125:199–208
Zheng SJ, Yang JL (2005) Target sites of aluminum phytotoxicity. Biol Plant 49:321–331
Acknowledgments
This work was supported by the National Science Foundation Plant Genome Project (award no. 0110206) and USDA-ARS. We thank Dr. Susan C. Miyasaka (University of Hawaii) for providing us with the media composition for Al-plate assays, Dr. David Galbraith (University of Arizona) for printing of microarrays, Dr. Dasharath P. Lohar (University of Minnesota) for microarray slide scanning and Dr. Judy Schnurr for assistance with RNA blots. We acknowledge support from the University of Minnesota Super Computing Institute for data analysis. This paper is a joint contribution from the Plant Science Research Unit, USDA, Agricultural Research Service and the Minnesota Agricultural Experiment Station. Mention of a trademark, proprietary product or vendor does not constitute a guarantee or warranty of the product by the USDA, and it does not imply its approval to the exclusion of other products and vendors that might also be suitable.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chandran, D., Sharopova, N., Ivashuta, S. et al. Transcriptome profiling identified novel genes associated with aluminum toxicity, resistance and tolerance in Medicago truncatula . Planta 228, 151–166 (2008). https://doi.org/10.1007/s00425-008-0726-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0726-0