Abstract
Flavonoids are a large family of polyphenolic compounds with manifold functions in plants. Present in a wide range of vegetables and fruits, flavonoids form an integral part of the human diet and confer multiple health benefits. Here, we report on metabolic engineering of the flavonoid biosynthetic pathways in apple (Malus domestica Borkh.) by overexpression of the maize (Zea mays L.) leaf colour (Lc) regulatory gene. The Lc gene was transferred into the M. domestica cultivar Holsteiner Cox via Agrobacterium tumefaciens-mediated transformation which resulted in enhanced anthocyanin accumulation in regenerated shoots. Five independent Lc lines were investigated for integration of Lc into the plant genome by Southern blot and PCR analyses. The Lc-transgenic lines contained one or two Lc gene copies and showed increased mRNA levels for phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), flavanone 3 beta-hydroxylase (FHT), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin reductases (LAR), anthocyanidin synthase (ANS) and anthocyanidin reductase (ANR). HPLC-DAD and LC-MS analyses revealed higher levels of the anthocyanin idaein (12-fold), the flavan 3-ol epicatechin (14-fold), and especially the isomeric catechin (41-fold), and some distinct dimeric proanthocyanidins (7 to 134-fold) in leaf tissues of Lc-transgenic lines. The levels of phenylpropanoids and their derivatives were only slightly increased. Thus, Lc overexpression in Malus domestica resulted in enhanced biosynthesis of specific flavonoid classes, which play important roles in both phytopathology and human health.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
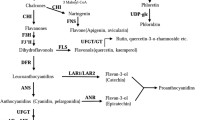
Flavonoids are plant polyphenolic compounds derived from the phenylpropanoid pathway. On the basis of their structure they can be classified into the classes flavanones, flavones, isoflavones, dihydroflavonols, flavonols, leucoanthocyanidins, flavan 3-ols (catechins and the polymeric proanthocyanidins) and anthocyanins. Representatives of most of these classes are also found in Malus domestica (Fig. 1). Chalcones are direct precursors of all flavonoids. The prominent dihydrochalcones present in M. domestica are chemical derivatives of them. Flavonoids have manifold functions in plants such as UV-protection, attraction of pollinators, warranty of pollen fertility, regulation of polar auxin transport (Taylor and Grotewold 2005), establishment of microbial symbioses and pathogen defence (Treutter 2005). In fruit crops such as M. domestica, flavonoids are of utmost importance as they impart much of the colour and contribute to the flavour of the apple fruit by astringency of proanthocyanidins. Phytopathological work demonstrated the significance of flavonoids, especially of flavan 3-ols, in resistance reactions to apple scab, caused by the fungus Venturia inaequalis (Cooke) G. Wint., the most important apple disease in all cultivating regions (Römmelt et al. 1999; Bazzi et al. 2003; Leser and Treutter 2005).
In addition to these implications in plant physiology and metabolism, flavonoids have attracted increasing interest as health promoting agents due to their antioxidant properties, their effects on mammalian enzymes (Geahlen et al. 1989; Cushman et al. 1991; Middleton et al. 2000), especially their actions at protein kinase and lipid kinase signalling pathways (reviewed by Williams et al. 2004). Results from several epidemiological studies revealed protective effects of flavonoids against cardiovascular diseases (Hertog et al. 1997), cancer (Knekt et al. 1997, 2000) and other age-related diseases such as dementia (Commenges et al. 2000) due to their anti-proliferative, anti-tumour and pro-apoptotic activities.
Apples are an important source of polyphenols in the human diet. These polyphenols include hydroxycinnamic acids (e.g. chlorogenic acid and p-coumaroyl glucoside), cyanidin-3-O-galactoside (idaein), flavan-3-ols/procyanidins (mainly catechin, epicatechin and procyanidins B2 and B5), flavonols (quercetin galactoside, glucoside, rhamnoside and arabinoside) and dihydrochalcones (mainly phloridzin and phloretin-2′-xyloglucoside; Treutter 2001; Tsao et al. 2005). Because of the high impact of flavonoids in plant defence and their great influence on quality and health of food and food products there is a growing interest in the development of agriculturally important food crops with optimized levels and composition of flavonoids.
Two classes of genes are determining the flavonoid pathway. Structural genes encode enzymes that directly participate in the formation of flavonoids and regulatory genes control the expression of the structural genes. Most known regulatory genes are homologous to myb and myc transcription factors, but some are also WD-repeat proteins or belong to other classes. One of the best characterized regulatory genes is the leaf colour (Lc) gene from maize (Ludwig et al. 1989). This gene, belonging to the myc type R gene family, encodes a protein with the basic-helix-loop-helix (bHLH) motif. Lc is one of the transcription factors which determines the amount, location and timing of anthocyanin accumulation in maize. The heterologous expression of Lc or combinations of Lc along with the myb type C1 (colourless) gene resulted in increased anthocyanin pigmentation in Nicotiana tabacum L., Arabidopsis (Lloyd et al. 1992), Lycopersicon esculentum Mill (Goldsbrough et al. 1996), a Petunia hybrid (Bradley et al. 1998), Medicago sativa L. (Ray et al. 2003) and Caladium bicolor (Li et al. 2005). Flavonoids of other classes have also been reported to accumulate after heterologous Lc expression. In the Petunia hybrid, an up-regulation of anthocyanin contents occurred accompanied by up-regulation of gene expression of most but not all enzymes of the flavonoid pathway (Bradley et al. 1998). Flavonol contents were positively and negatively influenced in a tissue-specific way. The coordinate expression of the maize regulatory genes Lc and C1 in transgenic L. esculentum resulted in an increased production of anthocyanins (in leaves) and of the flavonol kaempferol (in leaves and fruits; Bovy et al. 2002). In M. sativa, the transgenic Lc gene caused induction of anthocyanins and proanthocyanidins correlated with a reduction of flavone concentrations (Ray et al. 2003).
The study presented here reports on the introduction of the maize regulatory gene Lc into the M. domestica Borkh. cultivar Holsteiner Cox (HC) under control of the constitutive cauliflower mosaic virus (CaMV 35S) promoter to enhance flavonoid biosynthesis. Effects on the expression of phenylpropanoid and flavonoid genes and on the metabolite levels of anthocyanins, monomeric and polymeric flavan 3-ols (catechins, proanthocyanidins), flavonols, dihydrochalcones, hydroxycinnamic acids and hydroxybenzoic acids were considered in detail. This work was performed to comprehensively differentiate the various consequences of the heterologous myc transcription factor in a relevant fruit crop.
Material and methods
Agrobacterium strain and binary vector
The Lc gene of maize (Genbank M26227) was kindly provided by Ryan Peeler (Plant Biology Department, University of Georgia, Athens, GA, USA). The gene along with the CaMV35S promoter, was introduced into the binary vector pBI121 (Genbank accession no. AF485783) by replacing the GUS (β-glucuronidase) gene by HindIII and EcoRI sites. Additionally, pBI121 carries on its T-DNA the nptII gene coding for neomycin phosphotransferase II driven by the nos promoter. pBI-Lc thus confers kanamycin resistance in plants and was introduced into Agrobacterium tumefaciens EHA105 (Hood et al. 1993) through electroporation.
Plant material
The plant material consisted of in vitro cultivated shoots of apple (M. domestica Borkh.) cv. HC. In vitro cultures were initiated from nodal segments. Plant material was obtained from the Chamber of Agriculture Lower Saxony, Germany. Shoot cultures were maintained as described elsewhere (Szankowski et al. 2003).
Transformation and regeneration
The transformation of apple was done as described by Szankowski et al. (2003). In brief: explants obtained from the four youngest unfolded leaves from 4-week-old micropropagated in vitro shoots were inoculated with agrobacteria (grown over night in YEP medium, centrifuged and resuspended in liquid MS medium to an OD of 0.8) and cocultivated in the dark on regeneration medium [MS salts and vitamins (Murashige and Skoog 1962), 3% sorbitol, 3 μM TDZ, 1 μM IBA and 0.3% gelrite; pH 5.7] for 3 days.
Adventitious shoot induction was performed on regeneration medium supplemented with 150 mg l−1 ticarcillin (agrobacteria elimination) and 50 mg l−1 kanamycin for selection of transgenic plant cells. Explants were cultured in the dark for 2 weeks at 24 ± 1°C, then under 16/8 h photoperiod (70 μmol m−2 s−1) at the same temperature. The medium was changed every 2 weeks and after 6 weeks on regeneration medium they were cultured on elongation medium (MS salts and vitamins, 4.4 μM BAP, 0.28 μM GA3, 3% sucrose and 0.8% plant agar, pH 5.7).
DNA analysis
For the detection of the transgenes in the genome of kanamycin selected, regenerated plants PCR and Southern blot analysis were performed. DNA was extracted from leaves of in vitro plantlets (Doyle and Doyle 1990). The PCR reactions contained 0.2 U of Taq DNA polymerase (Genecraft, Lüdinghausen, Germany), 16 mM (NH4)2SO4, 67 mM Tris–HCl (pH 8.8), 1.5 mM MgCl2, 200 μM of each dNTP and 0.4 μM of each primer in a volume of 25 μl.
The primers 5′-ATG GCG CTT TCA GCT TCC CGA-3′ and 5′-TGT ACC AAG CTC AAG CAC GCC-3′ were used to amplify a specific Lc-fragment with the size of 583 bp under following conditions: 94°C for 1 min, followed by 30 cycles at 94°C for 45 s, 60°C for 45 s, 72°C for 45 s and with a final extension at 72°C for 5 min. The presence of the nptII gene was confirmed by using the primers 5′-GAT TGA ACA AGA TGG ATT-3′ and 5′-CAT TTT CCA CCA TGA TAT TC-3′, which amplify a 600 bp nptII specific fragment. The PCR reaction conditions were 95°C for 3 min, followed by 35 cycles at 94°C for 90 s, 63°C for 90 s, 72°C 150 s and with a final extension at 72°C for 10 min.
Southern blot analyses were performed to verify the integration of the transgenes. Twenty micrograms of genomic DNA were restricted with HindIII (single restriction site within the T-DNA) at 37°C overnight. DNA was separated in a 0.8% agarose gel and blotted on a positively charged nylon membrane (Boehringer, Mannheim, Germany). Membranes were hybridized with the digoxygenin-labelled PCR probes for Lc, incubated with alkaline phosphate conjugated anti-digoxigenin followed by the chemiluminescent substrate CDP-Star and finally exposed to X-ray film. All steps were performed following the supplier’s instructions (Roche, Mannheim, Germany). Primers and PCR conditions for probe generation were the same as mentioned above.
Transcription profiling and real-time PCR
Three young in vitro leaves were randomly selected from each transgenic line and the untransformed control line and frozen in liquid nitrogen. The total RNA from these leaves was extracted using the Invisorb® Spin Plant-RNA Mini Kit (Invitek, Berlin, Germany). One μg of total RNA was treated with Dnase I (MBI Fermentas, St. Leon-Rot, Germany) to eliminate genomic DNA contaminations. Presence of DNA residues was tested using 1 μl total RNA as template in a 25 μl standard PCR reaction with primers specific for an apple gene encoding the elongation factor EF1 alpha. The PCR mixture contained 1× NH4-buffer, 12 mM MgCl2, 25 mM dNTPs, 12.5 mM of each primer (EF1forw 5′-TAC TGG AAC ATC ACA GGC TGA C-3′ and EF1rev 5′-TGG ACC TCT CAA TCA TGT TGT C-3′) and 0.625 U of Taq DNA polymerase (Invitek). The PCR reaction was performed by denaturation at 94°C for 5 min, followed by 30 cycles of 1 min denaturation at 94°C, 30 s annealing at 65°C and 1 min extension at 72°C. After a final extension at 72°C for 5 min the amplified fragments were separated on a 0.8–1% agarose gel containing 0.5 μg ml−1 ethidiumbromid in 1× TAE buffer.
The remaining RNA was reverse transcribed using oligo(dT)18 primers and the RevertAid™ First Strand cDNA Synthesis Kit (MBI Fermentas) according to the manufacturer’s instruction. The success of reverse transcription was verified by PCR of 1 μl cDNA using primers EF1_F1 and EF1_R1 as described. Quantitative real-time PCR was performed using the iQTM SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and first-strand cDNA as template on an iCycler iQ real-time PCR Detection System (Bio-Rad).
Gene specific amplification was evaluated by melt curve analysis and agarose gel electrophoresis. Single melt peaks were obtained for each gene. No primer dimers or other PCR artefacts were detected. To determine the amplification efficiencies and correlation efficiencies of each PCR reaction, 15, 30, 60 and 120 ng cDNA of the transgenic line T1 were analysed. The PCR efficiencies were used to transform the Ct-values into raw data for relative quantification. The determination of PCR efficiency and the calculation of the mRNA transcript levels were done using the Gene Expression Macro™ Version 1.1 (Bio-Rad). The methodology of this macro is based on the algorithms outlined by Vandesompele et al. (2002). The Lc gene transcription of each line was quantified relatively using the same primers and conditions as described. All samples were normalized using ribulose-1,5-bisphosphate carboxylase/oxygenase activase mRNA and mRNA of the M. domestica cDNA clone Mdfw2033f21.y1 (similar to the RNA polymerase subunit II) as internal control samples for each line. As no Lc gene transcription was expected in the non-transgenic HC the scaling of the Lc gene transcript levels was performed in relation to the mRNA expression level of the reference genes. The scaling of the genes involved in the flavonoid biosynthetic pathway of apple was performed relative to the mRNA expression level of the non-transgenic HC set to be one.
LC-MS
A Bruker Daltonics esquire 3000plus ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany) connected with an Agilent 1100 HPLC system (Agilent Technologies, Waldbronn, Germany) equipped with a quaternary pump and a variable wavelength detector was used for LC-MS analysis. Components were separated on a Phenomenex, Aschaffenburg, Germany, Luna C-18 column (150 mm long × 2.0 mm inner diameter, particle size 5 μm) which was held at 25°C. The LC parameters went from 0% acetonitrile and 100% water (acidified with 0.05% formic acid) to 40% acetonitrile and 60% acidic water in 40 min, in further 5 min to 100% acetonitrile and remained for 10 min at these conditions with a flow rate of 0.2 ml min−1. The detection wavelength was 280 nm. The electrospray ionization voltage of the capillary was set to −4,000 V and the end plate to −500 V. Nitrogen was used as dry gas at a temperature of 300°C and a flow rate of 10 l min−1. The full scan mass spectra were measured in a scan range from 50 to 800 m/z with a scan resolution of 13,000 m/z/s until the ICC target reached 20,000 or 200 ms, whatever was achieved first. Tandem mass spectrometry was carried out using helium as collision gas (3.56 × 10−6 mbar) with the collision voltage set at 1 V. Spectra were acquired in the positive and negative ionization mode. Auto-tandem mass spectrometry was used to break down the most abundant [M+H]+, [M-H]− or the [M+HCOO]− ion of the different compounds of the extracts. Data analysis was performed using the DataAnalysis 3.1 software (Bruker Daltonics) and MZmine (http://www.mzmine.sourceforge.net; Katajamaa et al. 2006). Metabolites were identified by their retention times, mass spectra and product ion spectra in comparison with the data determined for authentic reference material. Signals of the compounds were integrated in their [M+H]+, [M-H]− or [M+HCOO]− ion traces. Induction factors were calculated from signal intensities.
Quantitative HPLC-DAD and identification of phenolic compounds
Analysis of leaves was done as follows: samples were lyophilized and ground in a ball mill. The extraction of phenolic compounds was done with methanol (100%) containing 6-methoxyflavone as an internal standard for 30 min in a cooled water bath during sonication. After centrifugation, the supernatant was evaporated and the residue redissolved in small quantities of methanol and injected for HPLC analysis. The phenolic compounds were separated on a column (250 × 4 mm2 i.d.) prepacked with Hypersil ODS, 3-μm particle size, following a stepwise gradient using mixtures of solvent A (formic acid, 5% in water) and solvent B (methanol, gradient grade) from 95:5 (v/v) to 10:90 (v/v) with a flow rate of 0.5 ml min−1 (Treutter et al. 1994). The gradient profile (% B in A) used was as follows: 0–5 min, isocratic, 5% B; 5–15 min, 5–10% B; 15–30 min, isocratic, 10% B; 30–50 min, 10–15% B; 50–70 min, isocratic, 15% B; 70–85 min, 15–20% B; 85–95 min, isocratic, 20% B; 95–110 min, 20–25% B; 110–140 min, 25–30% B; 140–160 min, 30–40% B; 160–175 min, 40–50% B; 175–190 min, 50–90% B. For the HPLC determination of the dihydrochalcone glycoside phloridzin the extract was diluted by 200 with methanol and the phloridzin was analysed using a short column (12.5 × 4 mm2 i.d.) prepacked with LiChrospher 100 RP18, 5-μm particle size, and a gradient range from 40 to 90% aqueous methanol. Hydroxycinnamic acids (p-coumaric acids and caffeic acid derivatives), the dihydrochalcone glycosides and the flavonols (quercetin glycosides), were detected at 320, 280 and 350 nm, respectively, whereas the monomeric flavan-3-ols, the procyanidins and the dihydrochalcone phloretin were estimated at 640 nm after postcolumn derivatization with 4-dimethylaminocinnamic aldehyde (DMACA; Treutter 1989). Peak identification is published elsewhere (Mayr et al. 1995; Römmelt et al. 2003a).
Results
Generation of Lc-transgenic apple lines
The maize Lc regulatory gene under the control of the CaMV 35S promoter was transferred into M. domestica cv. HC via Agrobacterium tumefaciens-mediated transformation. The shortened 2.2 kbp variant of the Lc gene lacking the inhibiting 200 bp of untranslated sequence was used (Lloyd et al. 1992). Explants inoculated with the agrobacteria and cultivated under kanamycin selection developed calli with red/purple groups of cells, later forming shoots. In regenerated shoots, the pigmentation was maintained (Fig. 2) which was neither observed in non-transformed control HC shoots nor in HC shoots transformed with other genes (Szankowski et al. 2003; Degenhardt et al. 2006; Rühmann et al. 2006). High levels of anthocyanins accumulated in leaves as well as in the stem (Fig. 2) of regenerated plantlets. Transgenic shoots developed much more callus on shoot propagation medium and this callus was dark red coloured. Contrarily, the callus of untransformed HC shoots was green. Furthermore, the growth of transgenic shoots was slower in comparison to non-transgenic shoots of the same cultivar.
a–e In vitro plants of Holsteiner Cox transformed with pBI-Lc (line T1) and regenerated under selective pressure with 50 mg l−1 kanamycin exhibit strong anthocyanin pigmentation in leaves (b, d) and stems (e, lower part, cross-section of a stem) compared to non-transformed leaves (a, c) and stems (e, upper part)
Integration analysis
Five transgenic lines were regenerated on kanamycin selective medium. Results of Southern blot analyses (Fig. 3) revealed the integration of at least one copy of the Lc gene in lines T1, T2, T3 and T4 and two gene copies in line T5. No hybridization signal occurred in the DNA sample of non-transformed control plants.
Southern hybridization of HindIII digested DNA isolated from Lc-transgenic apple plants and respective control plants. Genomic DNA was hybridized to a 583 bp Lc-probe. Lanes: M molecular-weight markers II, DIG labelled (Roche), HC negative control (DNA from non-transformed Holsteiner Cox plants), T1–T5 DNA isolated from transgenic lines
Transcription profiling
Real-time RT-PCR using gene specific primers (Table 1) was performed to obtain transcription profiles for the transgenes nptII and Lc as well as for the genes of the flavonoid biosynthetic pathway encoding PAL, CHS, CHI, FHT, FLS, DFR, LAR1+2, ANS, ANR and FGT. The transcripts of nptII, Lc (Fig. 4), and structural flavonoid genes were detected in tissues of the transgenic lines T1-T5. No nptII or Lc expression was observed in non-transgenic control samples, while rubisco expression was detected in all cDNA samples (Fig. 4). The Lc gene transcription levels of the transgenic lines ranged between 137 (line T4) and 291 (line T2; Figs. 5, 6).
mRNA expression levels of genes involved in flavonoid biosynthetic pathway in Lc transgenic apple lines (T1–T5, white bars from left to right) determined by real-time PCR. The values are expressed relative to the level of transcripts in the untransformed Holsteiner Cox (dark grey) set to be one. Values are the mean of three replicates ±SE. PAL phenylalanine ammonia lyase, CHS chalcone synthase, CHI chalcone isomerase, FHT flavanon 3-beta-hydroxylase, FLS flavonol synthase, DFR dihydroflavonol 4-reductase, LAR leucoanthocyanidin reductase, ANS anthocyanidin synthase, ANR anthocyanidin reductase, FGT, UDP-glucose:flavonoid 3-O-glucosyltransferase
The relative expression levels of the structural genes revealed that almost all Lc transgenic lines showed significantly increased transcription levels for pal, chs, fht, dfr, lar, ans and anr in comparison to the non-transgenic wild type (Fig. 6). Especially ans transcription was strongly induced in all lines. In contrast, fgt was clearly induced in only some lines. Neither chi nor fls were induced by the Lc transgene.
Metabolic profiling
Metabolic profiling of polyphenolic secondary metabolites was performed by quantitative HPLC-DAD and LC-MS analysis to obtain absolute compound concentrations as well as mass spectral data. A number of anthocyanins, monomeric and polymeric flavan 3-ols (catechins, proanthocyanidins), flavonols, dihydrochalcones, hydroxycinnamic acids and hydroxybenzoic acids were identified and quantified from leaves of untransformed in vitro plants and the five in vitro lines transgenic for 35S-Lc (Table 2, Fig. 7). Generally, members of almost all analysed polyphenol classes were induced by Lc. Strongest rises were observed for the anthocyanin idaein (12-fold), epicatechin (14-fold), catechin (41-fold) and proanthocyanidins (Fig. 7). Remarkably, only two distinct proanthocyanidins, namely B2 and a yet unidentified proanthocyanidin were selectively induced by mean factors of 58 and 134, respectively (Fig. 7). The concentration of hyperin (quercetin-3-O-galactoside) as the prominent M. domestica flavonol was only slightly increased while levels of other flavonol glycosides remained unchanged or were even reduced (Table 2). Also within the large pool of dihydrochalcones (phloretin, phloridzin and phloretin 2′-xyloglucoside) as the dominant M. domestica polyphenol class, the levels in the transgenic lines increased only slightly (up to threefold). Some hydroxycinnamic acids, hydroxybenzoic acids and their derivatives showed rises up to a mean factor of about 2.
a, b Lc-dependent increase in the levels of flavonoids (a) and other phenolic secondary metabolites (b) determined by LC-MS. Values are expressed as fold induction of metabolite levels in transgenic lines relative to metabolite levels in the untransformed controls. The black bar represents the mean ± SD of values obtained from all transgenic lines (T1–T5)
Discussion
Phenotype
The recombinant expression of the maize Lc regulatory gene enhances flavonoid biosynthesis in transgenic M. domestica. Lc transgenic HC shoots were phenotypically distinguishable from non-transgenic plantlets by strong anthocyanin accumulation in leaves and stems. Lc-transgenic shoots also grew slower in comparison to non-transformed shoots. Similar growth inhibiting effects due to Lc expression were observed in a Petunia hybrid (Bradley et al. 1998), in L. esculentum and in Arabidopsis transgenic lines, where calli failed to give rise to shoots or the shoots failed to root (Quattrocchio et al. 1993). Since flavonoids can negatively regulate polar auxin transport in vivo (Taylor and Grotewold 2005), disturbances in transport of endogenous auxins might be the reason.
Induction of structural gene expression
Phenotypic colour change is in accordance with transcription analyses which confirmed an accompanied increase in transcription levels of most anthocyanin structural genes except chi (Fig. 6). Especially expression of anthocyanidin synthase (ANS), the enzyme converting leucoanthocyanidins into anthocyanidins, dramatically increased in Lc-transgenic lines. Similar expression pattern changes, especially the unaffected CHI expression, were also found in Lc/C1 transgenic L. esculentum (Bovy et al. 2002). Gene expression of the diverging pathways to epicatechin (ANR) or catechin (LAR) was induced in Lc overexpressing M. domestica plants, too, but not the FLS responsible for the divergent pathway to the flavonols. The coordinate transcriptional regulation of ANR and LAR1 by a bHLH transcription factor was also found in transgenic Lotus corniculatus expressing the maize bHLH Sn gene (Paolocci et al. 2007).
Metabolites
The Lc protein caused pronounced enhancement of accumulation of anthocyanins, of monomeric flavan 3-ols (catechin, epicatechin) and of some polymeric flavan 3-ols (proanthocyanidins). Generally, this corresponds well with the observed changes in transcription levels of structural genes responsible for anthocyanin and flavan 3-ol biosynthesis (Figs 1, 6). However, the reason for the strongly increased levels of distinct proanthocyanidins (B2 and another unidentified dimer), and catechin as one of their supposed precursors remains unknown. While the level of procyanidin B2 (epicatechin-(4β-8)-epicatechin) was 58-fold increased in the Lc transgenic lines the concentration of B5 (epicatechin-(4β-6)-epicatechin) was only sevenfold higher. From the current data, no obvious correlation of transcript levels with either the stereochemistry of the respective monomers or the types of inter-flavonoid linkage can be seen. The most likely explanation seems the existence of specific condensing enzymes that are differentially regulated and, thus, differentially influenced by the transgenic Lc factor. However, only recently, the ANS catalysed dimerization of catechin monomers has been described (Wellmann et al. 2006). Thus, it is also conceivable that ANS, which is highly expressed in the Lc transgenic lines, plays an important role in the formation of selected proanthocyanidins.
The concentration of hyperin (quercetin 3-O-galactoside) is increased in the transgenic lines, but other quercetin glycosides are reduced and therefore the total amount of flavonols is unchanged. UDP-glycosyl:flavonoid 3-O-glycosyltransferases often show broad substrate specificity for anthocyanidins as well as for flavonols (Ford et al. 1998). Therefore, the induced UDP-galactosyl:flavonoid 3-O-galactosyltransferase from anthocyanin (idaein) biosynthesis may cause this shift towards galactosylated quercetin at the cost of the other quercetin glycosides. Furthermore, the levels of some hydroxycinnamic acids and some hydroxybenzoic acids are slightly increased by Lc.
The enhanced PAL expression, but also the rise in some hydroxycinnamic acids, argues for a rise in p-coumaroyl-CoA substrate supply for flavonoid biosynthesis. The alternative explanation, a drain of substrates from the major pool of dihydrochalcones can hardly be tested for, as the levels of dihydrochalcones in the Lc lines are almost unaffected. With respect to the enormous concentrations of the dihydrochalcones, their biological variation would hide what can be expected for a substrate drain explaining the observed rises in anthocyanins and flavan 3-ols. Nevertheless, this substrate drain may also take place. It appears that dihydrochalcones synthesizing enzymes are not induced by Lc, this could indicate a generally different regulation for them in comparison to the regulation of flavonoid structural genes. The closely related pear (Pyrus communis L.), which is also a member of the Maloidae, does not contain dihydrochalcones (Challice and Williams 1968; Andreotti et al. 2006). This hints to a phylogenetically young pathway in M. domestica.
Mode of transcription factor action
In maize and many other plants, the presence of both myb and bHLH (myc) type transcription factors is required for anthocyanin expression (e.g. Grotewold 2005; Heim et al. 2003). In HC apple, constitutive Lc expression was sufficient to strongly enhance anthocyanin, catechins and specific proanthocyanidin accumulation, making it likely that interacting myb-type transcription factors are expressed in the cultivar under the described experimental conditions. Recently, the light induced transcription factors MdMYB1 and MdMYB10, determining apple fruit colour, were described (Espley et al. 2006; Takos et al. 2006). In MdMYB10 a signature motif for the interaction with bHLH proteins was identified (Espley et al. 2006). It may be expected that the Lc protein interacts with or even induces myb-type transcription factors, which leads to the observed induction of anthocyanins and other phenylpropanoids. Further work is required to confirm this.
Perspective
In this study we have shown that heterologous expression of maize Lc enhanced flavonoid biosynthesis in leaf tissues of apple. Whether the effect will also be true for the fruit appears to be strongly influenced by and depending on expression patterns of other transcription factors and can only be determined when plants have fruited.
Flavonoids have become targets for genetic plant modification to modify flower colour and to improve forage traits, crop resistance and healthiness of the human diet. Especially flavan-3-ols and their derivatives have significant health promoting effects and show even pharmacological activities. The role of flavonoids in pathogen resistance is well documented (Jambunathan et al. 1990; Byrne et al. 1996; Römmelt et al. 1999, 2003b; Halbwirth et al. 2003). Studies on apple provided evidence for the significance of flavonoids in resistance to apple scab (V. inaequalis). Flavan 3-ols accumulate at the infection site (Treutter and Feucht 1990) and their concentration is associated with field resistance (Mayr et al. 1997). Inhibition of PAL, catalysing the first step of the phenylpropanoid pathway and supplying the precursors for flavonoid biosynthesis, led to severe apple scab symptoms on leaves of a resistant cultivar (Mayr et al. 1997), whilst vice versa enhanced biosynthesis improved resistance (Römmelt et al. 1999; Bazzi et al. 2003; Leser and Treutter 2005). Flavonoids also play a major role in post-harvest resistance of fruits and vegetables (Lattanzio et al. 1994; Lattanzio 2003). Based on these studies it can be assumed, that Lc-transgenic apple plants may exhibit increased pathogen resistance.
Abbreviations
- ANS:
-
Anthocyanidin synthase
- ANR:
-
Anthocyanidin reductase
- BAP:
-
6-Benzylaminopurine
- CaMV:
-
Cauliflower mosaic virus
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- DAD:
-
Diode array detection
- DFR:
-
Dihydroflavonol 4-reductase
- FGT:
-
UDP-Glucose:flavonoid 3-O-glucosyltransferase
- FHT:
-
Flavanon 3 beta-hydroxylase
- FLS:
-
Flavonol synthase
- GA:
-
Gibberellic acid
- HPLC:
-
High performance liquid chromatography
- HC:
-
Holsteiner Cox
- IBA:
-
Indole-3-butyric acid
- LAR1+2:
-
Leucoanthocyanidin reductase
- Lc :
-
Maize leaf colour
- LC-MS:
-
Liquid chromatography/mass spectroscopy
- MS:
-
Murashige and Skoog
- PAL:
-
Phenylalanin ammonia-lyase
- TDZ:
-
Thidiazuron
- YEP:
-
Yeast extract broth
References
Andreotti C, Costa G, Treutter D (2006) Composition of phenolic in pear leaves as affected by genetics, ontogenesis and the environment. Sci Hortic 109:130–137
Bazzi C, Messina C, Tortoreto L, Stefani E, Bini F, Brunelli A, Andreotti C, Sabatini E, Spinelli F, Costa G, Hauptmann S, Stammler G, Doerr S, Marr J, Rademacher W (2003) Control of pathogen incidence in pome fruits and other horticultural crop plants with prohexadione-Ca. Eur J Hortic Sci 68:108–114
Bovy A, de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, Santos-Buelga C, van Tunen A (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14:2509–2526
Bradley JM, Davies KM, Deroles SC, Bloor SJ, Lewis DH (1998) The maize Lc regulatory gene up-regulates the flavonoid biosynthetic pathway of Petunia. Plant J 13:381–392
Byrne PF, McMullen MD, Snook ME, Musket TA, Theuri JM, Widstrom NW, Wiseman BR, Coe EH Jr (1996) Quantitative trait loci and metabolic pathways: genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc Natl Acad Sci USA 93:8820–8825
Challice JS, Williams AH (1968) Phenolic compounds of the genus Pyrus—II. A chemotaxonomic survey. Phytochemistry 7:1781–1801
Commenges D, Scotet V, Renaud S, Jacqmin-Gadda H, Barberger-Gateau P, Dartigues JF (2000) Intake of flavonoids and risk of dementia. Eur J Epidemiol 16:357–363
Cushman M, Nagarathnam D, Burg DL, Geahlen RL (1991) Synthesis and protein-tyrosine kinase inhibitory activities of flavonoid analogues. J Med Chem 34:798–806
Degenhardt J, Poppe A, Montag J, Szankowski I (2006) The use of the phosphomannose-isomerase/mannose selection system to recover transgenic apple plants. Plant Cell Rep 25:1149–1156
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2006) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Ford CM, Boss PK, Høj PB (1998) Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 Locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem 273:9224–9233
Geahlen RL, Koonchanok NM, McLaughlin JL, Pratt DE (1989) Inhibition of protein-tyrosine kinase activity by flavonoids and related compounds. J Nat Prod 52:982–986
Goldsbrough AP, Tong Y, Yoder JI (1996) Lc as a non-destructive visual reporter and transposition excision marker gene for tomato. Plant J 9:927–933
Grotewold E (2005) Plant metabolic diversity: a regulatory perspective. Trends Plant Sci 10:57–62
Halbwirth H, Fischer TC, Römmelt S, Spinelli F, Schlangen K, Peterek S, Sabatini E, Messina C, Speakman JB, Andreotti C, Rademacher W, Bazzi C, Costa G, Treutter D, Forkmann G, Stich K (2003) Induction of antimicrobial 3-deoxyflavonoids in pome fruit trees controls fire blight. Z Naturforsch 58c:765–770
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20:735–747
Hertog MGL, Feskens EJ, Kromhout D (1997) Antioxidant flavonols and coronary heart disease risk. Lancet 349:699
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium vectors for plant transformation. Transgenic Res 2:208–218
Jambunathan R, Kherdekar MS, Bandyopadhay R (1990) Flavan 4-ols concentration in mold susceptible and mold resistant Sorghum at different stages of grain development. J Agric Food Chem 38:545
Katajamaa M, Miettinen J, Oresic M (2006) MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 22:634–636
Knekt P, Isotupa S, Rissanen H, Heliovaara M, Jarvinen R, Hakkinen S, Aromaa A, Reunanen A (2000) Quercetin intake and the incidence of cerebrovascular disease. Eur J Clin Nutr 54:415–417
Knekt P, Järvinen R, Seppänen R, Heliövaara M, Teppo L, Pukkala E, Aromaa A (1997) Dietary flavonoids and the risk of lung cancer and other malignant neoplasms. Am J Epidemiol 146:223–230
Lattanzio V, Cardinali A, Palmieri S (1994) The role of phenolics in the postharvest physiology of fruits and vegetables: browning reactions and fungal diseases. Ital J Food Sci 1:3–22
Lattanzio V (2003) Bioactive polyphenols: their role in quality and storability of fruit and vegetables. J Appl Bot 77:128–146
Leser C, Treutter D (2005) Effects of nitrogen supply on growth, contents of phenolic compounds and pathogen (scab) resistance of apple trees. Physiol Plant 123:49–56
Li SJ, Deng XM, Mao HZ, Hong Y (2005) Enhanced anthocyanin synthesis in foliage plant Caladium bicolor. Plant Cell Rep 23:716–720
Lloyd AM, Walbot V, Davis RW (1992) Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258:1773–1775
Ludwig SR, Habera LF, Dellaporta SL, Wessler SR (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA 86:7092–7096
Mayr U, Treutter D, Santos-Buelga C, Bauer H, Feucht W (1995) Developmental changes in the phenol concentrations of ‘Golden delicious’ apple fruits and leaves. Phytochemistry 38:1151–1155
Mayr U, Michalek S, Treutter D, Feucht W (1997) Phenolic compounds of apple and their relationship to scab resistance. J Phytopathol 145:69–75
Middleton E Jr, Kandaswami C, Theoharides TC (2000) The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol Rev 52:673–751
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15:473–497
Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5:1497–1512
Paolocci F, Robbins MP, Madeo L, Arcioni S, Martens S, Damiani F (2007) Ectopic Expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis. Structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus. Plant Physiol 143:504–516
Ray H, Yu M, Auser P, Blahut-Beatty L, McKersie B, Bowley S, Westcott N, Coulman B, Lloyd A, Gruber MY (2003) Expression of anthocyanins and proanthocyanidins after transformation of alfalfa with maize Lc. Plant Physiol 132:1448–1463
Römmelt S, Zimmermann N, Rademacher W, Treutter D (2003a) Formation of novel flavonoids in apple (Malus domestica) treated with the 2-oxoglutarate-dependent dioxygenase inhibitor prohexadione-Ca. Phytochemistry 64:709–716
Römmelt S, Fischer TC, Halbwirth H, Peterek S, Schlangen K, Speakman JB, Treutter D, Forkmann G, Stich K (2003b) Effect of dioxygenase inhibitors on the resistance-related flavonoid metabolism of apple and pears: chemical, biochemical and molecular biological aspects. Eur J Hortic Sci 68:129–136
Römmelt S, Treutter D, Speakman JB, Rademacher W (1999) Effects of prohexadione-Ca on the flavonoid metabolism of apple with respect to plant resistance against fire blight. Acta Hortic 489:359–363
Rühmann S, Treutter D, Fritsche S, Briviba K, Szankowski I (2006) Piceid (resveratrol glucoside) synthesis in stilbene synthase transgenic apple fruit. J Agric Food Chem 54:4633–4640
Szankowski I, Briviba K, Fleschhut J, Schönherr J, Jacobsen HJ, Kiesecker H (2003) Transformation of apple (Malus domestica Borkh.) with the stilbene synthase gene from grapevine (Vitis vinifera L.) and a PGIP gene from kiwi (Actinidia deliciosa). Plant Cell Rep 22:141–149
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. doi:10.1104/pp.106.088104
Taylor LP, Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8:317–323
Treutter D (1989) Chemical reaction detection of catechins and proanthocyanidins with 4-dimethylaminocinnamaldehyde. J Chromatogr A 467:185–193
Treutter D, Santos-Buelga C, Gutmann M, Kolodziej H (1994) Identification of flavan-3-ols and procyanidins by highperformance liquid chromatography and chemical reaction detection. J Chromatogr A 667:290–297
Treutter D (2001) Biosynthesis of phenolic compounds and its regulation in apple. Plant Growth Regul 34:71–89
Treutter D (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol 7:581–591
Treutter D, Feucht W (1990) The pattern of flavan-3-ols in relation to scab resistance. J Hortic Sci 65:511–517
Tsao R, Yang R, Xie S, Sockovie E, Khanizadeh S (2005) Which polyphenolic compounds contribute to the total antioxidant activities of apple? J Agric Food Chem 53:4989–4995
Vandesompele J, De PK, Pattyn F, Poppe B, Van RN, De PA, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:1–12
Wellmann F, Griesser M, Schwab W, Martens S, Eisenreich W, Matern U, Lukačin R (2006) Anthocyanidin synthase from Gerbera hybrida catalyzes the conversion of (+)-catechin to cyanidin and a novel procyanidin. FEBS Letters 580:1642–1648
Williams RJ, Spencer JPE, Rice-Evans C (2004) Flavonoids: antioxidants or signalling molecules? Free Radical Biol Med 36:838–849
Acknowledgements
We thank Ryan Peeler and Sue Wessler (Plant Biology Department, University of Georgia, Athens, GA, USA) for providing the Lc-gene. This work was partially supported by the Ministry of Science and Culture (MWK) of the state Lower Saxony and the Federal Ministry for Education and Research (BMBF, project number 0312638C), Germany.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, H., Flachowsky, H., Fischer, T.C. et al. Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 226, 1243–1254 (2007). https://doi.org/10.1007/s00425-007-0573-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-007-0573-4