Abstract
Arabidopsis shoots regenerate from root explants in tissue culture through a two-step process requiring preincubation on an auxin-rich callus induction medium (CIM) followed by incubation on a cytokinin-rich shoot induction medium (SIM). During CIM preincubation, root explants acquire competence to respond to shoot induction signals. During CIM preincubation, pericycle cells in root explants undergo cell divisions and dedifferentiate, losing the expression of a pericycle cell-specific marker. These cells acquire competence to form green callus only after one day CIM preincubation and to form shoots after 2–3 days CIM preincubation. Reversible DNA synthesis inhibitors interfered with the acquisition of competence to form shoots. Genes requiring CIM preincubation for upregulation on SIM were identified by microarray analysis and included RESPONSE REGULATOR 15 (ARR15), POLYGALACTURONASE INHIBITING PROTEIN 2 (PGIP2) and WUSCHEL (WUS). These genes served as developmental markers for the acquisition of competence because the CIM preincubation requirements for ARR15 and PGIP2 upregulation correlated well with the acquisition of competence to form green callus, and the CIM preincubation requirements for WUS upregulation matched those for shoot formation. Unlike ARR15, another cytokinin inducible, A-type ARR gene, ARR5, was upregulated on SIM, but the induction did not require CIM preincubation. These findings indicate that competencies for various events associated with shoot regeneration are acquired progressively during CIM preincubation, and that a set of genes, normally upregulated on SIM, are repressed by a process that can be relieved by CIM preincubation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant organs can be regenerated from vegetative tissue in culture by treatment with the appropriate plant hormones. Species, cultivars and varieties differ in the efficiency of organ formation and whether organogenesis is direct or indirect (Hicks 1994), i.e., whether callus production is required for shoot or root induction. Shoot or root organogenesis from Convolvulus leaf explants or tobacco pith are examples of indirect organogenesis in which prior callus production is required (Thorpe 1993). In general, callusing is undesirable for the regeneration of elite material because of the genetic instability associated with the undifferentiated growth (Lee and Phillips 1988). Hence, there is interest in reducing the callusing stage and in understanding the critical events during callus formation that empower shoot formation.
In Arabidopsis, procedures for the production of shoots from root and or hypocotyl explants typically involve indirect organogenesis (Valvekens et al. 1988). Explants are preincubated on an auxin-rich callus induction medium (CIM) and then are transferred to a cytokinin-rich shoot induction medium (SIM) for shoot formation. During CIM preincubation, root explants “acquire competence” to respond to shoot induction signals and to form shoots during subsequent incubation on SIM (Christianson and Warnick 1983). What the acquisition of competence is in cellular or molecular terms is not known. It has been generally thought that preincubation on CIM promotes dedifferentiation of tissues that ultimately redifferentiate into organs (Gautheret 1966; Hicks 1980).
Shoot regeneration has many features in common with lateral root primordia (LRP) formation. Lateral roots derive from the pericycle and more specifically from cell files adjacent to the xylem poles (Casimiro et al. 2003). LRPs in Arabidopsis arise acropetally along the root from clusters of about 11 pericycle founder cells (Laskowski et al. 1995). Sites of LRP formation are marked by clusters of shorter cells formed by anticlinal divisions that are both polarized and asymmetric occurring in pericycle cell files adjacent to the xylem pole and in the two flanking cell files (Casimiro et al. 2003). Later stages of LRP development involve periclinal divisions, cell differentiation and activation of the root meristem (Malamy and Benfey 1997).
Lateral root formation can be dramatically induced by auxin (Celenza et al. 1995; Torrey 1950). The formative divisions of the LRP depend on basipetal transport of auxin (Bhalerao et al. 2002; Casimiro et al. 2001), and Himanen et al. (2002) used this information to establish a synchronous lateral root inducible system. They applied the auxin transport inhibitor, NPA (N-1-naphthylphthalamic acid) to block lateral root initiation and subsequently administered the auxin, NAA (1-naphthalene acetic acid), to reactivate the process. By observing a CycB1;1-GUS reporter, they found that auxin activated the entire pericycle which was blocked at the G1–S boundary.
Cytokinin, on the other hand, has an inhibitory effect on LRP formation (Aloni et al. 2006; Kuroha and Satoh 2007; Li et al. 2006; Werner et al. 2001, 2003). Werner et al. (2001, 2003) genetically engineered tobacco and Arabidopsis plants to overexpress cytokinin oxidase to reduce the levels of endogenous cytokinin. In doing so, they found increased lateral root formation indicating cytokinins inhibited root branching. Li et al. (2006) found that cytokinins interfere with LRP initiation by blocking the G2–M transition. They argued that cytokinins reduce the levels of cyclins required for the transition (CYCA2;1. CYCB1;1, CYCB2;1 and CYCB2;3). Of interest they found that once LRPs had been established that cytokinins no longer had an inhibitory effect.
The contrasting roles of cytokinins and auxin in LRP formation creates an interesting framework for the study of shoot regeneration in which root explants are treated sequentially with an auxin-rich CIM followed by a cytokinin-rich SIM. In this report, we characterized events that occur during CIM preincubation and their effects on the gene expression program that unfolds subsequently during shoot development on SIM. To do so, we identified genes that require CIM preincubation for expression and used these genes to mark developmental events that occur during the acquisition of competence. We found that various competencies are progressively acquired during preincubation on CIM and that some of these events are associated with cell cycle transitions.
Materials and methods
Plant material and RNA extraction
Plant material (Arabidopsis thaliana) and procedures for RNA extraction are described in Che et al. (2006).
Microarray and statistical analysis
Gene expression profiling using Affymetrix Arabidopsis (ATH1) DNA chips and statistical analysis were as described in Che et al. (2006). To identify genes upregulated during the acquisition of competence, a single contrast of time point means (4 days CIM vs. 0 time) was implemented for each gene as part of our analysis of variance. Gene-specific t-tests with 4 degrees of freedom each were used to identify genes downregulated in the CIM dropout experiment. In all cases P-values were converted to q-values using the method of Storey and Tibshirani (2003) to obtain approximate control of the false discovery rate.
Semiquantitative RT-PCR analysis was performed as described in Che et al. (2006). The primers used for these analyses were as follows:
At1g74890: ARR15RTF 5′-GTATAGAACAATGTATGATA-3′, ARR15RTR 5′-TAGACTCTAATTTGATCCTC-3′; At2g40670: ARR16RTF 5′-GCTTCTGCAGTTCATGAGATGACA-3′, ARR16RTR 5′-CTTGTTGCAAAGTGACAACAGCAG-3′; At3g48100: ARR5F 5′-CTCACGAGTCACGATCCTACTC-3′, ARR5R 5′-GGTTCTATCAGCAAAAGAAGCC-3′; At4g17980: NAMCIMF 5′-GAGTTGCCAGGTAAATCTTT-3′, NAMCIMR 5′-CTTCTTAACTACGCGACATA-3′; At1g75040: PR5rtF 5′-TTCATCACAAGCGGCATTGC-3′, PR5rtR 5′-GATCCTCCGGATGGTCTTAT-3′; AT1G52400: BGL1rtF 5′-AGCATTTCAGGTTGAAGGAG-3′, BGL1rtR 5′-AGTAAGTTGTGACTGACTTG-3′; At5g57260: BGL2F 5′-AACCACACAGCTGGACAAAT-3′, BGL2R 5′-GTATCAGTGGTGGTGTCAGT-3′; At3G62950: GLUTLrtF 5′-ATGGAGAGAATAAGAGATTT-3′, GLUTLrtR 5′-ATATTTATCATCTATGCGCT-3′; At3G62960: GLUTF 5′-ATGGACAAGGTTATGAGAAT-3′, GLUTR 5′-CTAGTTATGAAATGACTGAT-3′; At5G24580: COPPERF 5′-CAAAATGAGAGGGGTCCAAACA-3′, COPPERR 5′-ATAGCCGTCATTTCATTGTCTC-3′; At3G54490: rbprtF 5′-AACTTTCCCGATTGAAGAT-3′, rbprtR 5′-CAACGAATTTGCCTTTCTTC-3′; At1G72110: 72110rtF TAGCATACTGGTGACTGGTCA-3′, 72110rtR 5′-AATGCCTCGGGATCACATGT-3′; At5g06870: PGIP1F 5′-AAGCTTACAGGTCCGATACC-3′, PGIP1R 5′-CCACACAGTCTGTTATAGCT-3′; At2g17950: WUSrtR 5′-GCTAGTTCAGACGTAGCTC-3′, WUSrtF2 5′-TGGATCTATGGAACAAGACT-3′; At5g53950: CUC2F 5′-AAGCTCCAAGGATGAATGGGT-3′, CUC2R 5′-GACGGCTGAATGAGTTAACGT-3′; At3g62250: UBQ5F 5′-CTTGAAGACGGCCGTACCCTC-3′, UBQ5R 5′-CGCTGAACCTTTCAAGATCCATCG-3′.
Protein extraction
Arabidopsis seedlings (0.1 g) were frozen in liquid N2 and homogenized with an Eppendorf pestle in 1.5 ml microcentrifuge tubes. The resulting powder was extracted at 4°C with 0.3 ml of extraction buffer (0.1 M Hepes–KOH pH 7.0, 20 mM 2-mercaptoethanol, 0.1 mg/ml phenylmethanesulphonylfluoride (PMSF), 0.1% (v/v) Triton X-100, 1 mM EDTA, and 20% (v/v) glycerol). The mixture was immediately centrifuged at 12,000g for 15 min at 4°C. The supernatant was recovered and frozen in liquid N2 and the pellet was discarded. Protein concentrations were determined by the Bradford method (BioRad, Hercules, CA, USA) using bovine serum albumin as a standard (Bradford 1976).
GUS constructs
An ARR15 promoter:GUS construct was generated as follows: 1.2 kb ARR15 promoter was amplified using a genomic DNA template and following primers:
-
ARR15pF 5′-cccaagcttgggCGATTGGTTTGTTTTATTTTGTTTTGAAAA-3′;
-
ARR15pR 5′-gctctagagcTGTTTTCTCTCGGGAAAGTAAACAACAAAAC-3′.
A transcriptional fusion linking the ARR15 promoter to GUS was created by inserting the promoter into HindIII and XbaI sites of pCAMBIA3300 GUS vector. GUS expression was localized in whole mounts or in histological sections as described in Che et al. (2006).
GUS activity was determined in extracts with a fluorometric assay essentially as described by Jefferson (1987). Plant samples were collected in triplicate and protein extracts prepared as described above. Ten microliter of protein extract were mixed with 50 μl GUS assay buffer [2 mM 4-methylumbelliferyl β-glucuronide (MUG) in extraction buffer] and incubated in 37°C. Aliquots of 10 μl were removed at timed intervals (generally 5, 15, 25, 35 and 45 min), and reactions were terminated by adding 90 μl of 0.2 M Na2CO3. The fluorescent product was quantified using a multi-detection microplate reader (BIO-TEK, Model Synergy HT). Excitation and emission wavelengths were 365 and 455 nm, respectively. Tissues from regenerated non-transformed plants were used to quantify background GUS activity.
GFP constructs
The enhancer-trap line J0121 expressing GFP in the pericycle and cell-wall GFP marker line CS84726 were obtained from the Arabidopsis Biological Resource Center (ABRC). For GFP visualization Arabidopsis root explants were stained for 2–10 min in μg/ml propidium iodide (PI) (Sigma, St Louis, MO, USA) rinsed and mounted in 0.7% low melting agarose with a #1 coverslip. Imaging was performed using Leica TCS/NT (Exton, PA) upright confocal system with 40× oil lens. GFP and PI fluorescence was excited with argon (488 nm) and krypton (568) lasers, respectively, along with DD 488/566 filter. Emission was filtered by using either 475–575 nm (for green) or 590+ nm (for red) band-pass filter. The confocal Z-stacks were aligned by using either Adobe Photoshop or MetaMorph imaging system.
Results
Proliferation and dedifferentiation of pericycle cells
A standard protocol for regenerating shoots from root explants of Arabidopsis seedlings involves preincubation on CIM (Valvekens et al. 1988) during which time explants acquire competence to form shoots on SIM. During the induction of LRPs, treatment of Arabidopsis roots with auxin stimulates anticlinal cell divisions in the pericycle layer of the root producing shorter cells (Himanen et al. 2002). Likewise, during CIM preincubation, pericycle cells in root explants undergo anticlinal divisions giving rise to shorter cells followed by periclinal cell divisions producing additional cell layers (Fig. 1a, b). At various points along the roots, more extensive cell divisions lead to the formation of cell masses or callus foci. The process appears much like lateral root formation, however, the cell masses at 4 days CIM are less well formed and spaced.
Cell proliferation and dedifferentiation during CIM preincubation. Confocal microscopy of root explants preincubating on CIM. a CS84726 is a cell wall marker that highlights root cell outlines with GFP. At 0 days CIM, the pericycle (arrow) is only one cell layer thick, and in this region of the root, the cells are very long (bracket). Root explants are counter-stained with propidium iodide. b At 4 days CIM, pericycle cells have undergone some anticlinal and periclinal cell divisions, and at sites where cell masses or foci of callus form (arrow), cells are shorter (brackets) and may be more than one cell-layer thick. c J0121 is a pericycle cell marker that marks xylem-pole pericycle cells with GFP (Laplaze et al. 2005). At 0 days CIM, the GFP-marked pericycle cells constitute a single-layered cell file adjacent to the xylem pole (arrow). d At 4 days CIM, GFP fluorescence is extinguished in cell masses (arrow). Bar 20 μM
During CIM preincubation, a fundamental change that occurs in the pericycle is the loss of pericycle-specific markers. J0121 is an enhancer-trap line that uses GAL4-mediated transactivation to express GFP specifically in the xylem-pole adjacent cell files of the pericycle (Laplaze et al. 2005) (Fig. 1c). We found that the GFP marker in J0121 was expressed in the pericycle, but was extinguished during CIM preincubation in the cell masses presumably derived from this cell layer. As a result, the cell masses were unmarked by the pericycle-specific marker (Fig. 1d). Note that pericycle-specific marker expression persisted outside of the cell masses even though these cells had undergone anticlinal, and at least one periclinal division. Thus, our findings indicate that extinguishment of this marker is not a prelude to early divisions of the pericycle, but occurs with more extensive cell proliferation in cell masses, some of which give rise to organs.
It appears that the loss of the pericycle-specific marker in cell masses is a process of pericycle cell dedifferentiation, rather than the proliferation of non-pericycle derived cells. Founder cells for LRP formation have been shown by others to be located in the pericycle by tracing the continuity of a cell lineage marker generated by heat shock excision of a transposon that activates the expression of a YFP marker gene (Kurup et al. 2005). Pericycle cells adjacent to the xylem poles are required for lateral root formation as found by Laplaze et al. (2005) who ablated these cell files by transactivating a diphtheria toxin chain A (DTA) effector transgene in the J0121 enhancer trap line. DTA is a ADP-ribosylase that targets eukaryotic elongation factor 2, and inhibits translation in a cell autonomous manner (Czako et al. 1992). We obtained a comparable, transactivatable DTA effector gene construct (pSDM7021) from R. Offringa (Weijers et al. 2003), introduced it into Arabidopsis and then transferred the construct into J0121 by crossing. We found that the DTA transgene did, indeed, ablate xylem-pole adjacent pericycle cell files as indicated by loss of GFP marker expression (Fig. 2a, b). F1 progeny seedlings expressing the construct produced roots that outwardly appeared normal, but were shorter and did not form lateral roots (Fig. 2c). Roots from the affected seedlings were explanted and subjected to our standard tissue culture regime for regenerating shoots. We found that pericycle ablation eliminated shoot regeneration on SIM (Fig. 2d) and callus formation on CIM (Fig. 2e). The latter was best demonstrated by continuing preincubation of root explants on CIM for 18 days at which time there was ample callus formation on wild type roots, but almost none on roots from seedlings expressing the pericycle-specific DTA transgene (Fig. 2e). We concluded that the xylem pole-adjacent cells of the pericycle are required, and are the likely source of cells for callus formation on CIM and for shoot formation on SIM as suggested by Atta et al. (2004).
Effect of ablation of xylem-pole pericycle cells by the action of a pericycle-specific diphtheria toxin chain A (DTA) transgene. a Light microscope and b epifluorescence images of roots from F1 progeny from a cross of DTA-1 × J0121 (upper) and wild-type (lower) seedlings. c F1 progeny expressing the DTA transgene (two seedlings on left) and wild type (two seedlings on right). Seedlings are 14-day-old. d Shoot regeneration in root explants from DTA-1 × J0121 F1 (left) and wild-type (right) seedlings after 4 days preincubation on CIM and 14 days incubation on SIM. e Callus formation in root explants from DTA-1 × J0121 F1 (left) and wild-type (right) seedlings after 18 days incubation on CIM. Bars in C, D and E = 2 mm
Time course for competence acquisition during CIM preincubation
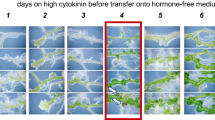
To define events during CIM preincubation as root explants acquire competence for shoot formation, samples were preincubated for various times in a culture scheme in which all samples were preincubated for the same total period of time either on CIM or on basal B5 medium (Fig. 3a). Without CIM preincubation, explants failed to form shoots efficiently on SIM (0 days, Fig. 3b, d). However, after only two days of CIM preincubation, explants rapidly acquired competence to form shoots, and competence continued to increase for the next two days (Fig. 3b). To demonstrate that CIM preincubation did not simply represent physical carryover of hormones from the CIM medium, explants were transferred from CIM to SIM after only 10 min CIM preincubation. Short exposure to CIM did not have a significant effect on green callus or shoot formation (10 min, Fig. 3b, c). We found that with only one day CIM preincubation, root explants formed green callus (after subsequent SIM incubation) as abundantly as they did when forming shoots (Fig. 3c, d). Therefore, we concluded that root explants first acquire competence to produce green callus, and then they acquire competence to form shoots.
Effect of CIM preincubation period length on green callus and shoot formation. a CIM preincubation period was varied between 0 and 4 days before transferring explants onto SIM for 10 days. (Explants were cultured on B5 medium without hormones when they were not preincubated on CIM, such that explants were all cultured for the same period of time.) b Average number of shoots formed in three replicate cultures after 10 days incubation on SIM depended on the time of CIM preincubation. Error bars represent ± SE. c Tissue culture dish showing explants after 10 days on SIM that had been preincubated previously on CIM for various times as indicated. d Close-up of root explants after 14 days incubation on SIM. Number of days of CIM preincubation is indicated. Bar 1 mm. e Effect of reversible cell cycle inhibitors on shoot formation. Explants were preincubated on CIM for times indicated in the presence or absence of 5 μg/ml aphidicolin (APH) or 100 mM hydroxyurea (HU). Number of shoots per explant scored in three replicate samples with no less that 20 root segments per sample. Error bars are ± SE
Effects of cell cycle inhibitors and on shoot regeneration
To determine whether cell cycling is required during CIM preincubation for the acquisition of competence to form shoots, root explants were treated with reversible cell cycle inhibitors. Root explants were preincubated on CIM containing either 5 μg/ml aphidicolin (APH) or 100 mM hydroxyurea (HU) to inhibit DNA synthesis. HU inhibits ribonucleotide diphosphate reductase, APH is a DNA polymerase α and δ inhibitor and both block the cell cycle in the G1–S transition (Planchais et al. 2000). Explants were transferred to SIM without inhibitors and scored for shoot formation 10–14 days later. To avoid long-term cytotoxic effects, explants were preincubated no longer than two days in the presence of the inhibitor. However, as a control, explants were incubated for more than a week on these inhibitors to show that the concentrations were effective in blocking cell divisions and callus growth (data not shown).
Treatment with 5 μg/ml APH for two days during preincubation on CIM significantly reduced shoot formation during subsequent incubation on SIM (Fig. 3e). Similar treatment with APH for one day had less effect because fewer shoots were formed in the untreated control with only one day CIM preincubation. However, treatment with 100 mM HU was harsher because 1 day HU treatment during CIM preincubation blocked all subsequent shoot formation (Fig. 3e). A two-way ANOVA was conducted to demonstrate the significance of the differences in time (P = 0.00635) and inhibitor treatments (P = 0.02410) and of the interaction between time and treatment (P = 0.01266). We conclude that the G1–S transition in pericycle cells during CIM preincubation is required for explants to acquire full competence to efficiently form shoots on SIM. Beeckman et al. (2001) demonstrated that pericycle cells leaving the root meristem are G1 arrested, and cells in presumptive LRP sites progress into G2. Thus, most of the pericycle cells from which regenerating shoots are derived are G1-arrested cells, and apparently, auxin treatment during CIM preincubation is a necessary step to engage that cell population.
Impact of CIM dropout on gene expression
In other studies, Che et al. (2002, 2006) described gene expression changes during shoot development; therefore, we investigated how the gene expression program was affected by omitting CIM preincubation. For this “CIM dropout” experiment, we preincubated explants as above on basal B5 medium without hormones for the same period of time as normal CIM preincubation. The dropout explants and normal CIM preincubated controls were then transferred to SIM, further incubated for 6 days at which time samples were taken for RNA extraction. The experiment was analyzed as a randomized block design with three biological replicates per treatment. The impact of the CIM dropout was profound in the sense that 57 genes were downregulated 20-fold or more when controlling the false discovery rate (FDR) at the 0.02 level (Storey and Tibshirani 2003) (Table 1). The five top genes include an AAA-type ATPase family protein (At3g28510), NAM (no apical meristem)-like transcription factor (At4g17980), IAA-amido synthase (At2g23170), a subtilisin-like serine protease (At1g01900) and cytokinin oxidase-like protein (At3g63440). Of possible consequence to shoot formation was the fact that three NO APICAL MERISTEM-like transcription factor genes (At4g17980, At5g46590 and At2g18060) and WUSCHEL (WUS, At2g17950) homeodomain factor gene were all more than 20-fold downregulated in the absence of CIM preincubation. WUS is required for specifying the stem cell niche in the shoot meristem (Baurle and Laux 2005; Laux et al. 1996; Mayer et al. 1998). Other genes of note are those normally upregulated on SIM (indicated by X’s, Table 1). Topping this list are genes encoding pathogenesis-related protein 5 (PR5, At1g75040), β-glucosidase (At1g52400), β-glucanase (At3g57260), glutaredoxin (At3g62950), copper-binding family protein (At5g24580), RNA polymerase 24KD subunit (At3g54490), an unknown protein (At1g72110), polygalactronase inhibiting protein (At5g06870), WUS (At2g17950) and ARABIDOPSIS RESPONSE REGULATOR 15 (ARR15, At1g74890). Thus, this analysis yielded a wealth of genes, which can be used as developmental markers to study the role of CIM preincubation.
Developmental markers for the acquisition of competence
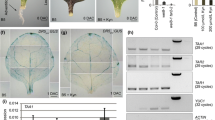
Of the genes most highly downregulated in the CIM dropout experiment, ARR15, encoding an A-type response regulator, caught our attention because the regulation of this gene had been well studied by others (Kiba et al. 2002, 2003). ARR15, like other A-type response regulator genes, such as ARR5, is rapidly induced in seedlings by cytokinin (Brenner et al. 2005). In our system, both A-type ARRs were upregulated during incubation on SIM, a cytokinin-rich medium (Fig. 4a). ARR5 had a fairly high basal level of expression, however, as reported by Che et al. (2002; 2006), it was upregulated 3–4 fold on SIM. ARR15 was upregulated more than 10-fold (Fig. 4a), but the upregulation was fairly gradual, taking days, not hours to reach peak levels (Fig. 4b). Although both A-type response regulator genes were upregulated during incubation on SIM, only ARR15 was dependent on CIM preincubation (Table 1). To determine whether the dependency of ARR15 on CIM preincubation is a transcriptional phenomenon, we developed an ARR15 promoter:GUS reporter construct and introduced it into transgenic plants. We observed that the promoter construct was upregulated on SIM and required CIM preincubation for upregulation (Fig. 4c). Therefore, we concluded that the major effect of CIM preincubation on subsequent ARR15 expression is, at least, partly transcriptional.
Expression profiles of ARR5 and ARR15 during normal shoot regeneration. a Root explants were preincubated on CIM for four days and then transferred to fresh CIM, SIM or RIM (root induction medium). RNA was extracted at various times and analyzed by semiquantitative RT-PCR analysis. b Time course for the expression of ARR5 and ARR15 during incubation on SIM as analyzed by semiquantitative RT-PCR analysis. UBIQUITIN5 (UBQ5) was used as a control. c GUS expression in roots from transgenic seedlings bearing ARR15promoter:GUS constructs. Top group of explants were preincubated for four days on B5 medium without hormones, while group below were preincubated for four days on CIM. Both were transferred to SIM and subsequently incubated on SIM for 6 days before histochemical staining
We examined the time course by which explants acquire competence to upregulate ARR15 expression following transfer to SIM (Fig. 5a). As with shoot formation, explants were preincubated on CIM for various periods of time, then transferred to SIM. Explants were analyzed for MUG expression six days later on SIM (Fig. 5b). Also, as before, all experimentals and controls were preincubated for the same amount of time (4 days), and to do so, explants were preincubated on basal B5 medium when not on CIM. Root explants from the transgenic plants gained full competence to express the ARR15 promoter:GUS reporter construct within one day of CIM preincubation (Fig. 5b). The various cell cycle inhibitors, including HU that blocked shoot formation when administered during CIM preincubation, were ineffectual in blocking ARR15 promoter expression (Fig. 5b).
Effect of CIM preincubation period length on ARR15 promoter activity. a CIM preincubation period was varied between 0 and 4 days before transferring explants onto SIM for 6 days. b Mean GUS activity as determined in MUG assays from three replicate samples of explants from ARR15promoter:GUS seedlings after 6 days incubation on SIM. Explants were also preincubated on CIM in the presence or absence of 5 μg/ml aphidicolin, APH, or 100 mM hydroxyurea, HU (shaded bars). Error bars are ± SE
CIM preincubation was also needed to accumulate ARR15 RNA transcripts as assayed by semiquantitative RT-PCR (Fig. 6). ARR15 required one day CIM preincubation for high-level transcript accumulation on SIM. Thus, the CIM preincubation time required for ARR15 expression closely corresponds to the acquisition of competence to form green callus. As indicated above, however, not all SIM upregulated A-type ARRs were dependent on CIM preincubation. ARR5, which is also upregulated during transfer to SIM, did not require CIM preincubation (Fig. 6). Several other genes that were highly dependent on CIM preincubation and upregulated during SIM incubation (At1g75040, At1g52400, At3g57260, At3g62950, At3g62960, At5g24580, At3g54490, At1g72110, At5g06870) required only one day CIM preincubation (Fig. 6). Thus, many SIM upregulated genes that are highly dependent on CIM preincubation required only one day of CIM preincubation, similar to the time required for green callus formation. One important exception was WUSCHEL (WUS, At2g17950) that required longer CIM preincubation. WUS expression during SIM incubation rose with 3–4 days CIM preincubation (Fig. 6). Therefore, WUS expression comes the closest of the genes on the CIM dropout list (Table 1) in representing the CIM preincubation time required for shoot development.
CIM preincubation time required for upregulation of developmental marker genes on SIM. CIM preincubation period was varied between 0 and 4 days before transferring explants onto SIM for 6 days. Semiquantitative RT-PCR analysis of the expression of genes requiring CIM preincubation from the CIM dropout experiment described in Table 1. FC is fold change
We searched further down the list of genes dependent on CIM preincubation for other genes associated with shoot apical meristem formation and noted CUP SHAPED COTYLEDON2 (CUC2, At5g53950). CUC2, an important shoot meristem identity gene, which acts with CUC1 in activating the expression of SHOOT MERISTEMLESS (STM) (Daimon et al. 2003) and in establishing boundaries in organ formation (Breuil-Broyer et al. 2004). CUC2 is highly upregulated during shoot development and incubation SIM (Che et al. 2002, 2006) but like most other SIM upregulated genes, CUC2 only requires a single day of CIM preincubation (Fig. 6). Thus, not all meristem identity genes have the same CIM preincubation requirements as does shoot development.
We examined preincubation time requirements for other genes that were highly CIM dependent, but not normally upregulated on SIM. A NO APICAL MERISTEM-like (NAM) gene (At4g17980) and a subtilisin-like serine protease (subtilase) gene (At1g01900) were not appreciably upregulated during SIM (Che et al. 2002, 2006), however, these genes were highly dependent on CIM preincubation (Fig. 6). But like ARR15 these genes required only one day CIM preincubation to acquire competence for high level SIM expression.
Discussion
Competence to form shoots on SIM is acquired progressively during CIM preincubation (Fig. 7). We described different events that were used to monitor progress during the period of competence acquisition, and these included shoot and green callus formation and the expression of various marker genes. Competence to express ARR5, an A-type response regulator that is upregulated during shoot development and functions as a feedback inhibitor of cytokinin responses (To et al. 2004) did not require CIM preincubation. Competence to form green callus and to express another marker gene ARR15, an A-type response regulator that also functions as a feedback inhibitor (Kiba et al. 2003), was acquired within one day preincubation without the need for cell divisions. Competence to form shoots necessitated two or more days of CIM incubation and required cell cycle transit beyond the G1–S boundary. Banno et al. (2001) found that the upregulation of ENHANCER OF SHOOT REGENERATION1 (ESR1) during shoot development required CIM preincubation. (ESR1 is not included on the ATH1 gene chip and was not included in our analysis.) ESR1 may play an important role in shoot development because overexpression of ESR1 in transgenic plants increases the efficiency of shoot regeneration.
Events during the acquisition of competence for shoot regeneration. Arabidopsis shoots are regenerated from root explants through a process that involves preincubation CIM followed by incubation on SIM, which leads to green callus formation and shoot development. During CIM preincubation competence to express different developmental marker genes and undergo various developmental step during subsequent SIM incubation are acquired progressively. Cell cycle progression during CIM preincubation is required for acquisition of some competencies, but not others
One model that might account for the effect of CIM preincubation on the different developmental markers is that expression of markers is under repression and that the mechanism of repression is inactivated during CIM preincubation. ARR15 would be a prime candidate for gene under repressive control, while ARR5, which does not require CIM preincubation for SIM upregulation, would not. It is known that several A-type ARR genes are upregulated by the action of B-type ARRs (Hwang and Sheen 2001; Sakai et al. 2001), and we have shown in other studies that ARR5 is a direct transcriptional target of ARR2, a major B-type ARR (P. Che, unpublished observations). We have observed that ARR15 is also directly targeted by ARR2, however, we speculate that its potential for upregulation might be offset by the action of some repressive mechanism that can be inactivated by CIM preincubation. We have investigated whether repression relief would be prevented by treatment with the proteasome inhibitor MG132, and so far we have observed that it is not (P. Che, unpublished observations).
ARR15 upregulation could be useful as a developmental marker for green callus formation. ARR15 is expressed in the pericycle (Kiba et al. 2002) and other vascular bundle tissues (P. Che, unpublished observations). Competence to form green callus and to upregulate ARR15 expression are both acquired only after one day preincubation on CIM and both are unaffected by the action of cell cycle inhibitor treatment during CIM preincubation. The requirements for WUS expression correlate well with those for shoot formation. For upregulation on SIM, WUS requires three days preincubation on CIM—similar to the requirements for shoot formation. WUS expression may be key to shoot regeneration because expression of the gene is required for specifying the stem cell niche in the shoot meristem (Baurle and Laux 2005; Laux et al. 1996; Mayer et al. 1998). Gallois et al. (2002) demonstrated that ectopic WUS and STM expression promotes shoot development. The subsequent interplay between WUS and the A-type ARR genes in shoot regeneration is probably very interesting, since the A-type ARRs are negative regulators of shoot development and WUS is a repressor of A-type ARR genes, such as ARR5 and -15 (Leibfried et al. 2005).
Fundamental to understanding the role of CIM preincubation is the question why a two-step process is required for shoot regeneration. The answer may relate to the counteracting effects of cytokinin and auxin on LRP formation. Li et al. (2006) reported that cytokinin blocks the formative divisions in LRP formation and that increasing concentrations of auxin were not able overcome the cytokinin effects. If cytokinin blocks the formative steps in shoot primordia formation, then that would justify sequential treatment with the two hormones. Justification for the two-step process may also relate to cell cycle events. Menges et al. (2002) reported that nearly 500 genes in Arabidopsis show significant fluctuation in expression in the cell cycle, and the CIM requiring genes in our study might be differentially inducible at different cell cycle stages. It is assumed that most of the pericycle cells from which shoots arise are arrested in the G1 phase of the cell cycle (Beeckman et al. 2001). Preincubation on an auxin-rich CIM medium could be the only treatment able to overcome the arrest of pericycle cells at the G1–S boundary.
The requirement for DNA synthesis or overcoming G1–S arrest during CIM preincubation does, indeed, distinguish various developmental events. ARR15 upregulation and green callus formation do not require DNA synthesis but shoot formation does. A possible interpretation of this finding is that cells in G1 are incapable of responding to signals required for shoot formation, however, G1 cells are capable of responding to signals required for ARR15 upregulation and green callus formation.
Abbreviations
- APH:
-
Aphidicolin
- ARR:
-
Arabidopsis response regulator
- CIM:
-
Callus induction medium
- DTA:
-
Diphtheria toxin chain A
- FDR:
-
False discovery rate
- GUS:
-
β-glucuronidase
- HU:
-
Hydroxyurea
- LRP:
-
Lateral root primordium
- MUG:
-
4-Methylumbelliferyl β-glucuronide
- NAA:
-
1-Naphthalene acetic acid
- NPA:
-
N-1-naphthylphthalamic acid
- PI:
-
Propidium iodide
- RT-PCR:
-
Reverse transcriptase polymerase chain reaction
- SIM:
-
Shoot induction medium
- YFP:
-
Yellow fluorescent protein
References
Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot (Lond) 97:883–893
Atta R, Guivarc H, Laurens L, Traas J, Giraudat-Pautot V, Chriqui D (2004) Totipotency of pericycle cells in Arabidopsis thaliana root and hypocotyl explants for both root and shoot regeneration. In: 15th international conference on Arabidopsis research. Berlin
Banno H, Ikeda Y, Niu QW, Chua NH (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13:2609–2618
Baurle I, Laux T (2005) Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17:2271–2280
Beeckman T, Burssens S, Inze D (2001) The peri-cell-cycle in Arabidopsis. J Exp Bot 52:403–411
Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29:325–332
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72:248–254
Brenner WG, Romanov GA, Kollmer I, Burkle L, Schmulling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44:314–333
Breuil-Broyer S, Morel P, de Almeida-Engler J, Coustham V, Negrutiu I, Trehin C (2004) High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J 38:182–192
Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8:165–171
Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, Bennett M (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13:843–852
Celenza JL Jr, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9:2131–2142
Che P, Gingerich DJ, Lall S, Howell SH (2002) Global and cytokinin-related gene expression changes during shoot development in Arabidopsis. Plant Cell 14:2771–2785
Che P, Lall S, Nettleton D, Howell SH (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141:620–637
Christianson ML, Warnick DA (1983) Competence and determination in the process of in vitro shoot organogenesis. Dev Biol 95:288–293
Czako M, Jang JC, Herr JM Jr, Marton L (1992) Differential manifestation of seed mortality induced by seed-specific expression of the gene for diphtheria toxin A chain in Arabidopsis and tobacco. Mol Gen Genet 235:33–40
Daimon Y, Takabe K, Tasaka M (2003) The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol 44:113–21
Gallois J, Woodward C, Reddy G, Sablowski R (2002) Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129:3207–3217
Gautheret RJ (1966) Factors Affecting differentiation of plant tissue grown in vitro. In: Beerman W, Nieuwkoop PD, Wolff E (eds) Cell differentiation and morphogenesis. North-Holland, Amsterdam, pp 55–95
Hicks GS (1980) Patterns of organ development in plant tissue culture and the problem of organ determination. Bot Rev 46:1–23
Hicks GS (1994) Shoot induction and organogenesis in vitro: a developmental perspective. In Vitro Cell Dev Biol 30P:10–15
Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inze D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14:2339–2351
Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413:383–389
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Kiba T, Yamada H, Mizuno T (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43:1059–1066
Kiba T, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T (2003) The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44:868–874
Kuroha T, Satoh S (2007) Involvement of cytokinins in adventitious and lateral root formation. Plant Root 1:27–33
Kurup S, Runions J, Kohler U, Laplaze L, Hodge S, Haseloff J (2005) Marking cell lineages in living tissues. Plant J 42:444–453
Laplaze L, Parizot B, Baker A, Ricaud L, Martiniere A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J (2005) GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56:2433–2442
Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM (1995) Formation of lateral root meristems is a two-stage process. Development 121:3303–3310
Laux T, Mayer KFX, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122:87–96
Lee M, Phillips RL (1988) The chromosomal basis of somaclonal variation. Ann Rev Plant Physiol 39:413–437
Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438:1172–1175
Li X, Mo X, Shou H, Wu P (2006) Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol 47:1112–1123
Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124:33–44
Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95:805–815
Menges M, Hennig L, Gruissem W, Murray JA (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277:41987–42002
Planchais S, Glab N, Inze D, Bergounioux C (2000) Chemical inhibitors: a tool for plant cell cycle studies. FEBS Lett 476:78–83
Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294:1519–1521
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100:9440–9445
Thorpe TA (1993) In vitro organogenesis and somatic embryogenesis: physiological and biochemical aspects. In: Roubelakis-Angelakis KA, Tran Thanh Van K (eds) Morphogenesis in plants- molecular approaches. Plenum, NewYork, pp 19–38
To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16:658–671
Torrey JG (1950) The induction of lateral roots by indoleacetic acid and root decapitation. Am J Bot 37:257–264
Valvekens D, Van Montagu M, Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85:5536–5540
Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R (2003) Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol 133:1882–1892
Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15:2532–2350
Werner T, Motyka V, Strnad M, Schmulling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98:10487–104
Acknowledgments
This study was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (2003-35304-13363), by the National Science Foundation (IBN-0236060), and by the Plant Sciences Institute at Iowa State University. We acknowledge the important contributions of Rhonda DeCook for her help with the bioinformatics analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Che, P., Lall, S. & Howell, S.H. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226, 1183–1194 (2007). https://doi.org/10.1007/s00425-007-0565-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-007-0565-4