Abstract
The microstructure and the connectivity of the pore space are important variables for better understanding of the complex gas transport phenomena that occur in plant tissues. In this study, we present an experimental procedure for image acquisition and image processing to quantitatively characterize in 3D the pore space of apple tissues (Malus domestica Borkh.) for two cultivars (Jonagold and Braeburn) taken from the fleshy part of the cortex using X-ray computer microtomography. Preliminary sensitivity analyses were performed to determine the effect of the resolution and the volume size (REV, representative elementary volume analysis) on the computed porosity of apple samples. For comparison among cultivars, geometrical properties such as porosity, specific surface area, number of disconnected pore volumes and their distribution parameters were extracted and analyzed in triplicate based on the 3D skeletonization of the pore space (medial axis analysis). The results showed that microtomography provides a resolution at the micrometer level to quantitatively analyze and characterize the 3D topology of the pore space in apple tissue. The computed porosity was confirmed to be highly dependent of the resolution used, and the minimum REV of the cortical flesh of apple fruit was estimated to be 1.3 mm3. Comparisons among the two cultivars using a resolution of 8.5 μm with a minimum REV cube showed that in spite of the complexity and variability of the pore space network observed in Jonagold and Braeburn apples, the extracted parameters from the medial axis were significantly different (P-value < 0.05). Medial axis parameters showed potential to differentiate the microstructure between the two evaluated apple cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The structural geometry of the pore space plays a fundamental role in governing fluid and gas transport through any cellular tissue (Celia et al. 1995; Dražeta et al. 2004). Particularly, in plant organs the gas-filled intercellular spaces are considered the predominant pathways for gas transport through the plant and are important in relation to the characteristics of gas exchange (Raven 1996; Kader 1988; Kuroki et al. 2004; Schotsmans et al. 2004). However, establishing the relationships between the geometrical properties of the pore structure of cellular materials such as plant materials, and the transport properties of the medium has been difficult for two main reasons: the difficulty in making direct measurements on the pore space and the random nature of its structure (Khan and Vincent 1990; Thovert et al. 1993; Maire et al. 2003). In addition, the majority of elements that critically participate in gas and fruit transport properties of plants are less than 100 μm in size (Aguilera 2005; Mebatsion et al. 2006a, b), requiring suitable techniques for quantitative analyses of their microstructures.
X-ray microtomography is a relatively new technique, which has found potential applications in plant science research and food quality evaluation. The technique allows visualization and analysis of the architecture of cellular materials with an axial and lateral resolution down to a few micrometers, and without sample preparation or chemical fixation (Maire et al. 2003; van Dalen et al. 2003; Lim and Barigou 2004). X-rays are short wave radiations, which can penetrate through plant tissue. The level of transmission of these rays depends mainly on the mass density and mass absorption coefficient of the material (Maire et al. 2001; Salvo et al. 2003). Its capabilities for biological plant research and quality evaluations in fruits and vegetables have been proven by a number of studies. Thus, using high resolution computed tomography Fromm et al. (2001) detected and measured xylem water content and wood density in spruce and oak trees. Barcelon et al. (1999) detected internal quality changes in peaches, and Lammertyn et al. (2003a, b) investigated core breakdown disorders in “Conference” pears based on their mass density variations during storage. Kuroki et al. (2004) obtained 3D spatial information about gas-filled intercellular spaces in sarcocarp tissue of cucumber fruit, and also investigated the structural changes of these spaces during storage. Recently, Babin et al. (2005) studied the microstructure of cellular cereal products captured by synchrotron radiation and the relationships with their mechanical properties based on modeling and simulations. However, X-ray applications for visualization and quantitative characterization of pore space microstructure of apple tissue have been restricted.
Apple fruit is mainly composed of the fleshy tissue of parenchyma cells permeated with vascular tissue and intercellular air spaces (Esau 1977). The volume of air increases during fruit growth and occupies a considerable proportion of the fruit at harvest (Harker and Ferguson 1988; Yamaki and Ino 1992). This increase in air space is accompanied by a proportional decline in fruit density while the density of the fruit cells themselves remains roughly constant (Westwood et al. 1967; Baoping 1999). In addition, the microstructure and fraction of intercellular air differs between cultivars (Baumann and Henze 1983; Vincent 1989). Larger fruits of the same cultivar have a higher proportion of air than smaller ones (Volz et al. 2004). In general, the knowledge of these intercellular air spaces based on microstructural properties and realistic percolation models has important applications since they are related to the understanding of postharvest quality and shelf life of the fruit. Fruit with greater fractional air volumes have been shown to be softer (Yearsley et al. 1997a, b; Volz et al. 2004), or more mealy (Harker and Hallet 1992; Tu et al. 1996) and to have greater internal gas diffusion rates (Rajapakse et al. 1990; Ho et al. 2006). Furthermore, the volume of these intercellular air spaces continues to increase during storage, and therefore, its measurement can be used to define the age of the fruit and also to characterize the effect of different storage conditions on its quality (Khan and Vincent 1990; Tu et al. 1996, Harker et al. 1999). All these features have been related to the known susceptibility of Braeburn apples for internal tissue browning and breakdown disorders (Cheng et al. 1998).
The objectives of this study were: (1) to implement an experimental procedure to obtain high quality radiotomographic images of porous media at micrometer resolution for 3D visualization and quantitative characterization of the topology of apple tissue using sophisticated image analysis techniques; (2) to evaluate the sensitivity of this technique on the computed porosity of apple tissue with respect to the image resolution and the volume size of the sample, and for determining a representative elementary volume (REV) over which statistical average parameters of its microstructure can be extracted; and finally (3) to compare the geometrical features of the pore structure of tissue from two apple cultivars (Malus domestica Borkh.): cv. Jonagold and cv. Braeburn.
Materials and methods
Apple samples
Apples (Malus domestica Borkh.) cultivars Jonagold (six apples, ∼7.5 cm diameter) and Braeburn (six apples, ∼5.6 cm diameter) were selected from a commercial orchard in Belgium and stored under controlled atmospheric conditions (0.8°C, 2.5% CO2, and 1% O2). For analysis, samples were taken in the fleshy part of the apple (cortical tissue) located at ∼10 and ∼7.5 mm from the skin to the axis of the tissue cylinder for Jonagold and Braeburn apples, respectively (as shown for a Jonagold apple in Fig. 1a). Each sample was cut into cylinders of 5 mm diameter and enclosed in a plastic tube to avoid dehydration without any special preparation. Only images of this region of the apple were chosen for comparison since the purpose of this paper was to assess the applicability of the implemented technique for the microstructural characterization of the fruit.
Schematic view of the experimental procedure for sample extraction and pre-processing of images using Jonagold apples. a Sampling of fleshy region of the apple at ∼10 mm from the skin to the axis of the tissue cylinder (showing the five extracted regions from each apple for the image resolution analysis). b Scanned region (the length and diameter of the scanned samples were ∼3 cm and 5 mm , respectively, and the analyzed region was 5.1 mm). c Reconstructed cross-sectional image with resolution of 8.5 m (black regions represent pores and gray regions represent cellular material) and showing the extracted region of interest (ROI, dashed line). d Stack of radiograph images (2563 pixels3 equivalent to 10.4 mm3) for 3D reconstruction of the sample using sliceomatic.m program from MATLAB 7.0
X-ray computed microtomography
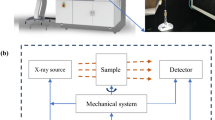
The samples were scanned using a SkyScan 1072 high resolution desk-top X-ray micro-CT system (SkyScan, Aartselaar, Belgium; http://www.skyscan.be), operating at a voltage of 63 kV, current of 156 μA, an exposure time of 8.4 s, and scanned over the interval 0°–180° using a 0.9° scan step. Each pixel represented a linear resolution of 8.5 μm. To account for the non-uniformities in the X-ray beam and non-uniform response of the CCD detector, the raw images were corrected for dark and white fields by taking the average of flat field correction references collected at the beginning of the experiment. These experimental conditions were optimized to allow high quality radiographic slices (free of ring artifacts and beam hardening) and based on the compromise of enhancing both the contrast and resolution of the images using the shortest scanning time (∼1 h 05 min) of the whole volume of the tissue cylinder. The X-ray shadow projections of the 3D object digitalized as 1,024 × 1,024 pixels size with 4,096 brightness gradations (12 bit) were processed to obtain reconstructed cross-section images of linear attenuation coefficient values with 256 gradations (8 bit) using a mathematical algorithm based on the back-projection procedure implemented in CONE_REC ver. 1.4.3 software (http://www.skyscan.be/next/downloads.htm). Thus, a complete stack of 2D cross-sections (made with 200 projections) of the object with an 8.5 μm interslice distance was obtained and processed for reconstruction and analysis of the full 3D pore structure of apple tissue. A schematic representation of this procedure is shown in Fig. 1.
Image processing
From the middle part of the scanned region (Fig. 1b), a region of interest (ROI) cube of each apple sample was extracted and subjected to image analysis. Images were pre-processed using the Image Processing Toolbox of MATLAB 7.0 (The MathWorks, Inc., Natick, MA, USA) to render the apple tissue microstructure as 2D stacks and 3D isosurface images, as shown in Fig. 1c and d, respectively. The 3D image consisted of a finite number of points or small voxels (i.e. 2563 volume elements, Fig. 1d) with a length voxel side of 8.5 μm. Quantitative analysis was carried out by using the code 3DMA-Rock Primer ver. 08/05 (Lindquist 1999) which was compiled under the Linux operating system. The procedure mainly consisted of the two following steps and algorithms:
Image segmentation
Segmentation is the process of converting the grey-scale image into a black and white image by determining the population assignment (void space or solid material) for each voxel in the image. For this, a common practice is to choice a simple global threshold, which often is set to match a predetermined bulk measurement of porosity. However, this procedure is subjective and may lead to biases when one is trying to segment a stack of X-ray images. The distinction between the void and solid phases in radiographic images is frequently not sharp (i.e., do not show a bimodal distribution) due to the amount of peak overlap in the attenuation coefficient histogram and the nature of X-ray tomography. Moreover, the resulting binary images of apple tissue using global thresholds are noisy and the average porosity is highly dependent on the selected threshold value. The attenuation coefficient describes the intensity fraction of a beam of X-rays that is transmitted per unit thickness of the material, which is calculated based on Beer’s law (Maire et al. 2001). Figure 2 plots the distribution of the transmitted X-ray intensities (solid line, right axis) measured for the reconstructed tomography images of apple tissue (corresponding to the 3D image of Fig. 1d), and also show the typical dependence of the porosity (open circles, left axis) of the resulting segmented image when a simple global threshold is chosen.
Transmitted X-ray intensity (scaled 0–255) probabilities for the entire apple sample (solid line, right axis) and the log scale cumulative probability (open circles, left axis) interpreted as percentage of void fraction (porosity) with the indicator kriging thresholds (40 and 70) indicated as dashed vertical lines
For these reasons, the thresholding method developed by Oh and Lindquist (1999) was used to perform segmentation of the apple tissue images into pores and cellular material. The algorithm is a non-parametric formulation able to analyze regions of uncertainty based on the estimation of the spatial covariance of the image in conjunction with indicator kriging to determine object edges (Mardia and Hainsworth 1988). Use of indicator kriging makes the thresholding local based on two threshold values, T 0 and T 1, and guarantees smoothness in the threshold surface. Implementation of the method requires a-priori population identification of some percentage of the image. The segmentation process is done fully in three dimensions, and not slice-wise. Thus, for the thresholding step of apple tissue images, the gray threshold values were set at 40 and 70. According to this thresholding window, a partial identification of the image was done as pores being voxels having gray values not greater than 40, and voxels of gray value greater than 70 being judged as cellular material (in general, non-edge). Finally, the remainder voxels of the population were identified by indicator kriging. Small disconnected void or solid phase components which may be either physical or arise from segmentation error, were cleaned up from each image using a morphological procedure called opening which considers a simple step of erosion followed by dilation (Russ 2005). Figure 3 compares the segmentation results in a representative 2D image using a simple global threshold and kriging-based algorithm followed by a clean up opening procedure.
Image segmentation of a 2D microtomography image of cv. Jonagold. a Original slice (black regions represent pores and gray regions cellular material). b Segmented image using a simple global threshold of 60. c Segmented image using kriging-based algorithm proposed by Lindquist et al. (1996) followed by the resultant clean up image (d) using the opening morphological procedure (erosion + dilation = opening)
Medial axis analysis
The medial axis of an object is the skeleton of the void space of an object running along its geometrical middle (Throvert et al. 1993; Lindquist et al. 1996). Once the stack of images are segmented, the medial axis is constructed using a burning algorithm developed by Lee et al. (1994) and modified by Lindquist et al. (1996). The burning algorithm is applied as follows (Fig. 4): each voxel in the solid phase is labeled with integer 0, voxels in the exterior region of the image are labeled with integer −1, and voxels in the void space are initially unlabeled. The algorithm proceeds by simultaneously assigning unlabeled voxels in the void space with an integer n + 1 for each voxel neighboring a solid-phase voxel, with n = 0 for this first iteration. This procedure continues at a rate of one layer per iteration until all voxels in the pore space are labeled. Then, the medial axis is determined by the voxels where different solid-pore boundaries intersect. There are two methods to define the 3D neighborhood of a voxel. The first is face touching, where each voxel can be neighbored by a maximum of six voxels. The second is boundary touching, where each voxel can be neighbored by a maximum of 26 voxels. For complex 3D systems, the 26-connectivity approach is required because it provides more accurate description of the connectivity. The latter method was used in this study. The result of this burning algorithm is a set of identified medial axis voxels, each with a unique burn number k, which is related to its distance from the closest cellular material voxels (Lindquist et al. 1996). In addition, for reducing surface noise in the construction of the medial axis of pore phase, all isolated clusters of pore and cellular material less than 1% volume size limit (an user set cutoff) were removed and re-identified as cellular phase and pore phase, respectively using the cluster-merge algorithm. Details of the skeletonization algorithm used in this work can be found in Thovert et al. (1993), Lee et al. (1994), Lindquist et al. (1996), and Lindquist (1999).
Two-dimensional illustration of medial axis determination using the burning algorithm described by Lindquist et al. (1996). Each voxel in the solid phase is labeled with integer 0 and voxels in the void space are labeled by a simultaneous assigning of an integer n + 1 for each voxel neighboring a solid-phase voxel, with n = 0 for this first iteration. This procedure continues at a rate of one layer per iteration until all voxels in the pore space are labeled. The medial axis voxels are determined by the voxels where different cell-pore phase boundaries intersect
Since the reconstructed 3D binary image consists of a cubic array of small voxels identified as either pore or cellular material, they can be used to calculate the bulk porosity of the sample. Porosity represents the void space as a fraction of the total volume of the sample, and is determined by simple count of voxels of the pore space divided by the total number of voxels. Similarly, in the medial axis construction, the spatial location and the burn number of each voxel are recorded and can be used as the base from which to calculate the spatial position of pores in the sample and other geometrical features such as: specific surface area and distributions of disconnected pore volumes. Specific surface area (SSA) represents a measure of the interstitial surface area of the void-solid phase per bulk volume, and it is calculated by summing the areas of all pore-cellular material voxel faces, and dividing this area by the total image volume. Here an approximation of this parameter is obtained from (Lindquist et al. 1996):
where N(1) is the total number of voxels having burn number k = 1 (Fig. 4), N(U) is the total number of voxels in the sample (U), and d is the linear dimensions of a voxel (assumed cubic).
Sensitivity analyses on porosity measurements of apple tissue using X-ray computed microtomography
In order to assess the sensitivity of this technique on the computed porosity in relation to the changes of the image resolution and volume size of the analyzed sample, two experiments were performed using Jonagold apples.
Image resolution analysis
The quality of low resolution images can be improved using dedicated image-enhancing filters such as difference-of-Gaussian (DoG) filter, Laplacian operator, sharpening filters, among others techniques such as deconvolution using the Fourier transform of the image, or also combination of them. However, they frequently make smooth surfaces of the objects in the image, which could affect in unpredictable ways the true structure of the sample, and consequently, the accuracy of the extracted parameters from it. Therefore, less processed images, which have been captured with an appropriate resolution should always be preferred in order to preserve both the finest details of the microstructure and the quality of the data. The classical approach for resolution tests could consider images of the same sample (scene) where only the viewpoints differ by small offsets in position and adjusting the parameters of the X-ray system. Using apple samples, however, this procedure is not possible since the dehydration of the tissue by longer times of scanning could seriously affect the porous microstructure of the tissue. Here, we assumed that in spite of the high variability among samples taken from the same apple and even from the same geometrical position, the average of three apple samples can give an objective idea of the resolution effect on the computed porosity and further on the extracted parameters from the medial axis analysis. In this first experiment series, the effect of five resolutions (8.5, 11.4, 13.7, 17.1, and 22.8 μm/pixel) of tissue samples taken around the same apple (as shown in Fig. 1a) on the porosity measurements was analyzed in triplicate (using different apples). Since the sample was placed in between the source and detector, and had to be moved closer to the detector to reach a lower resolution (to decrease the magnification), in this particular experiment, the acquisition conditions were re-adjusted to improve the definition of the images. For this, only the voltage of the X-ray source adjusting the contrast of the image, and the exposure time of the CCD adjusting the camera sensitivity to the X-ray illumination conditions, were decreased in a small step for each of the different resolutions.
Representative elementary volume
Considering that the complexity of the analysis and computation time using 3D images are dependent on the size of the analyzed images, it is much desirable for geometrical medial axis analysis to use a sub-region of this original stack of radiographic images (in a form of a cylinder or a box). This sub-volume should be small enough so that it can be treated as a mathematical point of the continuum scale, i.e., large enough to provide representation of the macroscopic properties of the porous medium. The REV is defined as the range of volumes over which a statistical average can be performed (Bear 1972). In this second experiment, in order to ensure that the sample of concern was representative in this respect, the effect on the porosity measurements of six volume sizes (2.6 × 10−3, 2 × 10−2, 0.16, 1.3, 10.4, and 20.6 mm3) extracted from the same stack of images was analyzed in triplicate (using different apples). Thus, from each apple, the different volumes were obtained from the same stack of images dividing the entire 3D image in a number of sub-volumes. The procedure was implemented in MATLAB and it works by increasing the side length of the voxel in all directions until lengths of 16, 32, 64, 128, 256, and 320 pixels (with 8.5 μm/pixel) per side are reached. Then the average porosity and standard deviations (STD) for this number of sub-volumes were calculated. Since the average porosities are computed using different number of sub-volumes, the standard error (SE) for each volume size was also estimated using the relation:
where SE represents the estimated standard error, STD is the standard deviation, and n the respective number of sub-volumes used in each evaluated size.
Porous medium characterization of Jonagold and Braeburn varieties
Finally, the microstructure of both cultivars using reconstructed REV volumes was visually compared. Averages of porosity, specific surface area and total pore number of disconnected volumes (26-connectivity assumed) for each variety were also calculated and compared using three apple samples. In addition, the microstructure of these cultivars was characterized in terms of the burn number k and disconnected medial axis distributions computed from the medial axis analysis.
Results
Interpretation of 3D reconstructed binary image and medial axis for the void space of apple tissue
An example of a reconstructed binary cube of Jonagold apple and its corresponding medial axis for the void space is shown in Fig. 5a and b, respectively. A smaller array was used for the example since the minimum REV of apple tissue was estimated to be 1283 pixels3 with a pixel length size of 8.5 μm (as shown later in the sensitivity analyses). The segmented 3D binary image can be interpreted as a cubic array of 1283 small voxels identified as either pore or cellular material from which realistic quantitative descriptions of void and cellular material geometry can be extracted. Bulk porosity estimations and medial axis construction are based on this spatial information. Furthermore, it is evident from Fig. 5a that using a small binary sub-volume of 1283 pixels3 (equivalent to 1.3 mm3) it is possible to visualize and characterize the pore size distribution, shape, and orientation upon each face of the cube, and in the volume of the cube (not shown). This simple 3D representation allows better understanding of the pore organization and complexity of the sample, and therefore serves as a first approach for characterization and comparison between different apple cultivars and also between different regions of the same cultivar.
a, b Three-dimensional visualization of pore structure for Jonagold apple (1283 pixels3 equivalent to 1.3 mm3). a Reconstructed binary cube image after clean-up procedure. b Medial axis of the pore space. Colors represent relative distance to the nearest cellular surface and indicate the local width of the pore. The intercellular space forms a complicated network inside the tissue
The medial axis or skeleton can be viewed as a graph of edges and vertices (Fig. 5b). The skeleton has a strict geometrical relationship to the object’s surface and preserves important geometrical properties of the object (Thovert et al. 1993). For a porous medium, it provides simple and compact information about the topology and geometry of the void space in the form of nodes connected to paths. Thus, for a digitalized pore network in 3D space, the medial axis consists of a network of tortuous paths and clusters. A path is defined as a connected string of filled voxels (3D pixels), each voxel having exactly two filled neighbors. A cluster is defined as a set of medial axis voxels, each with at least three neighbors lying in the same cluster. Thus, a vertex on the medial axis, where several pore paths meet, corresponds to the location of a nodal pore. The number of paths meeting at a cluster (nodal pore) is termed the coordination number. Further, visualization of the 3D percolating backbone of pore space in the fleshy part of Jonagold tissue (Fig. 5b) revealed that the intercellular spaces are highly interconnected and form a complicated network inside the tissue.
Image-resolution analysis
Spatial resolution is the ability to resolve close and high contrast features in the image. Two objects will not be separable in an image if the spatial resolution of the image is larger than the distance between them. Figure 6 shows representative cross-sectional microtomography images of Jonagold apples at different resolutions with a length side of 2.72 mm (equivalent to 320 pixels/side), and Fig. 7 presents their average porosities and standard deviations computed from three apple samples. As expected, low resolutions increase the scattering of the images, making the visualization and analytical determination of the smallest microstructures of the sample more difficult (Fig. 6). It can also be observed that the quality of the images is drastically affected by the resolution when magnifications lower than 13.7 μm/pixel are used for scanning of apple tissue. These images appear blurred and have a reduced contrast. Also, using the lowest resolution (22.8 μm/pixel) the image is observed pixeled.
The computed porosities (Fig. 7) confirmed the high sensitivity to the resolution changes. The porosities systematically decreased and the standard deviation increased when the resolution was diminished. Analysis of variance showed that resolution does have a significant effect (P-value < 0.05) on porosity measurement. When comparing the average porosity at every resolution with that at the higher level (8.54 μm/pixel), statistical differences were found only for resolutions lower than 13.7 μm/pixel. These results suggest that there is a critical resolution around 13.7 μm/pixel, which could be used as the limit for suitable geometrical measurements in apple tissue using a X-ray micro CT system SkyScan 1072.
Representative elementary volume analysis (REV)
Figure 8 shows the average porosity computed from three stacks of images of Jonagold apples in relation to the volume size of the sample. With the exception of the largest volume (20.4 mm3, equivalent to 320 pixels/side), the rest of analyzed sub-volumes do not show statistical differences between their average porosities, however, their estimated variability expressed as standard error (Eq. 2) is affected strongly by the size of the image. As expected, this variability decreases when the size of the volume is increased. For the smallest volume (2.6 × 10−3 mm3, equivalent to 16 pixels/side) the estimated standard error was 2.1%, while for a volume of 10.4 mm3 (equivalent to 256 pixels/side) it was only 0.7%. However, it can be observed that for the largest volume (20.4 mm3, equivalent to 320 pixels/side) the porosity slightly decreases and the variability increases. This may be due to the fact that these stacks of images consider regions closer to the boundaries of the sample, which might be damaged during the sample preparation, resulting in macroscopic heterogeneity of the analyzed sample.
Representative elementary volume analysis (REV) using three stacks of images of Jonagold apples. Bars represent the estimated standard error of the computed average porosity (Eq. 2)
From this analysis the minimum REV for a continuum assumption of apple tissue therefore appears to be 1.3 mm3 (equivalent to 128 pixels/side), which shows a reasonable standard deviation of 2.7% using sub-volumes of three apples (12 samples) and an estimated standard error of 0.7%. This image size was used for image processing and geometrical computations in further analyses.
Porous medium characterization of Jonagold and Braeburn varieties
Figure 9 shows representative REV cubes of 1.3 mm3 of Jonagold and Braeburn tissue taken from the fleshy region of each apple. Both cultivars are clearly characterized by different porous structures. In Jonagold apple, the pores appear larger, and dependent of the observed face of the cube, elongated and somehow oriented. However, pores from Braeburn tissue appear smaller and circulars in the three faces of the cube, which agrees with the observed pattern seen by Dražeta et al. (2004) for the same cultivar. Also, heterogeneity in the porosity distribution of the bulk apple tissue samples is evident. These 3D observations confirm the existence of cell anisotropy in apple tissue, and also show differences in the degree of anisotropy between both cultivars.
Quantitative analysis of three apple samples for each cultivar confirmed the geometrical differences. The average porosity for Jonagold and Braeburn tissue was estimated to be 28.6 ± 3.2% and 21.7 ± 2.9%, and the average specific surface to be 1.82 × 10−2 ± 2.3 × 10−3 μm−1 and 1.42 × 10−2 ± 2.6 × 10−3 μm−1, respectively. The two geometrical parameters of both cultivars were significantly different (P-value < 0.05).
In the following, we compare the microstructure of these cultivars in terms of the pore burn number distributions. The distribution of burn numbers shows the relative number of voxels in each layer [N(k)] with the same burn number (i.e., the kth layer from the cell–pore interface). Superimposed upon the histogram is the least-squares straight line fit. Figure 10 plots, on a log-square scale, the normalized histogram of average burn numbers from three samples contained in the void space during the construction of the pore medial axis of Jonagold and Braeburn varieties. The agreement is excellent with correlation coefficients (R 2) close to 0.99, indicating an exponential decay distribution of their burn numbers expressed as:
where N(k) represents the relative number of voxels in each burn layer, A is the amplitude, k is the layer, and λ the respective decay lengths. Both cultivars showed similar distribution patterns; however, notable differences were found between the two burn number measurements. As seen in Fig. 10, the respective decay lengths, λ, for the medial axis distributions of Jonagold and Braeburn showed notable statistical differences with average values of 6.34 (±0.4) and 3.69 (±0.3), respectively. The difference in slope (P-value < 0.05) between Jonagold and Braeburn distributions is indicative of inherent geometric differences in the void structure.
Log-square scale histogram of pore burn numbers for Jonagold and Braeburn samples showing the least squares fit predictions for sample burn number distributions. The distribution of pore burn numbers shows the relative number of voxels in each layer with the same burn number where N(k) represents the relative number of voxels in each burn layer and k the layer
A voxel’s burn number has a physical interpretation; it is the radius (in units of number of voxels on the medial axis) of the largest cube centered at that voxel, which just fits inside the void space. Thus, in addition to the geometrical portrait of the skeleton of the void space, we have some measure of the smallest cross sectional area of the void space at each point along the spine (Lindquist et al. 1996). The estimated value of λ from normalized exponential distributions based on burn number analysis is, hence, another feature of the void space of apple tissue.
Another quantitative measure of the topology of porous media is the connectivity of the sample. A simpler measure of this connectivity is the number of disconnected medial axes found in the sample, which are related to the number and size of disconnected void spaces. The distribution of volumes (measured as a percentage of the total pore volume) of the disconnected medial axis fragments of the two apple cultivars is shown in Fig. 11.
Distribution of number of disconnected medial axes occurring versus medial axis volume for the two data samples (symbols multiple sign, plus, open circles, and closed circles represent the three repetitions for each variety). The solid line is the least square fit to the range of shortest disconnected volumes considered (from 4.7 × 10−5 to 1.1 × 4.7 × 10−3 mm3)
For both apple samples, the data seem to suggest a composite distribution. The number of smallest medial axis fragments [N(v)] are distributed as a power-law
where the range among v 0 and v 1 (expressed as a fraction of the total pore volume) for both samples were set to 4.7 × 10−5 and 1.1 × 10−3 using a REV cube of 1.3 mm3. This is followed by uniform distributions of medial axes for the intermediate and largest lengths
The average exponent of the power law distribution (a) of the medial axis volumes for the smallest disconnected void spaces for Jonagold and Braeburn samples were significally different (P-value < 0.05) with average values of −1.27 ± 0.03 and −1.11 ± 0.08, respectively. By contrast, the uniform part of the distribution for both samples was characterized by the absence of largest length medial axis (with v volumes highest than 20 and 32% for Braeburn and Jonagold apples, respectively), which may be due to the complex nature of the sample.
These results provide a picture of how the pore structure of apple tissue can vary between cultivars. The connectivity analysis suggests that the medial axis volumes associated with the smallest disconnected void spaces are distributed according to a power-law distribution whose exponents also showed to be able to differentiate between the cultivars. The microstructural parameters from the medial axis analysis may be used to differentiate these two analyzed cultivars.
Discussion
A range of techniques has been developed to measure intercellular spaces in apple fruit. Many of them based on Archimedes’ principle combined with a range of sampling methods including vacuum pressures, treatment durations, and infiltration media (Harker and Ferguson 1988; Vincent 1989; Rajapakse et al. 1990; Yamaki and Ino 1992; Yearsley et al. 1996; Dražeta et al. 2004). Other used a gasometric technique that measured the amount of air extracted from samples when subjected to vacuum (Calbo and Sommer 1987), and also using stereological approaches to provide more realistic representation of the porous media and hence the pore space measure (Soudain and Phan Phuc 1979; Ruess and Stösser 1993; Goffinet et al. 1995). However, estimates of air volume in apple in the studies above varied widely (2.5–41%). Particularly, Dražeta et al. (2004) using an accurate gravimetric approach based on Archimedes’ principle has reported for cv. Braeburn fractional air volumes of 22.2 ± 2.5% in the range of 6–9 mm from the skin. This estimation is closer to the porosity measured here for the same cultivar at a distance of 7 mm from the skin (21.7 ± 2.9%).
For quantitative image analysis, special precautions should be taken to optimize the statistical reliability of the medial axis data. As it is defined relative to the cell-pore interface, construction of the medial axis is sensitive to surface noise. Surface noise occurs in two forms, irregularities in an otherwise smooth digitized void-cellular surface, and disconnected clusters of void or cellular voxels, which occur due to voxel misidentification. Irregularities in the void-cellular surface can have the same effect as dead-end pores, producing extraneous branches on the void medial axis. These extraneous branches must be identified and removed before the medial axis can be used to characterize the object. We have employed a user set cutoff on the length of such branches to distinguish between true dead end pore paths and those produced by surface irregularities. If only that portion of the void space, which supports fluid flow, is to be investigated, all dead-end branches can be trimmed from the void space axis using the cluster-merge algorithm. Isolated clusters of void phase voxels will appear as disconnected objects and isolated clusters of cellular voxels will appear as cavities within the void space, each one with their own skeleton piece. Since isolated regions of void phase may indeed be physical, it is much more difficult to distinguish between small isolated void space clusters which are physical and those which are artifacts, all isolated void and solid clusters less than 1% volume size limit were removed and re-identified as cellular phase and void phase, respectively. This produces error in the medial axis at the edges of the imaged volume in proportion to the ratio of boundary area to volume of the imaged region, but has the advantage of producing a medial axis for the void space that is only a union of one-dimensional curves (Lindquist 1999).
Results from image resolution analysis indicate that adequate image resolution is a key determinant of high-quality pore geometry and topology data of apple samples. Adequate resolution can provide well-defined images to facilitate not only the visualization of the smallest microstructures of the sample, but also the image segmentation process and medial axis analysis. Here, apple tissue images captured with a resolution of 8.5 μm/pixel (the highest analyzed resolution) have confirmed to be a relevant scale for a clear 3D representation of the tissue pore network and further geometrical analysis. However, selection of the resolution for imaging apple tissue may depend on the particular scientific problem under investigation and the scanning technique used. Other equipments with higher resolutions such as synchrotron X-ray microtomography (synchrotron radiation) which allows high intensity (number of photons per seconds) and high degree of collimation (source divergence leads to image blur) could provide not only more accurate calculations of the estimated properties, but also the capability to differentiate and quantify both air and water phases inside of the segmented pore spaces by using an appropriate monochromator. However, although SEM images of apple tissue frequently show sharp contrast among pores and cellular material even using resolutions lower than 13.7 μm/pixel, these images do not provide 3D information in the same way as X-ray microtomography does, and therefore, the segmentation processes and quantitative description of the apple pore microstructure could be difficult and limited. Porosity values frequently differ very slightly for larger differences in threshold numbers. This is not the case for a stack of radiographic images, which have indistinct pore-cellular material boundaries and therefore require a more complicated thresholding technique.
REV is frequently expressed as a range of averaging volumes within which the characteristic property remains quasi constant and thus isotropic. Below the admissible REV range, the averaging procedure causes significant fluctuations for the considered property and thus can no longer be used (Helmig et al. 2006). In this investigation the evaluated range of volume sizes using a resolution of 8.5 μm/pixel varied from 2.6 × 10−3 to 20.6 mm3. The minimum REV was determined to be 1.3 mm3. The maximum REV, however, could not be determined due to the limitations of the equipment used.
It should be also noted that a representative volume could be defined for different parameters ranging in complexity from porosity measurements to residual non-aqueous phase liquids characteristics (Bear 1972). Here, porosity was chosen as a quality index of the extracted geometrical data since the accuracy in the medial axis construction is highly dependent not only on the adequate resolution of the images, but also of the suitability of the segmentation technique to define the void space from which the bulk porosity is computed.
The heterogeneity in the porosity distribution for Jonagold and Braeburn cultivars is visually evident from Fig. 9. Therefore, apple flesh cannot be regarded as a homogeneous material. The differences in the size, orientation, and organization of the air channels strongly contribute to the anisotropy and heterogeneity of apple flesh tissue (Vincent 1989; Khan and Vincent 1990, 1993a). The implications of anisotropy of apple flesh are wide ranging; it has been frequently assumed to contribute to the particular textural properties of each fruit (Khan and Vincent 1990, 1993b).
The connectivity has a critical influence on transport properties such as permeability, and its quantification is important for understanding microstructure-transport property relationships (Celia et al. 1995; Dražeta et al. 2004). However, the topological characteristics of porous media and interconnections using image analysis techniques depend on the spatial resolution of the available data. This is true if pores exist which are in the size range of the resolution or smaller. Then, the connectivity of the porous structure is expected to change with resolution because more connections or more isolated features appear.
In spite of this, the implemented experimental procedure for image acquisition and image processing has significant benefits for qualitative and quantitative analysis of apple images, allowing conclusions to be drawn on the exact topography of the porous space of apple tissue. This experimental procedure and image analysis technique should provide valuable opportunities for further research in other plant materials.
Conclusions
Three-dimensional microtomography image of apple tissue at micron scale resolution provide rich data sets for use in extracting realistic quantitative description of the pore space. In this article, we have implemented an experimental procedure to obtain high quality radiotomographic images of porous media for 3D visualization and quantitative characterization of the microstructure of apple tissue using sophisticated image analysis techniques and without recourse to geometrical assumptions. A sensitivity analysis revealed a larger dependence of the computed porosity on the resolution and the size of the processed image. Resolutions higher than 13.7 μm/pixel were found to be suitable for visual and quantitative evaluations of apple tissue. The minimum REV of apple tissue was estimated to be 1.3 mm3.
Comparisons among the two cultivars of apple tissue (cv. Jonagold and cv. Braeburn) using a resolution of 8.5 μm with a minimum REV cube showed that in spite of the complexity and variability of the pore space network observed in both cultivars, the extracted parameters from the medial axis were significantly different (P-value < 0.05). Medial axis parameters showed potential to differentiate the microstructure between the two evaluated apple cultivars.
However, while the microstructure of the pore space of two cultivars of apple tissue was quantitatively compared based on bulk average quantities (i.e., porosity and specific surface area) and distributions of their disconnected volumes and burn numbers (k), for a complete characterization of the random nature of the pore space requires understanding of the spatial distribution (autocovariance) of geometric descriptive parameters and of the correlations between parameters. For flow properties of pore networks, the important geometry characterizations, and essential input required for network models able to directly measure the stochastic nature of the void space, are pore coordination number, pore size and throat size distributions, pore body-to-pore throat size and pore body-to-pore body aspect ratios. We are currently in the process of analyzing some of these geometrical features in different cultivars of apple tissue and also in different regions within an apple.
Abbreviations
- CCD:
-
Charge-coupled device
- CT:
-
Computed tomography
- REV:
-
Representative elementary volume
- ROI:
-
Region of interest
- SSA:
-
Specific surface area
- STD:
-
Standard deviation
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
References
Aguilera JM (2005) Why food microstructure? J Food Eng 67:3–11
Babin P, Della Valle G, Dendievel R, Lassoued N, Salvo L (2005) Mechanical properties of bread crumbs from tomography based finite element simulations. J Mat Sci 40:5867–5873
Baoping J (1999) Nondestructive technology for fruits grading. In: Proceedings of 99th international conference on agricultural engineering Beijing, December, pp IV127–IV133
Barcelon EG, Tojo S, Watanabe K (1999) X-ray computed tomography for internal quality evaluation of peaches. J Agric Eng Res 73:323–330
Baumann H, Henze J (1983) Intercellular space volume of fruit. Acta Hortic 138:107–111
Bear J (1972) Dynamics of fluids in porous media. Dover, New York
Calbo AG, Sommer NF (1987) Intercellular volume and resistance to air flow of fruits and vegetables. J Am Soc Hortic Sci 112:131–134
Celia M, Reeves P, Ferrand L (1995) Recent advances in pore scale models for multiphase flow in porous media. Rev Geophys Suppl 33:1049–1057
Cheng Q, Banks NH, Nicholson SE, Kingsley AM, Mackay BR (1998) Effects of temperature on gas exchange of ‘Braeburn’ apples. NZ J Crop Hortic Sci 26:299–306
Dražeta L, Lang A, Alistair JH, Richard KV, Paula EJ (2004) Air volume measurement of ‘Braeburn’ apple fruit. J Exp Bot 55:1061–1069
Esau K (1977) Anatomy of seed plants. Wiley, New York
Fromm JH, Sautter I, Matthies D, Kremer J, Schumacher P, Ganter C (2001) Xylem water content and wood density in spruce and oak trees detected by high-resolution computed tomography. Plant Physiol 127:416–425
Goffinet MC, Robinson TL, Lakso AN (1995) A comparison of ‘Empire’ apple fruit size and anatomy in unthinned and handthinned trees. J Hortic Sci 70:375–387
Harker FR, Ferguson IB (1988) Calcium ion transport across discs of the cortical flesh of apple fruit in relation to fruit development. Physiol Plant 74:695–700
Harker FR, Hallet IC (1992) Physiological changes associated with development of mealiness of apple fruit during cool storage. HortScience 27:1291–1294
Harker FR, Watkins CB, Brookfield PL, Miller MJ, Reid S, Jackson PJ, Bieleski RL, Bartley T (1999) Maturity and regional influences on watercore development and its postharvest disappearance in ‘Fuji’ apples. J Am Soc Hortic Sci 124:166–172
Helmig R, Miller CT, Jakobs H, Class H, Hilpert M, Kees CE, Niessner J (2006) Multiphase flow and transport modeling in heterogeneous porous media. In: Di Bucchianico A, Mattheij RMM, Peletier MA (eds) Progress in industrial mathematics at ECMI 2004. Springer, Heidelberg, pp 449–485
Ho QT, Verlinden BE, Verboven P, Vandewalle S, Nicolaï BM (2006) A permeation–diffusion-reaction model of gas transport in cellular tissue of plant materials. J Exp Bot (in press)
Kader AA (1988) Respiration and gas exchange of vegetables. In: Weichmann J (ed) Postharvest physiology of vegetables. Marcel Dekker, New York, pp 25–43
Khan AA, Vincent JFV (1990) Anisotropy of apple parenchyma. J Sci Food Agric 52:455–466
Khan AA, Vincent JFV (1993a) Compressive stiffness and fracture properties of apple and potato parenchyma. J Texture Stud 24:423–435
Khan AA, Vincent JFV (1993b) Anisotropy in the fracture properties of apple flesh as investigated by crack-opening tests. J Mat Sci 28:45–51
Kuroki S, Oshita S, Sotome I, Kawagoe Y, Seo Y (2004) Visualization of 3-D network of gas-filled intercellular spaces in cucumber fruit after harvest. Postharvest Biol Technol 33:255–262
Lammertyn J, Dresselaers T, Van Hecke P, Jancsók P, Wevers M, Nicolaï B (2003a) Analysis of the time course of core breakdown in ‘Conference’ pears by means of MRI and X-ray CT. Postharvest Biol Technol 29:19–28
Lammertyn J, Dresselaers T, Van Hecke P, Jancsók P, Wevers M, Nicolaï B (2003b) MRI and X-ray CT study of spatial distribution of core breakdown in ‘Conference’ pears. Magn Reson Imaging 21:805–815
Lee T-C, Kashyap R, Chu C-N (1994) Building skeleton models via 3-d medial surface/axis thinning algorithms. CVGIP Graph Models Image Process 56:462–478
Lim KS, Barigou M (2004) X-ray micro-computed tomography of aerated cellular food products. Food Res Int 37:1001–1012
Lindquist WD (1999) 3DMA general users manual. State of New York at Stony Brook, New York
Lindquist WD, Lee S-M, Coker D, Jones K, Spanne P (1996) Medial axis analysis of three dimensional tomographic images of drill core samples. J Geophys Res 101B:8297–8310
Mardia KV, Hainsworth TJ (1988) A spatial thresholding method for image segmentation. IEEE Trans Pattern Anal Mach Intell 6:919–927
Maire E, Buffière J-Y, Salvo L, Blandin JJ, Ludwig W, Létang JM (2001) On the application of X-ray microtomography in the field of material science. Adv Eng Mater 3:539–546
Maire E, Fazekas A, Salvo L, Dendievel R, Youssef S, Cloetens P, Letang J.M (2003) X-ray tomography applied to the characterization of cellular materials. Related finite element modeling problems. Compos Sci Technol 63:2431–2443
Mebatsion HK, Verboven P, Verlinden BE, Ho QT, Nguyen TA, Nicolaï B (2006a) Microscale modeling of fruit tissue using Voronoi tessellations. Comput Electron Agric 52:36–48
Mebatsion HK, Verboven P, Ho QT, Mendoza F, Verlinden BE, Nguyen TA, Nicolaï B (2006b) Modeling fruit microstructure using novel ellipse tessellations algorithm. Comp Modeling Eng Sci 14:1–14
Oh W, Lindquist W (1999) Image thresholding by indicator kriging. IEEE Trans Pattern Anal Mach Intell 21:590–602
Rajapakse NC, Banks NH, Hewett EW, Cleland DJ (1990) Development of oxygen concentration gradients in flesh tissues of bulky plant organs. J Am Soc Hortic Sci 115:793–797
Raven JA (1996) Into the voids: the distribution, function, development and maintenance of gas spaces in plants. Ann Bot 78:137–142
Ruess F, Stösser R (1993) Untersuchungen über das interzellularsystem bei apfelfrüchten mit methoden der digitalen bildverarbeitung. Gartenbauwissenschaft 58:197–205
Russ JC (2005) Image analysis of food microstructure. Boca Raton, London
Salvo L, Cloetens P, Maire E, Zabler S, Blandin JJ, Buffière JY, Ludwig W, Boller E, Bellet D, Josserond C (2003) X-ray micro-tomography an attractive characterisation technique in material science. Nucl Instrum Methods Phys Res B200:273–286
Schotsmans W, Verlinden BE, Lammertyn J, Nicolaï BM (2004) The relationship between gas transport properties and the histology of apple. J Sci Food Agric 84:1131–1140
Soudain P, Phan Phuc A (1979) La diffusion des gaz dans les tissus vegetaux en rapport avec la structure des organes massifs. In: Perspectives nouvelles dans la conservation des fruits et legumes frais. Seminaire International, Centre de Recherches en Sciences Appliquées à l’Alimentation, L’Université du Québec à Montreal, Canada, pp 67–86
Thovert J, Salles J, Adler P (1993) Computerized characterization of the geometry of real porous media: their discretization, analysis and interpretation. J Microsc 170:65–79
Tu K, De Baerdemaeker J, Deltour R, de Barsy T (1996) Monitoring post-harvest quality of Granny Smith apple under simulated shelf-life conditions: destructive, non-destructive and analytical measurements. Int J Food Sci Technol 31:267–276
van Dalen G, Blonk H, van Aalst H, Hendriks CL (2003) 3D imaging of foods using X-ray microthomography. GIT Imaging Microsc 3:18–21
Vincent JFV (1989) Relationships between density and stiffness of apple flesh. J Sci Food Agric 31:267–276
Volz RK, Harker FR, Hallet IC, Lang A (2004) Development of texture in apple fruit—a biophysical perspective. Acta Hort 636:473–479
Westwood MN, Batjer LP, Billingsley HD (1967) Cell size, cell number and fruit density of apples as related to fruit size, position in the cluster and thinning method. Proc Am Soc Hortic Sci 91:51–62
Yamaki S, Ino M (1992) Alteration of cellular compartmentation and membrane permeability to sugars in immature and mature apple fruit. J Am Soc Hortic Sci 117:951–954
Yearsley CW, Banks NH, Ganesh S, Cleland DJ (1996) Determination of lower oxygen limits for apple fruit. Postharvest Biol Technol 8:95–109
Yearsley CW, Banks NH, Ganesh S (1997a) Temperature effects on the internal lower oxygen limits of apple fruit. Postharvest Biol Technol 11:73–83
Yearsley CW, Banks NH, Ganesh S (1997b) Effects of carbon dioxide on the internal lower oxygen limits of apple fruit. Postharvest Biol Technol 12:1–13
Acknowledgments
The authors wish to thank K.U. Leuven (Project OT 04/31) and the Fund for Scientific Research Flanders (FWO—Vlaanderen, Project G.0200.02) for financial support for this investigation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mendoza, F., Verboven, P., Mebatsion, H.K. et al. Three-dimensional pore space quantification of apple tissue using X-ray computed microtomography. Planta 226, 559–570 (2007). https://doi.org/10.1007/s00425-007-0504-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-007-0504-4