Abstract
A full-length cDNA gene, designated Oryza sativa chymotrypsin inhibitor-like 1 (OCPI1), was characterized in rice. The predicted protein of OCPI1 shows very high sequence identity to reported chymotrypsin inhibitors from various plant species. Northern-blot analysis showed that the expression of OCPI1 was strongly induced by dehydration stresses and abscisic acid (ABA). The expression of beta-glucuronidase (GUS) reporter gene under the control of OCPI1 promoter transformed into rice was strongly induced by drought and salt stresses. Interestingly, strong dehydration stress-induced GUS activity was also detected in the transgenic rice containing the reverse sequence of OCPI1 promoter fused to GUS gene, suggesting of a bidirectional transcriptional activity in the OCPI1 promoter. OCPI1 gene was over-expressed in japonica cv. Zhonghua 11 and transgenic plants containing single copy of transgene were tested for drought resistance at reproductive stage. The positive transgenic plants (OCPI1 was over-expressed) had significantly higher grain yield and seed setting rate than the wild type and the negative transgenic control (no over-expression of the transgene) under the severe drought stress conditions, whereas the potential yield of transgenic plants under normal growth conditions was not affected. Chymotrypsin-inhibitor activity assay showed that the crude protein of the positive transgenic plants had stronger inhibitory activity than the negative control. Transgenic plants had less decrease of total proteins than the wild type under drought stress. Taken together, these data indicate that OCPI1 might potentially be useful in the genetic improvement of drought resistance in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant growth is greatly affected by environmental abiotic stresses such as drought, high salinity and low temperature. Severe drought stress can lead to dramatic suppression of plant growth and development and cause large loss of productivity. Rice (Oryza sativa L.) is one of the major crops planted worldwide with an annual production of about 608 million metric tons including 549 million metric tons in Asia (IRRI 2004). Therefore, developing rice cultivars with improving drought resistance has been well recognized as one of the most promising and effective strategy to alleviate food insecurity caused by drought and water shortage.
To date, however, limited success has been achieved in improving stress resistance because most stress resistance related traits have complex genetic basis which is controlled and influenced by differential expression of a network of genes (Shinozaki and Yamaguchi-Shinozaki 1997; Shinozaki et al. 2003). Several groups have conducted researches on the drought tolerance of rice using molecular markers to identify and utilize quantitative trait loci (QTL) of drought resistance related traits in rice breeding programs (Lilley et al. 1996; Price et al. 2000; Tripathy et al. 2000; Zhang et al. 2001; Yue et al. 2006). However, few of the QTLs have been finely mapped mainly because of the difficulties in accurate phenotyping of the quantitative traits in a large population, which has largely limited the cloning and utilization of the drought resistance-related QTL genes.
Increasing evidence suggests that transgenic engineering is an advantageous option to improve a single desired trait in plants (Dunwell 2000). Several transformation protocols have been established or optimized successfully for transforming foreign genes into rice (Hiei et al. 1994; Lin and Zhang 2005). So far, array of stress related genes, such as HVA1 (Xu et al. 1996), OsCDPK7 (Saijo et al. 2000), OsMAPK5 (Xiong and Yang 2003), OsDREB1 (Ito et al. 2006), and SNAC1 (Hu et al. 2006) have been transformed into rice to test their effect on improving drought resistance.
Proteinase inhibitors (PI) constitute a large and complex group of plant proteins and have an enormous diversity of function by regulating the proteolytic activity of their target proteinases, resulting in the formation of a stable protease inhibitor complex (Leung et al. 2000). PIs were classified into non-specific and class-specific superfamilies and the later was subcategorized into several families including serine proteinase inhibitor, aspartic proteinase inhibitor, metalloproteinase inhibitor, and cysteine proteinase inhibitor (Hibbetts et al. 1999). Serine proteinase appears to be the largest family of proteinase and plant serine-proteinase inhibitors have been classified into several subfamilies including soybean (Kunitz)-, Bowman-birk-, potato I-, potato II-, squash-, barley-, cereal-, ragi A- and thaumatin-PR like inhibitors (Haq et al. 2004). The expression of many plant PIs is detected in specific tissues and is developmentally regulated (Habu et al. 1996; Clark et al. 1997). Plant PI proteins have been shown to have functions in various physiological and developmental processes (Mosolov and Valueva 2005). Reports on proteinase inhibitor II (PIN2), a serine-proteinase inhibitor which occurs in Solanaceae plants, show that it could play endogenous roles in environmental responses and development (Hendriks et al. 1991; Pena-Cortes et al. 1991; Sin and Chye 2004; Sin et al. 2006). A soybean cysteine PI has been designated a novel role in modulating the programmed cell death (Solomon et al. 1999).
Many reports suggest that plant PIs may also be involved in the responses to various biotic and abiotic stresses. Soybean cysteine-proteinase inhibitor genes were reported for their expression level changes during wounding and methyl jasmonate treatment (Botella et al. 1996). PI proteins of the Kunitz family were identified from salt-treated radish (Lopez et al. 1994) and drought-stressed Arabidopsis thaliana (Gosti et al. 1995). The NGP1-1 encoding Nicotiana glutinosa proteinase inhibitor II was rapidly activated by pathogen and wound-related stresses (Choi et al. 2000). A cysteine-proteinase inhibitor gene from chestnut, designated CsC, was strongly induced in the roots and leaves of chestnut plantlets subjected to cold- and saline-shocks, and also in the roots after heat stress (Pernas et al. 2000). In rice, a wound-, jasmonate- and ethylene-induced proteinase inhibitor gene, OsBBPI, was also identified (Rakwal et al. 2001). So far, only a few drought stress-induced serine PIs were reported in plant. In Brassica napus plants, under water-deficient condition, an induced 22 kDa PI belonging to the Kunitz family was identified (Downing et al. 1992). A Brassica PI containing a motif for Kunitz-type proteinase inhibitor was induced by drought- and heat-stresses (Satoh et al. 2001). Kang et al. (2002) reported a 27 kDa potato Kunit-type PI that was induced by ABA and water deficit. To date, serine PI involved in dehydration responses has not been characterized in rice.
In this study, a putative chymotrypsin inhibitor gene (OCPI1), which belongs to the serine PI family, was isolated from rice. The expression of OCPI1 gene was responsive to different abiotic stresses and the stress-induced expression level was further investigated by using a fusion construct of OCPI1 promoter::GUS (beta-glucuronidase) gene. The OCPI1 gene was over-expressed in rice to evaluate the effect of over-expressing OCPI1 on improving drought resistance of transgenic rice under the drought-stressed field conditions. The chymotrypsin-inhibitor activity was also analyzed in the transgenic plants. Our results suggested that over-expression OCPI1 had significant effect on improving drought resistance at the reproductive stage of rice.
Materials and methods
Constructs and rice transformation
To make an over-expression construct, the full-length cDNA of OCPI1 was identified from a cDNA library (Chu et al. 2003) and amplified by primers att-B1-T7 and att-B2-SP6. T7 (5′-TAATACGACTCACTATAGGG-3′) and SP6 (5′-ATTTAGGTGACACTATAG-3′) are the primers flanking the cDNA in pSPORT (the vector used for cDNA library construction). The att-B1 and att-B2 are the two commercialized adaptor sequences (Invitrogen, Carlsbad, CA, USA) for recombination cloning. The purified PCR band was recombined into the vector pDONR207 by a BP recombination reaction and then recombined into the destination vector pC2004H by a LR reaction following the manufacture (Invitrogen). The destination vector pC2004H was constructed by inserting the CaMV 35S:R1-ccdB-R2-Terminator cassette into vector pCAMBIA-1301 (provided by CAMBIA, accession no. AF234297). The R1-ccdB-R2 fragment was amplified from the vector pDEST 17 (Invitrogen) using primers 5′-ACTACCATCACCATCACCAT-3′ and 5′-TTTGTTAGCAGCCTCGAAT-3′. A fragment (1341 bp in length) containing the OCPI1 promoter region was amplified from the genomic DNA of an upland rice cultivar IRAT109 (Oryza sativa L. ssp japonica, developed in Cote d’Ivoire and provided by Shanghai Agriculture Gene Center) with a sense primer (5′-TAGGATCCAAAATTCACGGATGTAAAGG-3′) containing BamHI restriction site (underlined) and an antisense primer (5′-TAGAATTCGCCTTATCTTTTCGCTTATG-3′) containing EcoRI site (underlined). The purified PCR product was confirmed by sequencing (ABI 3730 sequencer, Applied Biosystem, Foster City, CA, USA) and inserted in front of the GUS reporter gene in the vector pCAMBIA-1391Z (provided by CAMBIA, accession no. AF234312). By swapping the two restriction sites of the two primers, the antisense fragment of OCPI1 promoter region was cloned and inserted in front of the GUS reporter gene.
All constructs were introduced into Agrobacterium tumefaciens strain EHA105 by electroporation and then transformed into japonica cv. Zhonghua 11 (developed and provided by Tianjin Agriculture Research Institute), a genotype with high efficiency of transformation (data not shown), using the Agrobacterium-mediated transformation method by Hiei et al. (1994). Transgenic calli and plantlets were selected based on the hygromycin (50 mg/l) resistance.
Stress treatment
Transgenic T1 seeds were dehulled and sterilized in 70% ethanol for 1 min and in 0.15% HgCl2 for 15 min, then washed with sterile water for 5 min and placed on MS medium containing 50 mg/l hygromycin. The positive transgenic seeds (being able to germinate and grow in the medium) were picked out for transplanting. 20 seedlings with uniform growth from each independent transgenic family and the wild type rice Zhonghua 11 were planted in a two-row plot in the water-managed paddy field (equipped with a moveable rain-off shelter) with three replicates. The distance between the plants was 16.5 cm. Irrigation was stopped at about 3 weeks before flowering, and the irrigation was resumed when the water content of the soil drooped to about 16% (w/w). The field had been thoroughly mixed with one-third of sand, such drought stress treatments in the testing site allowed drought stress to develop rather uniformly at the flowering stage, a critical stage for yield stability in rice. The same set of transgenic families was planted in the full-irrigated field as control by following the same experimental design. Seed setting rate (or spikelet fertility) was investigated to evaluate the drought resistance. In each plot, the 16 plants in the middle were investigated.
For detecting the expression level of the gene under dehydration stress conditions, 2 week old seedlings of upland cultivar IRAT109 were cultured in greenhouse with the 14 h light/10 h dark cycle. Drought stress was applied by stopping watering and re-watering, when leaves became completely rolled. For salinity stress, roots of the seedlings were immersed in a nutrition solution containing 200 mM sodium chloride. Abscisic acid (ABA) treatment was conducted by spraying the seedling leaves with 0.1 mM ABA.
Southern-blot and RNA gel-blot analysis
For Southern-blot analysis, 3 μg of genomic DNA from the transgenic rice was digested with EcoRI restriction enzyme for overnight, fractionated in 0.8% agarose gel, alkali-transferred onto Hybond nylon membrane and hybridized with α-32P-labeled hygromycin phosphotransferase (Hpt) gene as the probe.
Total RNA was isolated from rice leaves using TRIzol reagent (Invitrogen). Fifteen micrograms of total RNA of each sample was resolved in 1.2% agarose gel containing 2% formaldehyde and blotted onto Hybond nylon membranes. RNA gel blot was hybridized with α-32P-labeled OCPI1 sequence-specific probe at 65°C for overnight. Blot was washed twice (once with 2 × SSC/0.1% SDS for 10 min and once with 1 × SSC /0.1% SDS for 5 min at 65°C), and then subjected to radiography.
ß-Glucoronidase (GUS) activity assay
Rice tissue of OCPI1 promoter:GUS transgenic plant was pulverized in liquid nitrogen and homogenized with extraction buffer (50 mmol/l Na2HPO4 pH 7.0, 10 mmol/l mercaptoethanol, 10 mmol/l Na2-EDTA, 0.1% Sarkosyl, and 0.1% Triton X-100) for 10 min on ice. The homogenous samples were centrifuged twice at 4°C with 8,000g for 15 min, and the supernatant was collected for quantification of total protein content in a spectrophotometer (Beckman DU-640) using bovine serum albumin as a standard protein. Fluorimetric GUS analysis was performed by using 4-methylumbelliferyl-β-d-glucuronide (4-MUG) as a substrate in Hoefer DyNA Quant 200 (Amersham Biosciences, San Francisco, CA, USA). Samples of various transgenic rice tissues were incubated in GUS staining buffer (50 mM Na-phosphate pH 7.0, 10 mM EDTA, 1% Triton X-100, 1 mg/ml X-Gluc, 100 μg/ml chloramphenicol, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, 20% methanol) at 37°C for overnight for observation of GUS expression.
Assay of chymotrypsin inhibitory and endogenous chymotrypsin activities
Plant tissues were pulverized in liquid nitrogen and crude total protein was extracted with extraction buffer containing 50 mM Tris (pH 8.1) and 20 mM CaCl2. The supernatant was used for spectrophotometric assay of chymotrypsin inhibitory activity as described previously (Xu et al. 2004), in which chymotrypsin inhibitory activity was estimated by the remaining esterolytic activity of bovine chymotrypsin using N-benzoyl tyrosine ethyl ester (BTEE; Sigma, St. Luis, MO, USA) as a substrate. A final volume of 1.5 ml mixture containing 50 μg leaf protein extract with 100 μl of bovine chymotrypsin (20 μg/ml in 1 mM HCl, Sigma) and assay buffer (100 mM Tris–HCl, pH 7.8; 100 mM CaCl2) was pre-incubated for 3 min at room temperature in a quartz cuvette. The reaction was initiated by adding 1.5 ml of substrate (1 mM BTEE in 50% [w/w] methanol) to the pre-incubated mixture. The absorbance at 256 nm was immediately measured at an interval of 30 s for 5 min. In the standard reaction, 100 μl of bovine chymotrypsin was used for analysis in the absence of leaf extract.
Endogenous chymotrypsin activities in leaves were determined using the same procedure described above with the omission of bovine chymotrypsin from the reaction.
Results
Identification of stress-responsive gene OCPI1
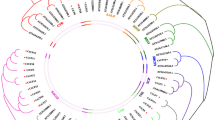
A full-length cDNA (designated OCPI1) encoding a putative chymotrypsin inhibitor was identified from a cDNA library of indica rice Minghui 63 (Chu et al. 2003). The cDNA sequence of OCPI1 showed 98.5% identity with the OsSCI3 (GenBank accession number AY878695, Zhao and Peng, unpublished). Protein sequence of OCPI1 showed 27–80% identity with various plant serine-proteinase inhibitors including the potato inhibitor I family (Fig. 1a). Using the protein sequence of OCPI1 to do BLASTP search against the rice annotation database (TIGR, http://www.tigr.org/tdb/e2k1/osa1/), at least 16 putative chymotrypsin inhibitor were browsed. Phylogenetic analysis of putative rice chymotrypsin inhibitors and a few chymotrypsin inhibitors from other species suggested that plant chymotrypsin inhibitors were largely diversified (Fig. 1b). Interestingly, quite a few putative rice chymotrypsin inhibitor genes are tandem located along with their highest homologs in the rice genome, such as OCPI1 (flanked by OCPI2), Os08g34249 (flanked by Os08g34258), Os02g03170 (flanked by Os02g03180 and Os02g03190), Os12g36210 (flanked by Os12g36220 and Os12g36240). This suggested that gene duplication might have contributed to the expansion and evolution of chymotrypsin inhibitor family in rice genome.
Sequence analysis of OCPI1. a Sequence alignment of OCPI1 with its homologs from rice and other species. The conserved residues are indicated with shadow. The homologous proteinase inhibitors from other species includes: NSTI-1 (GI: 547743) from tobacco, CI-1 (GI: 124125) from barley, DI-1 (GI: 124120) from tomato, ASI-I (GI: 124121) from adzuki bean, CMTI-V (GI: 1431715) from pumpkin, ATSI (GI: 461598) from amaranth, and BGIA (GI: 114950) from bitter gourd. Rice homologs of OCPI1 were from TIGR annotation database based on BLASTP search. b Phylogenetic relationship of OCPI1 and other Potato inhibitor I family members based on the conversed region by Maximum-likelihood tree. The bootstrap values for 100 trials are indicated at each fork
The OCPI1 gene was first identified for its induction by drought stress in our cDNA microarray experiment (our data not shown). RNA-gel blot was then performed with total RNA isolated from rice leaves after drought, salt, and ABA treatments (Fig. 2). In the drought treatment, very strong induction of OCPI1 was detected in the partially rolled leaves and its expression was decreased in the fully rolled leaves. When the plants were re-watered for 1 day, the expression level of OCPI1 dropped to the level similar as in the non-stressed leaves. The OCPI1 transcript level was rapidly increased shortly after salt treatment and maintained at high level of induction throughout the development of stress. In the treatment of ABA, the transcript level of the gene was increased shortly after the treatment and peaked at 12 h. These results suggested that the expression of OCPI1 was strongly induced by dehydration stresses (such as drought and salinity) and was responsive to ABA.
Northern-blot analysis of OCPI1 expression level under different abiotic stresses. Lane CK no stress, lane 1 of drought treatment, slight stress (about 10% leaves were rolled), lane 2 mild stress (about 50% leaves were rolled), lane 3 severe stress (100% leaves were rolled), lane RW recovery for 1 day by re-watering after stresses. Time courses of salt and ABA treatments are indicated under the lanes
Bidirectional stress-inducible activity of OCPI1 promoter
Strong induction of OCPI1 by dehydration stresses prompted us to analyze the promoter of this gene. A DNA fragment (1341 bp in length) covering the promoter region of OCPI1 (Fig. 3a) was amplified from upland rice IRAT109 and sequenced. Putative cis-element prediction by searching the PLACE database (http://www.dna.affrc.go.jp/place/; Higo et al. 1999) suggested that the sequence of this fragment contained putative CAAT boxes, TATA boxes, and quite a few putative stress-responsive cis-elements such as MYB1AT (Abe et al. 2003), MYB2AT and MYBCORE (Urao et al. 1993), MYCATERD1 (Tran et al. 2004), MYCATRD22 (Busk and Pages 1998), and ABRELATERD1 (Simpson et al. 2003) in both sense and antisense strands (Fig. 3b).
Genomic organization of OCPI1 and its promoter. a Diagram of the location of OCPI1, OCPI1 promoter, and another chymotrypsin inhibitor-like gene OCPI2. The transcription start site was marked as “+1”. Exons were shown in filled black rectangles (arrows indicating transcription direction). The fragment covering the OCPI1 promoter region was shown below the gene structure. b Predicted stress responsive elements were marked as frame (forward direction) and shadow (reverse direction). Core sequences for stress and responsive cis-elements are as follows: MYB1AT: WAACCA; MYB2AT: TAACTG; MYBCORE: CNGTTR; MYCATERD1: CATGTG; MYCATRD22: CACATG; ABRELATERD1: ACGTG
To investigate the transcriptional activity of the OCPI1 promoter in rice, the fragment of OCPI1 promoter was inserted in front of the GUS reporter gene (construct OCPI-F, Fig. 4a) and the construct was introduced into rice Zhonghua 11. By histochemical assay, slight GUS expression was detected in callus, leaf, root, stem, sheath, ligule, auricle, glume, rachilla, pistil, and stamen of transgenic rice (data not shown), suggesting that the endogenous OCPI1 gene may express in these tissues or organs with relatively low level under normal growth conditions. GUS activity of the crude protein extract from drought-stressed (Fig. 4b) and salt-stressed (Fig. 4c) transgenic leaves was significantly (P < 0.01) higher than the non-stressed transgenic samples and the stressed control plants (empty vector-transformed plants). These results further confirmed that the OCPI1 promoter was strongly induced by dehydration stress.
Transcriptional activity assay of OCPI1 promoter. a Constructs for testing the activity of OCPI1 promoter. CK, the backbone vector pCAMBIA-1391Z (pC1391Z); OCPI-F, the reporter gene GUS under the control of OCPI1 promoter (the fragment shown in Fig. 3a) in sense direction; OCPI-R, the reporter gene GUS under the control of the antisense strand of OCPI1 promoter. HPT hygromycin phosphotransferase gene. LB and RB are left and right borders of the T-DNA, respectively. GUS assay of transgenic plants under drought stress (air-dried for the time indicated) (b) and salt stress (200 mM NaCl) (c), respectively. GUS activity was given in picomole of 4-MU generated per minute per microgram protein. Each value represents the mean of three independent transgenic plants containing similar GUS activity pre-checked by GUS-staining (three repeats for each sample)
Putative stress-responsive cis-elements in the reverse strand of OCPI1 promoter prompted us to test if the antisense strand of OCPI1 promoter has promoter activity. The GUS gene under the control of the antisense strand of OCPI1 promoter (construct OCPI-R, Fig. 4a) was also transformed into rice and slight GUS activity was detected in callus, leaf, root, stem, sheath, glume, pistil, and stamen (data not shown). Interestingly, strongly induction of GUS activity was detected in the drought- and salt-stressed transgenic leaves (Fig. 4b, c). These results suggest that OCPI1 promoter has a bidirectional stress-inducible activity. As a fact, another putative chymotrypsin inhibitor-like gene (designated as OCPI2), located at the immediate upstream of the OCPI1 promoter fragment with reverse transcription direction to that of OCPI1 gene (Fig. 3a), was predicted in the genome annotation database (http://www.tigr.org/tdb/e2k1/osa1/). The OCPI2 gene is supported by a full-length cDNA (accession number AK062495) and is also induced by drought and salt stress based on our cDNA microarray profiling data (data not shown).
Generation and drought resistance testing of OCPI1-overexpression transgenic rice
The full-length cDNA for OCPI1 under the control of CaMV 35S promoter (Fig. 5a) was transformed into rice Zhonghua 11. A total of 26 independent transgenic plants of transgenic plants were generated. Leaves from 23 T0 transgenic plants and the wild type (WT) rice (Zhonghua 11) were sampled for detecting the expression level of the OCPI1. RNA-blot analysis showed that more than 50% transgenic plants had obviously higher level of OCPI1 transcript (reflecting the total transcript of both endogenous OCPI1 and the transgene) than WT (Fig. 5b). The OCPI1-overexpresed transgenic plants contained one to several copies of the transgene based on Southern-blot analysis (Fig. 5c). The progenies of three independent single copy plants, designated TL-4, 20, 25, and a non-overexpression transgenic family (TL-21) were selected for drought resistance testing.
Identification and drought testing of OCPI1-overexpressing rice. a Overexpression construct for rice transformation. b RNA gel-blot analysis of part of transgenic plants and the wild type (WT). c Southern-blot analysis of part of transgenic plants overexpressing the transgene. The total DNA was digested with EcoRI. d Performance of transgenic family and WT after severe drought stress. Picture was taken just 1 day before re-irrigation at 6 pm
To assess the effect of OCPI1-overexpression on drought resistance, positive transgenic plants (selected based on hygromycin resistance during seed germination) were grown in the water-managed field facilitated with a moveable rain-off shelter. Irrigation was stopped at 3 weeks before flowering, which allowed a severe drought stress to develop at flowering stage. Although the time of first appearance of leaf rolling showed no obvious difference between transgenic plants and WT, the OCPI1-overexpressed plants had less died leaves than WT did at the severe drought stress stage (Fig. 5d). The grain yields of the three OCPI1-overexpressed families (5.4–6.1 g per plant) were significantly (LSD test, P < 0.05) higher than the negative transgenic control and WT (1.9–2.1 g per plant) under the severe drought stress conditions (with soil water content about 16%). The significant less yield-reduction of transgenic plants was mainly due to significant (LSD test, P < 0.05) higher seed setting rates of the OCPI1-overexpressed transgenic families than the controls (Table 1) since spikelets per plant and kilo-grain weight, the other two yield components, had no significant difference between the transgenic and control plants (data not shown). In addition, no obvious difference was observed between the positive transgenic plants and WT for grain yield and seed setting rate (Table 1) and other morphologic traits (such as plant height, tillers, biomass, leaf area and root volume, data not shown) under normal growth conditions. These results clearly suggest that overexpression of OCPI1 gene can significantly improve drought resistance without obvious negative effect on growth and potential yield in rice.
Chymotrypsin-inhibitory activities and chymotrypsin-like activities in the OCPI1-overexpressing transgenic plants
To examine the effect of over-expressing OCPI1 on chymotrypsin proteinase inhibitory (PI) activity, crude proteins were extracted from the over-expression families TL-20 and TL-25 and the negative transgenic control (TL-21) plants for PI activity assay against bovine chymotrypsin. The standard reaction with bovine chymotrypsin only showed significantly (P < 0.01, t-test of dA256/min value) lower chymotrypsin activity than both the over-expression and non-overexpression transgenic plants (Fig. 6a), indicating that endogenous chymotrypsin-like activity existed in the crude protein from plants. However, the increase of absorbance (dA256/min) in the over-expression transgenic families was less than that in the negative transgenic control, suggesting a stronger chymotrypsin inhibitory activity in the OCPI1-overexpressed transgenic plants than in the negative transgenic control.
Assays of chymotrypsin-inhibitory activity and chymotrypsin activity in protein extracts of rice leaves. Crude total proteins were extracted from 8 week old control (the negative transgenic family TL-21) and positive transgenic (TL-20, TL-25) T1 plants. The activity was determined by measuring the increase of absorbance at 256 nm during hydrolysis of substrate. a Chymotrypsin-inhibitory activity assay (CI) with bovine chymotrypsin. Chymotrypsin activity assay of rice leaf samples from normal growth (b) and drought stress (c). Leaf extract (50 μg) was incubated with substrate without bovine chymotrypsin in b and c. Drought stress was carried out by stopping watering and the completely rolled leaves were sampled. Each value in a–c represents the mean ± SE of three plants
To further evaluate the chymotrypsin inhibitory activity in the transgenic plants, the endogenous chymotrypsin-like activities were determined in the transgenic plants using the same procedures as in the chymotrypsin inhibitory activity assay but with omission of bovine chymotrypsin from the reaction. The increase of A256 of the two transgenic families was significantly (P < 0.01, t-test) lower than the negative transgenic control under both normal (Fig. 6b) and drought stress (Fig. 6c) conditions. The decrease of endogenous chymotrypsin activity in transgenic family TL20, which showed better drought resistance than TL-25, was higher than that in TL25 under both normal and drought stress conditions. These results suggested that the endogenous chymotrypsin-like activity in the OCPI1-overexpressed transgenic plants in rice was significantly inhibited.
The concentrations of crude total proteins from leaves and young panicles (5–10 cm in length) were also compared between transgenic and WT plants. As shown in Table 2, transgenic plants had slightly higher concentrations of total proteins from leaves and panicles than WT plants under both normal and severe drought stress (with relative water content of leaves dropped to 75–78%) conditions. More importantly, the percentage of decreased total protein after drought stress was significantly lower in the transgenic plants than WT (Table 2), suggesting less protein degradation in transgenic plants than in WT under the severe drought stress.
Discussion
In this study, a stress responsive gene, OCPI1, encoding a putative chymotrypsin inhibitor was identified in rice. The most likely physiological function of PIs in plant is to regulate cell proteolysis by inhibiting endogenous proteinases and hence control protein turnover and metabolism (Ryan 1989). It has been suggested that proteinases activity can be considered as a regulatory mechanism in plants (Callis 1995). Protein breakdown and recycling, which depend on the levels of proteolytic enzymes, are essential parts of the plant responses to environmental stresses (Ingram and Bartels 1996; El Maarouf et al. 1999). To date, knowledge of the influence of water deficit on proteinases in plants is mainly limited to the changes of endoproteolytic activities against a few substrates (Zagdanska and Wisniewski 1996). Environmental stresses such as drought have been shown to cause changes in the expression level of genes encoding cysteine proteinases. The cysteine proteinase inhibitor (cystatin) mRNA accumulated in the vegetative tissues of barley plants submitted to anaerobiosis, darkness and cold shock (Gaddour et al. 2001), and cystatin in improving drought tolerance at the cellular level also has been reported (Diop et al. 2004). In this report, a putative chymotrypsin inhibitor gene OCPI1 strongly induced by dehydration stresses was identified and the endogenous chymotrypsin activity was increased in the rice plants treated by drought stress (control plants in Fig. 6b, c). Thus, our results have expanded the spectrum of proteinase inhibitors involved in the stress responses.
It is generally accepted that some specific alterations, such as an increase of proteolytic activities, occur during senescence of plant organs (Huffaker 1990). Drought stress can lead to senescence and differential expression of proteinases and PIs may play important roles in the control of proteolysis by allowing selective mobilization of proteins during senescence (Guo and Gan 2005). Increased cysteine proteinase activities were reported in the senescent carnation flower petals (Jones et al. 1995), drought-induced senescent tomato leaves (Harrak et al. 2001), and senescent leaves of sweet potato (Chen et al. 2002). Furthermore, genes showing high homology with proteinase inhibitor were induced in both dark-induced and natural senescent leaves of barley (Kleber-Janke and Krupinska 1997) and up-regulated in natural senescent leaves of sweet potato (Huang et al. 2001). Our results showed that the expression level of OCPI1 was strongly increased in rice plants subjected to the drought stress (Fig. 2), and the endogenous chymotrypsin activity was also increased in the drought-stressed plants (Fig. 6c). However, we did not observe any difference of senescence (both natural senescence and chlorophyll degradation of detached leaves by dark treatment, data not shown) between the transgenic plants and WT. This may suggest that OCPI1 is specifically involved in stress responses, which is still under investigation.
Despite a proteolytic regulatory system that has been recognized in stress responses of plants (Huffaker 1990; Vierstra 1996), the effect of over-expressing a proteinase inhibitor gene in improving drought resistance of plants growing under the field conditions has not been reported previously. Our data suggest that transgenic rice over-expressing OCPI1 can significantly improve drought resistance in terms of yield loss under severe drought stress conditions in the field. Since the field has a very good homogeneity (plant samples of WT rice from different sites of the field have no significant difference in yield) and the transgenic plants show no differences in plant and root volumes and leaf size, the field variation and drought avoidance (often caused by different plant sizes and root volumes) could be very limited in this experiment. The OCPI1-overexpressing transgenic families also showed somewhat increased tolerance to salt stress under hydroponic culture condition (data not shown) and these families will be further tested in the natural salinity field.
To investigate whether the increased drought resistance of OCPI1-overexpressing transgenic rice was due to an inhibition of endogenous chymotrypsin activity, proteins extracts from transgenic rice were assayed for chymotrypsin activity. Compared to the negative transgenic plant in which OCPI1 was not overexpressed, chymotrypsin activity was inhibited in the OCPI1-overexpressing transgenic plants that showed improved drought resistance, supporting the notion that OCPI1 may have a role in regulating the activity of endogenous proteinases and conferring drought tolerance in rice. Significant less decrease of total protein by drought stress in the transgenic plants provided another supporting evidence for the role of OCPI1 in regulating endogenous proteinases. We also noticed that the inhibition of chymotrypsin-like activity was stronger in the transgenic family TL-20 than TL-25, whereas TL-20 performed better than TL-25 under the drought stress conditions. More transgenic families are under investigation to establish the relationship of inhibited chymotrypsin activity and drought resistance.
Drought stress has emerged as a significant problem in agriculture. Plant engineering has great potentiality in increasing stress tolerance. The significant less yield loss of the OCPI1-overexpressed plants suggests a promising usefulness of OCPI1 in genetic improvement of drought resistance in rice, which, however, needs more trials with more replicates under different stress conditions. Constitutive over-production of a functional molecule sometimes causes abnormalities in plants under normal conditions (though it is not the case for OCPI1 under the control of CaMV 35S promoter in this study). Thus, using a stress-inducible promoter may be desirable. This has been demonstrated by the stable transformation of an ABA-inducible expression system in rice (Su et al. 1998; Kasuga et al. 1999). Cis- and trans-acting elements involved in dehydration-induced gene expression have been extensively analyzed. Many stress-inducible genes are also induced by exogenous application of ABA treatment. These genes contain potential ABA responsive elements (ABREs) in their promoter regions (Ingram and Bartels 1996). The OCPI1 promoter contains several putative stress-responsive cis-acting elements (Fig. 3b). Interestingly, this promoter has strongly stress-inducible activities in both the sense and the antisense strands. Such a promoter with inducible activity in both directions by abiotic stresses may be potentially useful in genetic engineering for stress resistance.
Abbreviations
- ABA:
-
Abscisic acid
- GUS:
-
Beta-glucuronidase
- Hpt:
-
Hygromycin phosphotransferase
- PI:
-
Proteinase inhibitor
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Barr HD, Weatherley PE (1962) A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci 15:413–428
Botella MA, Xu Y, Prabha TN, Zhao Y, Narasimhan ML, Wilson KA, Nielsen SS, Bressan RA, Hasegawa PM (1996) Differential expression of soybean cysteine proteinase inhibitor genes during development and in response to wounding and methyl jasmonate. Plant Physiol 112:1201–1210
Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37:425–435
Callis J (1995) Regulation of protein degradation. Plant Cell 7:845–857
Chen GH, Huang LT, Yap MN, Lee RH, Huang YJ, Cheng MC, Chen SC (2002) Molecular characterization of a senescence-associated gene encoding cysteine proteinase and its gene expression during leaf senescence in sweet potato. Plant Cell Physiol 43:984–991
Choi D, Park JA, Seo YS, Chun YJ, Kim WT (2000) Structure and stress-related expression of two cDNAs encoding proteinase inhibitor II of Nicotiana glutinosa L. Biochim Biophys Acta 1492:211–215
Chu ZH, Peng KM, Zhang LD, Zhou B, Wei JaW SP (2003) Construction and characterization of a normalized whole-life-cycle cDNA library of rice. Chin Sci Bull 48:229–235
Clark AM, Jacobsen KR, Bostwick DE, Dannenhoffer JM, Skaggs MI, Thompson GA (1997) Molecular characterization of a phloem-specific gene encoding the filament protein, phloem protein 1 (PP1), from Cucurbita maxima. Plant J 12:49–61
Diop NN, Kidric M, Repellin A, Gareil M, d’Arcy-Lameta A, Pham Thi AT, Zuily-Fodil Y (2004) A multicystatin is induced by drought-stress in cowpea (Vigna unguiculata (L.) Walp.) leaves. FEBS Lett 577:545–550
Downing WL, Mauxion F, Fauvarque MO, Reviron MP, de Vienne D, Vartanian N, Giraudat J (1992) A Brassica napus transcript encoding a protein related to the Kunitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J 2:685–693
Dunwell JM (2000) Transgenic approaches to crop improvement. J Exp Bot 51 Spec No:487–496
El Maarouf H, Zuily-Fodil Y, Gareil M, d’Arcy-Lameta A, Pham-Thi AT (1999) Enzymatic activity and gene expression under water stress of phospholipase D in two cultivars of Vigna unguiculata L. Walp. differing in drought tolerance. Plant Mol Biol 39:1257–1265
Gaddour K, Vicente-Carbajosa J, Lara P, Isabel-Lamoneda I, Diaz I, Carbonero P (2001) A constitutive cystatin-encoding gene from barley (Icy) responds differentially to abiotic stimuli. Plant Mol Biol 45:599–608
Gosti F, Bertauche N, Vartanian N, Giraudat J (1995) Abscisic acid-dependent and -independent regulation of gene expression by progressive drought in Arabidopsis thaliana. Mol Gen Genet 246:10–18
Guo Y, Gan S (2005) Leaf senescence: signals, execution, and regulation. Curr Top Dev Biol 71:83–112
Habu Y, Fukushima H, Sakata Y, Abe H, Funada R (1996) A gene encoding a major Kunitz proteinase inhibitor of storage organs of winged bean is also expressed in the phloem of stems. Plant Mol Biol 32:1209–1213
Haq SK, Atif SM, Khan RH (2004) Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: natural and engineered phytoprotection. Arch Biochem Biophys 431:145–159
Harrak H, Azelmat S, Baker EN, Tabaeizadeh Z (2001) Isolation and characterization of a gene encoding a drought-induced cysteine protease in tomato (Lycopersicon esculentum). Genome 44:368–374
Hendriks T, Vreugdenhil D, Stiekema WJ (1991) Patatin and four serine proteinase inhibitor genes are differentially expressed during potato tuber development. Plant Mol Biol 17:385–394
Hibbetts K, Hines B, Williams D (1999) An overview of proteinase inhibitors. J Vet Intern Med 13:302–308
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Huang YJ, To KY, Yap MN, Chiang WJ, Suen DF, Chen SC (2001) Cloning and characterization of leaf senescence up-regulated genes in sweet potato. Physiol Plant 113:384–391
Huffaker RC (1990) Proteolytic activity during senescence of plants. New Phytol 116:199–231
Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47:377–403
IRRI (2004) World Rice Statistics. International Rice Research Institute
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Jones ML, Larsen PB, Woodson WR (1995) Ethylene-regulated expression of a carnation cysteine proteinase during flower petal senescence. Plant Mol Biol 28:505–512
Kang SG, Choi JH, Suh SG (2002) A leaf-specific 27 kDa protein of potato Kunitz-type proteinase inhibitor is induced in response to abscisic acid, ethylene, methyl jasmonate, and water deficit. Mol Cells 13:144–147
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Kleber-Janke T, Krupinska K (1997) Isolation of cDNA clones for genes showing enhanced expression in barley leaves during dark-induced senescence as well as during senescence under field conditions. Planta 203:332–340
Leung D, Abbenante G, Fairlie DP (2000) Protease inhibitors: current status and future prospects. J Med Chem 43:305–341
Lilley JM, Ludlow MM, McCouch SR, O’Toole JC (1996) Locating QTLs for osmotic adjustment and dehydration tolerance in rice. J Exp Bot 47:1427–1436
Lin YJ, Zhang Q (2005) Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23:540–547
Lopez F, Vansuyt G, Derancourt J, Fourcroy P, Casse-Delbart F (1994) Identification by 2D-page analysis of salt-stress induced proteins in radish (Raphanus sativus). Cell Mol Biol (Noisy-le-grand) 40:85–90
Mosolov VV, Valueva TA (2005) Proteinase inhibitors and their function in plants: a review. Prikl Biokhim Mikrobiol 41:261–282
Pena-Cortes H, Willmitzer L, Sanchez-Serrano JJ (1991) Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor II gene family. Plant Cell 3:963–972
Pernas M, Sanchez-Monge R, Salcedo G (2000) Biotic and abiotic stress can induce cystatin expression in chestnut. FEBS Lett 467:206–210
Price AH, Steele KAB, Moore J, Barraclough PP, Clark LJ (2000) A combined RFLP and AFLP linkage map of upland rice (Oryza sativa L.) lised to identify QTLs for root-penetration ability. Theor Appl Genet 100:49–56
Rakwal R, Kumar Agrawal G, Jwa NS (2001) Characterization of a rice (Oryza sativa L.) Bowman-Birk proteinase inhibitor: tightly light regulated induction in response to cut, jasmonic acid, ethylene and protein phosphatase 2A inhibitors. Gene 263:189–198
Ryan CA (1989) Proteinase inhibitor gene families: strategies for transformation to improve plant defenses against herbivores. Bioessays 10:20–24
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Satoh H, Uchida A, Nakayama K, Okada M (2001) Water-soluble chlorophyll protein in Brassicaceae plants is a stress-induced chlorophyll-binding protein. Plant Cell Physiol 42:906–911
Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115:327–334
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J 33:259–270
Sin SF, Yeung EC, Chye ML (2006) Downregulation of Solanum americanum genes encoding proteinase inhibitor II causes defective seed development. Plant J 45:58–70
Sin SF, Chye ML (2004) Expression of proteinase inhibitor II proteins during floral development in Solanum americanum. Planta 219:1010–1022
Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11:431–444
Su J, Shen Q, David Ho TH, Wu R (1998) Dehydration-stress-regulated transgene expression in stably transformed rice plants. Plant Physiol 117:913–922
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Tripathy JN, Zhang J, Robin S, Nguyen HT (2000) QTLs for cell-membrane stability mapped in rice (Oryza sativa L.) under drought stress. Theor Appl Genet 100:1197–1202
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Vierstra RD (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32:275–302
Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15:745–759
Xu D, Duan X, Wang B, Hong B, Ho T, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110:249–257
Xu ZF, Teng WL, Chye ML (2004) Inhibition of endogenous trypsin- and chymotrypsin-like activities in transgenic lettuce expressing heterogeneous proteinase inhibitor SaPIN2a. Planta 218:623–629
Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q (2006) Genetic basis of drought resistance at reproductive stage in rice: Separation of drought tolerance from drought avoidance. Genetics 172:1213–1228
Zagdanska B, Wisniewski K (1996) Endoproteinase activities in wheat leaves upon water deficit. Acta Biochim Pol 43:515–519
Zhang J, Zheng HG, Aarti A, Pantuwan G, Nguyen TT, Tripathy JN, Sarial AK, Robin S, Babu RC, Nguyen BD, Sarkarung S, Blum A, Nguyen HT (2001) Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor Appl Genet 103:19–29
Acknowledgments
This research was supported by grants partially from the National Basic Research Program of China, the National Natural Science Foundation of China, Commission of the European Communities (Contract No. INCO-015468) and the Rockefeller Foundation (2004FS070).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, Y., Xiao, B. & Xiong, L. Characterization of a stress responsive proteinase inhibitor gene with positive effect in improving drought resistance in rice. Planta 226, 73–85 (2007). https://doi.org/10.1007/s00425-006-0469-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0469-8