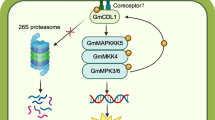
Abstract
Plants recognize microbial pathogens by discriminating pathogen-associated molecular patterns from self-structures. We study the non-host disease resistance of soybean (Glycine max L.) to the oomycete, Phytophthora sojae. Soybean senses a specific molecular pattern consisting of a branched heptaglucoside that is present in the oomycetal cell walls. Recognition of this elicitor may be achieved through a β-glucan-binding protein, which forms part of a proposed receptor complex. Subsequently, soybean mounts a complex defense response, which includes the increase of the cytosolic calcium concentration, the production of reactive oxygen species, and the activation of genes responsible for the synthesis of phytoalexins. We now report the identification of two mitogen-activated protein kinases (MAPKs) and one MAPK kinase (MAPKK) that may function as signaling elements in triggering the resistance response. The use of specific antisera enabled the identification of GmMPKs 3 and 6 whose activity is enhanced within the signaling pathway leading to defense reactions. Elicitor specificity of MAPK activation as well as the sensitivity against inhibitors suggested these kinases as part of the β-glucan signal transduction pathway. An upstream GmMKK1 was identified based on sequence similarity to other plant MAPKKs and its interaction with the MAPKs was analyzed. Recombinant GmMKK1 interacted predominantly with GmMPK6, with concomitant phosphorylation of the MAPK protein. Moreover, a preferential physical interaction between GmMKK1 and GmMPK6 was demonstrated in yeast. These results suggest a role of a MAPK cascade in mediating β-glucan signal transduction in soybean, similar to other triggers that activate MAPKs during innate immune responses in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are able to defend themselves against potential pathogens. Mounting of multicomponent plant defenses is facilitated through non-self-recognition systems for numerous pathogen-derived compounds called elicitors and through subsequent signal transduction pathways, which mediate the activation of defense mechanisms in either a cultivar-specific or non-cultivar-specific mode depending on the type of elicitor. Among the latter group are the hepta-β-glucoside fragment from oomycete cell wall polysaccharides, fungal cell wall chitin fragments, the Pep-13 motif from Phytophthora transglutaminases, and flg22, a peptide within the most conserved domain of the N-terminal part of bacterial flagellin (Ebel and Mithöfer 1998; Felix et al. 1999; Nürnberger and Scheel 2001; Brunner et al. 2002). Sensing of elicitors by proposed receptors is generally followed by early changes in levels of free calcium ions in the cytoplasm and the production of reactive oxygen species and nitric oxide (Nürnberger and Scheel 2001). Elicitor signal transduction has been proposed to involve protein phosphorylation (Ebel and Mithöfer 1998; Nürnberger and Scheel 2001), suggesting that protein kinases are activated or deactivated during the process. Pharmacological studies using inhibitors implicated that defense responses are likely to be controlled by a balanced action of protein kinases and phosphoprotein phosphatases (Nürnberger and Scheel 2001). Serine/threonine kinase inhibitors did not only block ion fluxes and protein phosphorylation, but also defense activation, while protein phosphatase inhibitors stimulated the inducible defenses in the absence of elicitors. In addition, salicylic acid (SA), jasmonic acid, and ethylene are mediators that can modulate responses and give rise to the observed specificity in plant-pathogen interactions by regulating the appropriate multicomponent defense reactions.
The induction of sets of mitogen-activated protein kinase (MAPK) cascades is a common response of plants to abiotic and biotic stimuli including pathogen infection and elicitor treatment (Romeis 2001; Zhang and Klessig 2001). Based on the analysis of the Arabidopsis thaliana genome sequence, at least 20 genes encoding putative MAPKs were identified which fall into 4–6 groups (Tena et al. 2001; Zhang and Klessig 2001; MAPK Group 2002). In all plant systems where elicitor- or infection-mediated effects on MAPK activity were studied, activation of closely related members of the AtMPK6 sub-group was found (Nürnberger and Scheel 2001; Zhang and Klessig 2001; Nakagami et al. 2005). This includes the responsiveness of tobacco SIPK to general elicitors, race-specific elicitation, and TMV infection, of alfalfa SIMK to carbohydrate elicitors and ergosterol, and of the A. thaliana AtMPK6 to bacterial elicitors. Members of a second group of MAPKs with homology to AtMPK3, such as tobacco WIPK and parsley ERM kinase (PcMPK3a) were also associated with pathogen defense. Recently, two separate, but interdependent, MAPK cascades were proposed to operate downstream of AvrPto-Pto defense signaling in tomato resistance to bacterial speck disease (del Pozo et al. 2004; Pedley and Martin 2004). The tomato studies confirmed the major findings in tobacco, showing that the orthologues of tobacco SIPK and WIPK, tomato MPK1, MPK2, and MPK3, are activated in this system (Pedley and Martin 2005).
Several MAPK kinases (MAPKKs) as members of MAPK pathways have been identified from different plants, including Arabidopsis, alfalfa, maize, tobacco, and tomato, and their involvement in pathogen defense has been established (MAPK Group 2002; Nakagami et al. 2005; Pedley and Martin 2005). The results further indicated that the activation of MAPKs was dependent not only on pair-wise affinities between MAPKKs and MAPKs but also on complex signal-dependent intracellular mechanisms. Through a combination of transient expression analyses, biochemical, and genetic approaches, AtMPK3 and AtMPK6 were placed into the first complete MAPK cascade identified in plants (Asai et al. 2002). In this system, following flg22 perception by the flagellin receptor FLS2 of Arabidopsis (Chinchilla et al. 2006) the MEKK1-MKK4/MKK5-MPK3/MPK6 module acts downstream of the receptor and upstream of the WRKY22, WRKY29, and defense-related genes (Asai et al. 2002). Transient overexpression of the MEKK1 kinase domain, constitutively active MKK4 and MKK5 or WRKY29 rendered Arabidopsis leaves resistant to infection by fungal and bacterial pathogens (Asai et al. 2002). By using similar approaches, support for a causal link between MAPK activation and defense gene induction has also been obtained for tobacco, tomato, and parsley (for review, see Nakagami et al. 2005; Pedley and Martin 2005). As MAPKs have also been implicated in relaying adaptation to many other stimuli in plants, MAPK cascades apparently constitute central components in complex signaling networks (Nürnberger and Scheel 2001; Nakagami et al. 2005). Evolutionary aspects of plant MAPK cascades have been described in greater detail (MAPK Group 2002) and may give a basis for a systematic analysis of plants not covered so far.
In soybean, two recent reports addressed the involvement of MAPKs in signaling events upon challenge by external stimuli. Wounding of soybean plants activated a MAPK that was recognized by an antibody against alfalfa SIMK (Lee et al. 2001a). Wound activation of the MAPK was prevented by an inhibitor of phosphatidic acid production, and exogenously applied phosphatidic acid activated the MAPK in soybean cell cultures. The correlation between activation of two MAPKs and of the oxidative burst was analyzed in soybean cells employing different elicitor fractions, including an oligogalacturonide preparation (Taylor et al. 2001). In the present biochemical study, we examined the effects and specificity of β-glucans, known as potent elicitors of a phytoalexin defense response in soybean, on MAPK activities and searched for a potential upstream MAPKK mediating signal transduction in soybean.
Materials and methods
Cell culture propagation, elicitor treatment, and extraction of phytoalexins
Soybean (Glycine max L. cv. Harosoy 63) cell suspension cultures were obtained from our culture collection, propagated in the dark, and treated with a β-glucan elicitor fraction from Phytophthora sojae (100 μg/ml glucose equivalents) as described earlier (Hille et al. 1982; Fliegmann et al. 2003). For immunoblot analyses and kinase assays, soybean cultures were sub-cultured in fresh medium (4 g fresh mass per 40 ml medium) in the dark for 2 h with shaking (110 rpm) and 1 h without. Aliquots of 500 μl were transferred into Eppendorf tubes and allowed to settle. The supernatants were removed and the cells were treated with glucan elicitors. After 20 min, the medium was removed with a syringe and the cells were frozen in liquid nitrogen. Other chemicals to be tested were added in water or dimethyl sulfoxide (final assay concentration <0.1%). Cell free protein extracts were prepared according to Suzuki and Shinshi (1995), modified after Bögre et al. (1997) as described (Fliegmann et al. 2003), and the protein concentration determined using the method of Bradford (1976). Phytoalexins were extracted from cell cultures after treatment with glucan elicitor for 48 h and were determined as described (Fliegmann et al. 2003).
Immunoblot analysis
Protein extracts containing 50 μg of total protein per lane were separated by SDS-polyacrylamide gelelectrophoresis (PAGE) (10% acrylamide in the separating gel), and the proteins were transferred onto nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany, or PALL) using a semi-dry electroblotting unit. Blocking was performed in 50 mM Tris–HCl, pH 7.8, containing 150 mM NaCl, 1 mM MgCl2 and 0.05% tergitol supplemented with 5% low-fat milk powder, for 1 h at room temperature. The blots were probed over night at 4°C with primary antibodies (M23 or M24, respectively, raised against C-terminal peptides of MMK1 (M23) or MMK4 (M24) of Medicago sativa L.; Cardinale et al. 2000) dissolved in blocking suspension (1:5,000 and 1:2,500, respectively). An anti-rabbit alkaline phosphatase-conjugated secondary antibody (1:20,000, Sigma St. Louis, MO, USA) was used to detect the antibody-protein complexes by the hydrolysis of the substrate, tetrazolium-5-bromo-4-chloro-3-indolyl phosphate.
Kinase assays
In-gel kinase activity assays (Suzuki and Shinshi 1995) were performed as described (Fliegmann et al. 2003). Relative kinase activity was analyzed by visualizing the phosphorylated myelin basic protein (MBP, Sigma) using a phosphorimager and was documented by autoradiography.
Immune complex-kinase assays of GmMPK3 and GmMPK6 were performed according to Jonak et al. (1996) with modifications by incubating protein extracts (50 μg total protein, unless otherwise noted) with anti-MAPK antiserum (M23 and M24) and protein A-Sepharose over night at 4°C. The precipitates were washed twice with 20 mM Tris–HCl, pH 7.5, 5 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 100 mM NaCl, 1 mM Triton X-100 and once with kinase buffer containing 20 mM Hepes, pH 7.5, 15 mM MgCl2, 5 mM EGTA, 1 mM dithiothreitol, and 0.2 mg/ml bovine serum albumin. The kinase reaction was started by adding 25 μl kinase buffer supplemented with 100 μg/ml MBP, 100 μM ATP, and 37 KBq [γ-32P]ATP. After 30 min at room temperature, the reaction was stopped by adding SDS sample buffer and heating to 100°C for 5 min. Kinase activity was analyzed after SDS-PAGE (15% acrylamide) as described above.
Phosphorylation of MBP by partially purified recombinant GmMPK3His6 and GmMPK6His6 was assayed by incubating 1 μg of protein after immobilized metal chelate affinity chromatography (IMAC) (see below) with 100 μM ATP and 37 KBq [γ-32P]ATP in 30 mM Hepes, pH 7.5, 40 mM KCl, 4 mM MgCl2, 5.2% glycerol in a total volume of 50 μl. After incubation for 30 min at room temperature, 1 μg of MBP was added. After a further 30 min at room temperature, proteins were precipitated by adding 1 ml of cold methanol. The samples were held at −20°C for 2 h and then centrifuged at 10,000 g for 15 min at 4°C. The proteins were dissolved in 20 μl of SDS sample buffer, heated for 5 min at 100°C, and separated by SDS-PAGE (15% acrylamide). Relative kinase activity was analyzed as described above.
Phosphorylation of partially purified recombinant GmMPK3His6 and GmMPKHis6 as substrate was studied with recombinant GmMKK1His6. The MAP kinases (equal amounts) were incubated with and without GmMKK1His6 and 50 μM ATP containing 148 KBq [γ–32P]ATP in 20 mM Tris–acetate, pH 7.5, 10 mM MgCl2, 1 mM NaF, 1 mM dithiothreitol, 2 μg aprotinin/ml in a total volume of 35 μl for 1.5 h at 30°C. After precipitation with cold methanol, the proteins were subjected to SDS-PAGE (10% acrylamide). Relative kinase activity was analyzed as described above.
Phosphoamino acid analysis of GmMPKHis6
The phosphoamino acid analysis was performed as published (Contor et al. 1987; Boyle et al. 1991). After individual phosphorylation assays with GmMPK6His6 as substrate in the presence or absence of GmMKK1His6, 20 samples were pooled and subjected to SDS-PAGE. Following transfer to a nitrocellulose membrane by using a semi-dry electroblotting unit, the proteins were localized by Ponceau S staining. The area of the membrane corresponding to the GmMPK6His6 band was cut out, the Ponceau S dye was removed, and the proteins were eluted over night in 100 mM ammonium acetate, pH 8.9, and 50% pyridin at 37°C. The samples were dried in a vacuum concentrator, hydrolyzed in 6 N HCl for 1 h at 110°C, and the hydrolysate was dried again. The residue was dissolved in 5 μl of 2.5% formic acid and 7.8% acetic acid, pH 1.9, containing phosphoamino acid standards (P-S, P-T, and P-Y), and the solution spotted onto a TLC plate (cellulose, 0.1 mm, POLYGRAM CEL 300, Macherey-Nagel, Düren, Germany). Electrophoresis was carried out in 2.5% formic acid with 7.8% acetic acid, pH 1.9, for 1.5 h at 500 V in the first dimension and in 5% acetic acid with 0.5% pyridine, pH 3.5, for 1.75 h at 500 V in the second dimension. The phosphoamino acid references were visualized by ninhydrin and the incorporation of radioactivity was analyzed by phosphorimaging.
MAP kinase and MAPK kinase cDNA selection
A soybean root cDNA library (Mithöfer et al. 2000) was screened by filter hybridization using the MMK4 cDNA clone of M. sativa L. as a probe (Jonak et al. 1996). Hybridization conditions were according to Fliegmann et al. (2003). Membranes were washed at high-stringency conditions for 20 min twice in 0.5× standard saline citrate (SSC), 0.1% (w/v) SDS at 65°C and autoradiographed between intensifying screens at −80°C. A positive clone (GmMPK3, AF104247) carrying an open reading frame of 1,116 bp was sequenced and shown to encode a protein of 371 amino acids with a calculated molecular mass of 42.7 kDa. The cDNA was cloned into pTrcHisC (Invitrogen, Carlsbad, CA, USA) using PCR in order to eliminate the start codon by introducing a XhoI site (forward primer: 5′-aactgatggctcgagttaat-3′; reverse primer: vector primer).
The cDNA encoding the soybean MMK1 homologue GmMPK6 (AF329506) was isolated by using PCR. A cDNA library from a non-elicited soybean cell suspension culture (5 days after sub-culturing) was used as a template. The degenerated forward oligonucleotide (5′-atactcgagaahaatggawggwggagswsmws-c-3′) was designed after comparing the cDNA sequences of known plant MMK1 homologues, the reverse primer (5′-tagaagcttctactgctgatactcagggttaaatgctag-3′) corresponded to the coding sequence (3′-end) of an expressed sequence tag (EST) clone from soybean (AW102298) that showed homology to MAP kinase MMK1 from M. sativa. The resulting PCR fragment was verified as MMK1 homologue (open reading frame of 1,176 bp) by sequencing and performing BLAST homology search and cloned into the expression vector pQE-31 (Qiagen, Crawley, UK). The calculated molecular mass of GmMPK6 consisting of 391 amino acids was 44.8 kDa.
The cDNA encoding the soybean GmMKK1 (AY070230) was isolated by screening a soybean root cDNA library (as above, Mithöfer et al. 2000; see Fliegmann et al. 2003) with the soybean EST clone (BE059041) as a probe that showed high homology to the 5′-sequence of the tobacco SIPKK in a BLAST homology search. The resulting clone (AY070230) was sequenced and verified as a SIPKK homologue by BLAST homology screening. The cDNA encoded a protein of 356 residues corresponding to 39.5 kDa and was cloned into the expression vector pTrcHisA (Invitrogen) using PCR in order to eliminate the start codon by introducing a XhoI site (forward primer: 5′-tactcgagaagaaaggaaacttggg-3′; reverse primer: vector primer).
Data retrieval and multiple sequence alignment
Amino acid sequences of MAPK and MAPKK used for comparisons were deduced from the following nucleotide sequence accessions: M. sativa MsMMK1 (SIMK, acc. X66469), MsMMK4 (SAMK, X82270), MsPRKK (AJ293275), and MsSIMKK (AJ293274); Nicotiana tabacum NtSIPK (U94192), NtWIPK (D61377), NtMEK1 (AJ302651), NtMEK2 (AF325168), and NtSIPKK (AF165186); A. thaliana AtMPK3 (D21839), AtMPK6 (D21842), and AtMKK2 (AB015313); Lycopersicon esculentum LeMEK1 (AJ000728); Petroselinum crispum PcMPK3a (Y12785), PcMPK3b (AY173415), PcMPK6 (AY173413), PcMKK2 (AY533301), and PcMKK5 (AY533302). Amino acid sequences were aligned using MultAlin (Corpet 1988) and optimized manually. Pair-wise identities using the optimized alignments were calculated using the BioEdit Sequence Alignment Editor (Hall 1999).
Production in E. coli and isolation of recombinant proteins
Escherichia coli strains SG13009 harboring the plasmid pQE-31/GmMPK6 and Bl21(DE2)pLysS harboring pTrcHisC/GmMPK3 or pTrcHisA/GmMKK1 were grown at 37°C until A600 ≈ 0.5 was reached, induced with 1 mM isopropyl-β-D-thiogalactopyranoside, and incubated for 4 h at 37°C. After centrifugation, the bacterial cells were resuspended in 25 mM Tris–HCl, pH 8.0, containing 5 mM MgCl2, 200 mM NaCl, and 0.5 mM phenylmethylsulfonyl fluoride and disrupted by sonication (SG13009 cells) or by a freeze-thaw cycle (Bl21(DE2)pLysS cells). After removing cellular debris by centrifugation (10,000 g for 10 min at 4°C), the recombinant fusion proteins containing N-terminal His6-tags were purified by IMAC using a Ni-NTA metal affinity matrix (Qiagen) according to the instructions of the manufacturer. The eluents were frozen immediately in liquid nitrogen and stored at −80°C.
Interaction of GmMPKHis6 with GmMKK1His6 in yeast
The ProQuest™ Two-Hybrid System (Invitrogen) was used to analyze the interaction of GmMPKs and GmMKK1 in Saccharomyces cerevisiae. Both MAP kinase cDNAs and the MAPKK cDNA were inserted into pDBLeu or pPC86 using SalI and NotI restriction sites. Oligonucleotides containing 5′ SalI and 3′ NotI sites were used to amplify the respective cDNAs. The yeast strain MaV203 was co-transformed with pDBLeu/GmMPK3 (or pDBLeu/GmMPK6) and pPC86/GmMKK1. Both combinations were examined for the activation of the HIS3 reporter gene and for the activation of lacZ in a membrane-based X-Gal-assay. To determine the threshold of resistance to 3-amino-1,2,4-triazole (3AT), a series of increasing 3AT concentrations up to 100 mM was included in plates lacking histidine. Activation of the HIS3 reporter gene was analyzed in the presence of 25 mM 3AT and the absence of histidine, leucine, and tryptophan in the growth medium. The strength of interaction was estimated by comparing the results of growth assays with the behavior of control strains under the same conditions.
Results
Phytophthora β-glucan elicitors induce protein kinase activation in cultured soybean cells
Among the multiple responses to elicitor compounds in plants, the induction of MAPKs was reported in different systems (Nürnberger and Scheel 2001). For the activation of protein kinases in soybean, suspension-cultured cells were treated with β-glucan elicitors from the oomycete pathogen of this plant, P. sojae. Cell samples were collected at different time points after onset of treatment and cell free extracts of the samples were analyzed by in-gel kinase assays using MBP as an artificial substrate. As shown in Fig. 1a, protein kinases with relative molecular masses between 43 and 45 kDa were rapidly activated in elicitor-treated cells. Activities increased at ∼5 min following onset of treatment, highest activity levels occurred at ∼20 min, and thereafter returned to basal levels at ∼120 min (Fig. 1b). These results show that P. sojae β-glucan elicitors induce rapid and transient protein kinase activation in soybean cells, reminiscent to several other rapid responses that have been observed after elicitor treatment in this plant (Mithöfer et al. 1997, 1999, 2001, 2005; Ebel and Mithöfer 1998).
Enhancement of MBP kinase activities by β-glucan elicitors in soybean cell cultures. a, b Time course of elicitor-induced activity changes. Soybean cells were treated with a β-glucan elicitor fraction from P. sojae (100 μg/ml glucose equivalents) for various periods of time. Cells were harvested, cell free extracts prepared, and kinase activities measured by in-gel kinase assays using MBP as substrate (a). b Kinase activities at each time point were determined in triplicate by phosphorimage analysis and plotted against length of treatment (filled circle β-glucan elicitor, open circle control using equivalent of water). c Concentration dependence of kinase activation by β-glucan elicitors. Cells were treated with various concentrations of β-glucan elicitor for 20 min. MBP kinase activities were measured as indicated above. Activity levels were plotted against the maximum response seen following treatment of cells with 1,000 μg/ml β-glucan elicitor. d Differential effects of various β-glucan fractions on kinase activity levels. Cells were treated with different β-glucan fractions (100 μg/ml of CE; 50 μM for all other glucan fractions) for 20 min. Kinase activities in response to each treatment were determined in triplicate as indicated above and plotted against the level found with crude β-glucan elicitor (CE crude elicitor; 100%). Abbreviations denote: c control, DP5 purified β-glucan fraction with degree of polymerization (DP) of 5, DP < 7 purified β-glucan fraction with DP smaller than 7, DP 7–15 (DP 15–25) purified β-glucan fractions with different size ranges, HG synthetic hepta-β-glucoside, cycl. cyclic β-glucan from Bradyrhizobium japonicum. β-Glucan sources were as reported earlier (Mithöfer et al. 1999)
The effectiveness of the β-glucan elicitors to cause protein kinase activation was analyzed by exposing soybean cells to different glucan concentrations for 20 min (Fig. 1c). Over a concentration range between 3 and 1,000 μg/ml of β-glucans, protein kinase activation differed greatly, with half-maximal activation occurring at about 60 μg/ml. Protein kinase activation also strongly depended on the chemical composition and the size of the β-glucan fragments (Fig. 1d). An unfractionated β-glucan extract from purified P. sojae cell walls caused an approximately tenfold enhancement of protein kinase activity level in soybean cells. Similarly effective were (1,3–1,6)-β-glucan fragments with a degree of polymerization (DP) >7, whereas little inducing capacity resided in fragments of DP = 5 or <7. The chemically defined hepta-β-glucoside (Sharp et al. 1984a, b) was as active as β-glucan fragments with DP > 7 of the natural polysaccharide. In contrast, the cyclic (1,3–1,6)-β-glucan from Bradyrhizobium japonicum, described as a suppressor of linear β-glucan-elicited defense in soybean (Mithöfer et al. 1996), possessed a low capacity to enhance protein kinase activity levels. The relative efficiency of the β-glucans to cause kinase activation is thus in good agreement with several other β-glucan-mediated responses in soybean cells (Cosio et al. 1990; Mithöfer et al. 1996, 1999, 2001).
Influence of inhibitors and antagonists on protein kinase activation
Various inhibitors and antagonists have earlier been found to affect several β-glucan elicitor-mediated responses in soybean cells to different degrees. In particular, putative anion channel inhibitors and calcium antagonists blocked rapid Ca2+ influx into the cells and subsequent phytoalexin production, whereas diphenyleneiodonium (DPI), a known inhibitor of mammalian NAD(P)H oxidase, inhibited an oxidative burst in soybean, but did not affect calcium flux and phytoalexin synthesis (Stäb and Ebel 1987; Mithöfer et al. 1997, 1999, 2001).
As demonstrated in Table 1, the putative anion channel inhibitors, anthracene-9-carboxylate (A9C) and 5-nitro-2-(3-phenylpropylamino)-benzoate (NPPB), efficiently blocked the activation of MBP kinase activity at low concentrations. The concentration causing 50% inhibition (IC50) of the MBP kinase response was 10 μM for A9C and 7 μM for NPPB. Likewise, the IC50 for the calcium antagonists, La3+ and EGTA, was 1 and 2.5 mM, respectively. Similar values for the calcium antagonists were found for causing inhibition of elicitor-mediated enhancement of the cytosolic Ca2+ concentration (Mithöfer et al. 1999). The comparatively low concentration of La3+ resulting in the inhibition of phytoalexin production (Table 1) may result, at least in part, from the long exposure time (96 h) of the cells to La3+ (Stäb and Ebel 1987) when compared with treatments for the MBP kinase assay. In contrast, DPI was inactive in the concentration range tested (Table 1). These data support the hypothesis that there exist two parallel pathways in soybean cells for triggering defense reactions in response to β-glucan signals, one leading to the oxidative burst, and a second being independent of this response and resulting in the activation of phytoalexin production concomitantly with calcium and MBP kinase(s) responses.
Use of M23 and M24 antibodies to recognize MAPKs and to reveal their β-glucan-induced activation
To investigate whether the 43- and 45-kDa protein kinases correspond to known MAPKs from alfalfa (M. sativa), protein extracts from β-glucan-treated and untreated soybean cells were subjected to immunoreactions with antibodies specifically recognizing SIMK (MMK1) and SAMK (MMK4) from alfalfa. Antibodies raised against C-terminal peptides of SIMK (MMK1, M23) and SAMK (MMK4, M24; Munnik et al. 1999; Cardinale et al. 2000) were used to probe proteins of cell extracts after SDS-PAGE and transfer to nitrocellulose membranes. As shown in Fig. 2a, a major crossreactive protein of ∼45 kDa could be visualized with antibody M23, whereas M24 detected a protein of ∼43 kDa. Antibody M23 appeared to be less specific than antibody M24 against soybean proteins as the former detected a protein of 43 kDa in addition to the 45-kDa protein. Immunodetection of the 45- and 43-kDa proteins could be inhibited by the peptides against which the corresponding antibodies were raised (Cardinale et al. 2000; Fig. 2a). As was shown later (see below), the cDNAs encoding the two 45- and 43-kDa protein kinases contained C-terminal peptides that are entirely conserved when compared with SIMK and SAMK, respectively. Peptide P13 specifically inhibited immunodetection of the 45-kDa protein by the M23 antibody. Peptide P14 competed the immunodetection of the 45- and 43-kDa protein by the M23 and M24 antibody, respectively. This cross competition might be explained by the high degree of similarity between the P13 and P14 peptides (Munnik et al. 1999; Cardinale et al. 2000).
Use of MAPK-specific antisera and immunoprecipitation/protein kinase assays to identify 43- and 45-kDa protein kinases as MAPKs. a Specificity of antibodies raised against peptide sequences contained in alfalfa MMK1 (SIMK) and MMK4 (SAMK), respectively. Antisera crossreactivity was tested by Western blotting following separation by SDS-PAGE of cell free protein extracts from cultured soybean cells and using antibodies M23 directed against MMK1 or M24 against MMK4 in the presence or absence of competitor peptides (20 μg/ml) corresponding to the C-termini of MMK1 (P13) and MMK4 (P14; Cardinale et al. 2000). An anti-rabbit alkaline phosphatase-conjugated secondary antibody was used to detect primary antibody-protein complexes. b Immune complex-protein kinase assays (upper part) and immune complex-in-gel kinase assays (lower part). Soybean cells were elicited with β-glucans, and cell free extracts containing 50 μg protein were incubated with the indicated antiserum (upper part) and protein A-Sepharose. The immune complexes adsorbed to protein A-Sepharose were subsequently tested for kinase activity by measuring incorporation of 32P from [γ32P]ATP into MBP visualized following separation of the reaction products by SDS-PAGE. Immune complex-in-gel kinase assays (lower part) were performed with protein fractions extracted from immune complexes adsorbed to protein A-Sepharose. In-gel kinase activity was monitored by phosphorimage analysis
To support the protein kinase function of the 45- and 43-kDa proteins, immunoprecipitation-protein kinase assays were performed (Fig. 2b). Equal amounts of protein extracted from β-glucan-treated and untreated cells were immunoprecipitated with the antibodies. The immunoprecipitated proteins were subjected to in vitro kinase assays using MBP as substrate, and the reaction products were analyzed by SDS-PAGE followed by autoradiography. As demonstrated in Fig. 2b (upper part), the precipitates with both antibodies showed kinase activity, which was higher in the extracts from elicitor-treated compared with extracts from untreated cells. The precipitate with the M23 antibody contained the 45-kDa kinase, as documented by a subsequent in-gel kinase assay (Fig. 2b, lower part). Although the M23 antibody also detected a 43-kDa protein (Fig. 2a) no 43-kDa kinase could be detected in the precipitate. A similar assay using the M24 antibody for the suspected 43-kDa kinase failed. This might have been due to a low recovery of the protein or to a loss of activity during the handling. These results indicate that soybean cells contain MAPKs whose activity is induced by β-glucan elicitor treatment of the cells and that these are related to MAPKs from alfalfa.
Isolation of GmMPK3, GmMPK6, and GmMKK1 cDNAs and interactions in the yeast two-hybrid system
To identify the MAPKs detected in the Western blotting, immunoprecipitation, and in-gel kinase assays, and to study an upstream MAPKK whose function might be correlated with elicitor signal transduction we decided to clone different cDNAs encoding MAPK members. Screening of a soybean root cDNA library (Mithöfer et al. 2000) with the cDNA encoding SAMK of M. sativa as a probe (Jonak et al. 1996) resulted in the identification of GmMPK3 cDNA. The open reading frame coded for a protein of 371 amino acids with a calculated molecular mass of 42.7 kDa. The cDNA encoding the soybean homologue (GmMPK6) of the alfalfa SIMK (Jonak et al. 1993) was isolated using PCR and a cDNA library from a non-elicited soybean cell culture as template. The forward oligonucleotide was designed according to the SIMK cDNA and the reverse primer corresponded to the coding sequence of an EST clone from soybean that showed homology to the alfalfa SIMK cDNA (3′-end). The open reading frame contained in the PCR fragment encoded a protein of 391 amino acids with a calculated molecular mass of 44.8 kDa for GmMPK6. Comparison of the deduced amino acid sequences of the encoded proteins showed a high level of sequence identity when compared with SIMK and SAMK and other related kinases of this group (Table 2).
The cDNA encoding a putative soybean MAPKK (GmMKK1) was isolated by screening a soybean root cDNA library (Mithöfer et al. 2000) with the soybean EST clone, BE059041, as a probe that showed high homology to the MAPKK amino acid sequences of alfalfa PRKK (Cardinale et al. 2002) and tobacco SIPKK (Liu et al. 2000). The resulting GmMKK1 clone was verified as an alfalfa PRKK homologue with a high level of sequence identity when compared with other known plant MAPKKs (Table 3).
To study protein-protein interactions in S. cerevisiae between the soybean MAPKs and the MAPKK, both MAPK cDNAs and the MAPKK cDNA were inserted into pDBLeu or pPC86, respectively. The yeast strain MaV203 was co-transformed with pDBLeu/GmMPK3 (or pDBLeu/GmMPK6) and pPC86/GmMKK1. The transformants were examined for the ability to confer histidine prototrophy to the yeast cells and for the activation of lacZ using an X-Gal assay. The results presented in Supplementary Fig. S1 demonstrate that the GmMKK1-GmMPK6, but not the GmMKK1-GmMPK3 combination resulted in the expression of the lacZ reporter gene. In the assay for histidine prototrophy, both kinase combinations indicated at least some degree of histidine independent growth.
Recombinant GmMKK1 is an active MAPKK and accepts GmMPK6 as substrate
To determine whether GmMKK1 encodes a functional MAPKK, the activity of recombinant GmMKK1 using the GmMPKs as putative substrates was examined. The cDNAs encoding the three kinases were expressed in E. coli as inducible fusion proteins containing N-terminal His6-tags. Following immobilized metal affinity chromatography, the recombinant kinase proteins showed the expected molecular masses as calculated for the fusion proteins when using the in-gel kinase assay and crossreacted with the antisera M23 and M24 directed against the M. sativa kinases (results not shown). When subjected to in-gel kinase assays, both GmMPK3 and GmMPK6 displayed some basal kinase activity with MBP as substrate. Phosphorylation of the recombinant MAPKs as substrate by GmMKK1 was analyzed by incubating equal amounts of the purified MAPKs with purified recombinant GmMKK1 in the presence of [γ-32P]ATP. After precipitation with methanol, the proteins were separated using SDS-PAGE, and the relative GmMKK1 activity analyzed using a phosphorimager. As shown in Fig. 3a, GmMKK1 caused a significant enhancement of the phosphate content of GmMPK6 but was inactive toward GmMPK3. To localize the radioactive phosphate group, samples of GmMPK6 before and after incubation with GmMKK1 were subjected to phosphoamino acid analysis. Following hydrolysis using 6 N HCl and separation of the amino acid mixture by two-dimensional electrophoresis, phosphotyrosine was the predominantly labeled amino acid (Fig. 3b). The results indicate that recombinant GmMKK1 is a functional MAPKK enzyme, which is able to preferentially catalyze the phosphorylation of one of the identified MAPKs, GmMPK6, in vitro.
GmMKK1 is a functional enzyme and catalyzes the differential phosphorylation of GmMPKs. a Recombinant GmMPK3 and 6 were used as substrates for recombinant GmMKK1. Proteins purified by IMAC were incubated in the presence of [γ32P]ATP for 1.5 h and the reactions terminated by adding methanol. After precipitation with methanol, the proteins were separated using SDS-PAGE, and the relative activity of GmMKK1 analyzed using a phosphorimager. b The phosphoamino acid composition of recombinant GmMPK6 was analyzed following incubation with [γ32P]ATP in the presence or absence of GmMKK1. Following SDS-PAGE and transfer of the proteins to a nitrocellulose membrane, the areas of the membrane corresponding to GmMPK6 were eluted and dried in a vacuum concentrator. After hydrolysis in 6 N HCl, the reaction products were mixed with phosphoamino acid standards and were separated by two-dimensional electrophoresis on a TLC plate coated with cellulose. The phosphoamino acid references were visualized by ninhydrin and the incorporation of radioactivity was analyzed by phosphorimaging. Abbreviations denote: P-S phosphoserine, P-T phosphothreonine, P-Y phosphotyrosine
Discussion
Remarkable similarities exist between mammals, insects, and plants for the systems by which microbial pathogens are sensed and defenses are activated to combat infections (Janeway and Medzhitov 2002; Nürnberger et al. 2004). In the well-studied cases of mammals and insects, pattern recognition receptors are biosensor proteins that interact with pathogen-associated molecular patterns and subsequently transduce signals necessary for activation of an appropriate innate immune response (Medzhitov and Janeway 1997). For non-host resistance, plants likewise employ sensitive perception systems to respond to pathogen-associated molecular structures, referred to as elicitors, with the transcriptional activation of defense-related genes and the synthesis of antimicrobial phytoalexins (Ebel and Mithöfer 1998; Nürnberger and Scheel 2001).
Our previous studies on the interactions of soybean with P. sojae supported the general conclusions about the analogy of the defense systems. Surface-exposed β-glucan elicitors representing a fraction of a structural polysaccharide of P. sojae, a typical component of oomycetal cell walls, induced several metabolic changes in cell cultures and other tissues of soybean, including rapid calcium fluxes (Mithöfer et al. 1999, 2005), the production of H2O2 (Mithöfer et al. 1997, 2001), changes in the phosphorylation status of proteins, and subsequently the synthesis of phytoalexins (for review, see Ebel and Mithöfer 1998). The results were compatible with the assumption that an oxidative burst-independent pathway controls the transcriptional activation of genes involved in phytoalexin synthesis which may be connected with the rapid transient increase in the cytosolic calcium concentration (Mithöfer et al. 1997, 2001; Ebel and Mithöfer 1998). The interaction of β-glucan elicitors with the β-glucan-binding protein, a proposed β-glucan receptor, has been suggested to be the critical event initiating defense reactions (Mithöfer et al. 2000; Fliegmann et al. 2004) implicated in the innate immune response of soybean.
Important components of the innate immune signaling pathway of plants are MAPK cascades (Lee et al. 2001b, 2004; Asai et al. 2002; He et al. 2006). In the present study, we have identified two β-glucan-responsive MAPKs and an upstream MAPKK, which might be members within a MAPK pathway involved in defense response signaling in soybean. GmMPK3 and GmMPK6 are activated by β-glucan elicitors of P. sojae under physiological conditions that are favorable for inducing phytoalexin production in soybean cell cultures and other tissues of this plant. The activation profile observed for the MAPKs when different size classes and structurally divergent β-glucan fractions were used as elicitor closely matched the relative activities obtained with the same fractions in ligand-binding assays with the proposed β-glucan receptor (Mithöfer et al. 2000), and in bioassays on phytoalexin production (Cosio et al. 1990). This close correspondence also included the dose-response relationships for the differential enhancement of the cytosolic calcium level by the glucan fractions (Mithöfer et al. 1999). Conversely, inhibitors, which are known to adversely interfere with the β-glucan-mediated stimulation of phytoalexin synthesis also prevented activation of the MAPKs, except for DPI that did not affect enhancement of kinase activity and phytoalexin accumulation but inhibited H2O2 production. MAPKK inhibitors, such as PD98095 or U0126, did not affect MAPK activation and phytoalexin synthesis in the soybean cell culture system (data not shown) and hence these inhibitors did not disclose the interrelationship between MAPK and defense activation. The time courses and the pattern of inhibitor interference recorded for β-glucan-induced enhancement in soybean cells of cytosolic calcium levels and MAPK activity levels indicate that these two metabolic changes are part of the same or of parallel signaling pathways mediating defense reactions, such as phytoalexin production. A similar correlation between elicitor-induced MAPK activation and calcium influx, which did not exist for elicitor-induced production of reactive oxygen species, has been reported for several plant systems (Nürnberger and Scheel 2001; Nakagami et al. 2005). In summary, the correlation between the enhancement of MAPK activity and the activation of defense-related metabolic changes suggests that these kinases could play a role in β-glucan-mediated defense signaling in soybean.
GmMPK3 and GmMPK6 were found to be homologous to MAPKs implicated in defense signaling in other plant species (Nürnberger and Scheel 2001; Zhang and Klessig 2001; MAPK Group 2002; Nakagami et al. 2005). Given the large difference in the relative degree of activation and the lack of further cross-reacting antisera we have at present no information on whether other β-glucan-responsive MAPKs exist in soybean. The β-glucan-responsive GmMPK3 and GmMPK6 were shown to be homologous to MsSAMK (Jonak et al. 1996), NtWIPK (Seo et al. 1995), AtMPK3 (Mizoguchi et al. 1993), PcMPK3a and 3b (Kroj et al. 2003) from alfalfa, tobacco, Arabidopsis, and parsley, respectively, and MsSIMK (Jonak et al. 1993), NtSIPK (Zhang and Klessig 1997), AtMPK6 (Mizoguchi et al. 1993), and PcMPK6 (Kroj et al. 2003) from the same plants, respectively. Each of these pairs of kinases has been shown to be activated following elicitation by microbial proteins or peptides (Nürnberger and Scheel 2001; Zhang and Klessig 2001; Nakagami et al. 2005). Only a few studies have been reported on the activation by carbohydrate-type elicitors. In alfalfa, chitin (oligomers) and laminarin-type β-glucans were shown to preferentially activate MsSIMK and MsMMK3 (Cardinale et al. 2000). Two suspected MAPKs were observed to respond to oligogalacturonide treatment of soybean (Taylor et al. 2001) and tobacco cell cultures (Lebrun-Garcia et al. 1998). Likewise, a 48 kDa kinase was found to be activated in tomato leaves in response to treatment with polygalacturonic acid and chitosan (Stratmann and Ryan 1997).
To gain knowledge about how β-glucan elicitors can activate a common signaling pathway, it is important to characterize MAPKKs as putative upstream activator(s) of GmMPK3 and GmMPK6 in soybean. The biochemical characteristics of GmMKK1, identified in the present studies, disclosed distinct properties with respect to its capability to accept putative downstream MAPKs as substrate in vitro. By using recombinant GmMKK1 as a potentially active kinase, recombinant GmMPK6 but not GmMPK3 could be phosphorylated on a tyrosine residue. This result suggested that GmMKK1 possessed substrate preference for one of the MAPKs. Preferential physical interaction of GmMKK1 with GmMPK6 protein was likewise indicated in S. cerevisiae. Although GmMPK6 represents the kinase isoform with the apparent mass of 45 kDa which responded to β-glucan treatment in vivo with a much stronger increase in activity when compared to GmMPK3 (43 kDa), the functional relationship of the GmMAPKs to GmMKK1, as suggested by the present in vitro studies, should be substantiated by complementary experiments in vivo, preferably using a β-glucan-responsive experimental system involving a model legume.
Upstream activators of MAPKs have been identified in A. thaliana following FLS2-mediated perception of bacterial flagellin (Asai et al. 2002), in tobacco in response to elicitin from Phytophthora (Yang et al. 2001), in parsley upon perception of Pep-13 elicitor from Phytophthora (Cardinale et al. 2002; Lee et al. 2004), and in the tomato HR triggered by AvrPto and AvrPtoB (del Pozo et al. 2004; Pedley and Martin 2005). GmMKK1 is the orthologue of members of MAPKK sub-group A, such as AtMKK2, MsPRKK, NtSIPKK, and PcMKK2. Several reports indicated that MAPKKs of this sub-group can function upstream of sub-group A MAPKs, whereas in other cases upstream activators of group C may dominate (Nakagami et al. 2005; Pedley and Martin 2005). Interestingly, MAPKKs of different sub-groups were proposed to function in the activation of the endogenous MAPKs from parsley (Lee et al. 2004) and of the alfalfa MAPKs, SIMK, and SAMK, when heterologously co-expressed in parsley protoplasts and subsequently treated with the Pep-13 elicitor (Cardinale et al. 2002). Another aspect concerns the mode of activation of MAPKs by MAPKKs. In general, MAPKKs are reported to be dual-specific kinases that phosphorylate threonine and tyrosine residues in the T-X-Y motif. In most cases, Thr alone or Thr and Tyr were shown to harbor the transferred phosphate group coinciding with the activation of plant MAPKs (Huang et al. 2000; Kiegerl et al. 2000; Nühse et al. 2000). Huang et al. (2000) concluded that AtMEK1 may not be a dual-specificity kinase. Although AtMPK4 required Tyr phosphorylation, it appeared to occur in vitro primarily from autophosphorylation. The mode of activation of the soybean MAPKs and their integration in the putative β-glucan elicitor-responsive MAPK pathway in vivo clearly deserves further analysis. Gain-of-function experiments overexpressing wild-type and mutant MAPKK or MAPKs, which have advanced our knowledge of the role of MAPK cascades in disease resistance of a number of plants (Yang et al. 2001; Asai et al. 2002; Ren et al. 2002; Kroj et al. 2003; He et al. 2006) may aid in the further characterization of defense signaling in legumes triggered by important pathogens, such as Phytophthora sp.
Abbreviations
- A9C:
-
Anthracene-9-carboxylate
- DP:
-
Degree of polymerization
- DPI:
-
Diphenyleneiodonium
- EGTA:
-
Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid
- EST:
-
Expressed sequence tag
- IMAC:
-
Immobilized metal chelate affinity chromatography
- MAPK:
-
(MPK) mitogen-activated protein kinase
- MAPKK:
-
(MKK) MAPK kinase
- MBP:
-
Myelin basic protein
- NPPB:
-
5-Nitro-2-(3-phenylpropylamino)-benzoate
- PAGE:
-
Polyacrylamide gelelectrophoresis
- SA:
-
Salicylic acid
- SSC:
-
Standard saline citrate
References
Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W-L, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983
Bögre L, Ligterink W, Meskiene I, Barker PJ, Heberle-Bors E, Huskisson NS, Hirt H (1997) Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9:75–83
Boyle WJ, van der Geer P, Hunter T (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol 201:110–149
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 282:152–160
Brunner F, Rosahl S, Lee J, Rudd JJ, Geiler C, Kauppinen S, Rasmussen G, Scheel D, Nürnberger T (2002) Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J 21:6681–6688
Cardinale F, Jonak C, Ligterink W, Niehaus K, Boller T, Hirt H (2000) Differential activation of four specific MAPK pathways by distinct elicitors. J Biol Chem 275:36734–36740
Cardinale F, Meskiene I, Ouaked F, Hirt H (2002) Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14:703–711
Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18:465–476
Contor L, Lamy F, Lecocq RE (1987) Use of electroblotting to detect and analyze phosphotyrosine containing peptides separated by two-dimensional gel electrophoresis. Anal Biochem 160:414–420
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucl Acids Res 16:10881–10890
Cosio EG, Frey T, Verduyn R, van Boom J, Ebel J (1990) High-affinity binding of a synthetic heptaglucoside and fungal glucan phytoalexin elicitors to soybean membranes. FEBS Lett 271:223–226
del Pozo O, Pedley KF, Martin GB (2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23:3072–3082
Ebel J, Mithöfer A (1998) Early events in the elicitation of plant defense. Planta 206:335–348
Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276
Fliegmann J, Mithöfer A, Wanner G, Ebel J (2004) An ancient enzyme domain hidden in the putative β-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J Biol Chem 279:1132–1140
Fliegmann J, Schüler G, Boland W, Ebel J, Mithöfer A (2003) The role of octadecanoids and functional mimics in soybean defense responses. Biol Chem 384:437–446
Hall TA (1999) BioEdit. A user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucl Acid Symp Ser 41:95–98
He P, Shan L, Lin N-C, Martin GB, Kemmerling B, Nürnberger T, Sheen J (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125:563–575
Hille A, Purwin C, Ebel J (1982) Induction of enzymes of phytoalexin synthesis in cultured soybean cells by an elicitor from Phytophthora megasperma f. sp. glycinea. Plant Cell Rep 1:123–127
Huang Y, Li H, Gupta R, Morris PC, Luan S, Kieber JJ (2000) AtMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol 122:1301–1310
Janeway CA Jr, Medzhitov R (2002) Innate immune recognition. Annu Rev Immunol 20:197–216
Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H (1996) Stress signaling in plants: a mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA 93:11274–11279
Jonak C, Pay A, Bögre L, Hirt H, Heberle-Bors E (1993) The plant homologue of MAP kinase is expressed in a cell cycle-dependent and organ-specific manner. Plant J 3:611–617
Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, Liwosz A, Eklöf S, Till S, Bögre L, Hirt H, Meskiene I (2000) SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12:2247–2258
Kroj T, Rudd JJ, Nürnberger T, Gäbler Y, Lee J, Scheel D (2003) Mitogen-activated protein kinases play an essential role in oxidative burst-independent expression of pathogenesis-related genes in parsley. J Biol Chem 278:2256–2264
Lebrun-Garcia A, Ouaked F, Chiltz A, Pugin A (1998) Activation of MAPK homologues by elicitors in tobacco cells. Plant J 15:773–781
Lee J, Klessig DF, Nürnberger T (2001b) A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13:1079–1093
Lee J, Rudd JJ, Macioszek VK, Scheel D (2004) Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem 279:22440–22448
Lee S, Hirt H, Lee Y (2001a) Phosphatidic acid activates a wound-activated MAPK in Glycine max. Plant J 26:479–486
Liu YH, Zhang SQ, Klessig DF (2000) Molecular cloning and characterization of a tobacco MAP kinase kinase that interacts with SIPK. Mol Plant-Microbe Interact 13:118–124
MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7:301–308
Medzhitov R, Janeway CA Jr (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell 91:295–298
Mithöfer A, Bhagwat AA, Feger M, Ebel J (1996) Suppression of fungal β-glucan-induced plant defence in soybean (Glycine max L.) by cyclic 1,3–1,6-β-glucans from the symbiont Bradyrhizobium japonicum. Planta 199:270–275
Mithöfer A, Daxberger A, Fromhold-Treu D, Ebel J (1997) Involvement of an NAD(P)H oxidase in the elicitor-inducible oxidative burst of soybean. Phytochemistry 45:1101–1107
Mithöfer A, Ebel J, Bhagwat AA, Boller T, Neuhaus-Url G (1999) Transgenic aequorin monitors cytosolic calcium transients in soybean cells challenged with β-glucan or chitin elicitors. Planta 207:566–574
Mithöfer A, Ebel J, Felle H (2005) Cation fluxes cause plasma membrane depolarization involved in β-glucan elicitor-signaling in soybean roots. Mol Plant-Microbe Interact 18:983–990
Mithöfer A, Fliegmann J, Daxberger A, Ebel C, Neuhaus U, Bhagwat AA, Keister DL, Ebel J (2001) Induction of H2O2 synthesis by β-glucan elicitors in soybean is independent on cytosolic calcium transients. FEBS Lett 508:191–195
Mithöfer A, Fliegmann J, Neuhaus-Url G, Schwarz H, Ebel J (2000) The hepta-β-glucoside elicitor-binding proteins from legumes represent a putative receptor family. Biol Chem 381:705–713
Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K (1993) AtMPKs: a gene family of plant MAP kinases in Arabidopsis thaliana. FEBS Lett 336:440–444
Munnik T, Ligterink W, Meskiene I, Calderini O, Beyerly J, Musgrave A, Hirt H (1999) Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J 20:381–388
Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAPK pathways in plant stress signaling. Trends Plant Sci 10:339–346
Nühse TS, Peck SC, Hirt H, Boller T (2000) Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK6. J Biol Chem 275:7521–7526
Nürnberger T, Scheel D (2001) Signal transmission in the plant immune response. Trends Plant Sci 6:372–379
Nürnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: similarities and obvious differences. Immunol Rev 198:249–266
Pedley KF, Martin GB (2004) Identification of MAPKs and their possible MAPKK activators involved in the Pto-mediated defense response of tomato. J Biol Chem 279:49229–49235
Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8:541–547
Ren D, Yang H, Zhang S (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277:559–565
Romeis T (2001) Protein kinases in the plant defence response. Curr Opin Plant Biol 4:407–414
Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y (1995) Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270:1988–1992
Sharp JK, McNeil M, Albersheim P (1984b) The primary structures of one elicitor-active and seven elicitor-inactive hexa(β-D-glucopyranosyl)-D-glucitols isolated from the mycelial walls of Phytophthora megasperma f. sp. glycinea. J Biol Chem 259:11321–11336
Sharp JK, Valent B, Albersheim P (1984a) Purification and partial characterization of a beta-glucan fragment that elicits phytoalexin accumulation in soybean. J Biol Chem 259:11312–11320
Stäb MR, Ebel J (1987) Effects of Ca2+ on phytoalexin induction by fungal elicitor in soybean cells. Arch Biochem Biophys 257:416–423
Stratmann JW, Ryan CA (1997) Myelin basic protein kinase activity in tomato leaves is induced systemically by wounding and increases in response to systemin and oligosaccharide elicitors. Proc Natl Acad Sci USA 94:11085–11089
Suzuki K, Shinshi H (1995) Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell 7:639–647
Taylor ATS, Kim J, Low PS (2001) Involvement of mitogen-activated protein kinase activation in the signal-transduction pathways of the soya bean oxidative burst. Biochem J 355:795–803
Tena G, Asai T, Chiu W-L, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4:392–400
Yang K-Y, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98:741–746
Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9:809–824
Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6:520–527
Acknowledgment
This work was supported by the Deutsche Forschungsgemeinschaft (SFB369).
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequences encoding the MAPKs and MAPKK1 from soybean can be accessed through the GenBank database under GenBank accession numbers AF104247, AF329506, and AY070230.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Daxberger, A., Nemak, A., Mithöfer, A. et al. Activation of members of a MAPK module in β-glucan elicitor-mediated non-host resistance of soybean. Planta 225, 1559–1571 (2007). https://doi.org/10.1007/s00425-006-0442-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0442-6