Abstract
Cyclic nucleotide gated channels (CNGCs) that are regulated by calmodulin (CaM) have been shown to play essential roles in signal transduction, metabolism, and growth in animals. By contrast, very little is known about the subcellular location and the function of these channels in plants. Here we report on the effects of antisense suppression of the expression of AtCNGC10, a putative K+ channel, and the immunolocalization of the protein using an AtCNGC10-specific antiserum. In Arabidopsis thaliana leaves, AtCNGC10 was localized to the plasma membrane of mesophyll and parenchyma cells. Antisense AtCNGC10 plants had 40% of the AtCNGC10 mRNA levels and virtually undetectable protein levels relative to wild type plants. Antisense expression of AtCNGC10 did not affect the mRNA levels of AtCNGC13, the most closely related CNGC family member in the genome. Relative to wild type Columbia, antisense AtCNGC10 plants flowered 10 days earlier, and had a 25% reduction in leaf surface area, thickness and palisade parenchyma cell length. Their roots responded more slowly to gravitropic changes and the chloroplasts accumulated more starch. We propose that AtCNGC10, through interactions with CaM and cGMP, modulates cellular K+ balance across the plasma membrane, and that perturbations of this K+ gradient affect numerous growth and developmental processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cyclic-nucleotide gated channels (CNGC) of plants resemble the Shaker superfamily of channels of animals (Varnum and Zagotta 1997; Fox and Guerinot 1998), which contain an N-terminal calmodulin (CaM)-binding domain followed by six conserved transmembrane domains (designated S1–S6), a hydrophilic C-terminal CN-binding domain and, in some cases, an ankyrin domain (Sentenac et al. 1992; Anderson et al. 1992; Ketchum and Slayman 1996; Finn et al. 1996; Grunwald et al. 1999). The plant CNGCs also contain the conserved transmembrane domains (Fig. 1a), but the CaM- and CN-binding domains overlap in the C-terminus (Schuurink et al. 1998; Arazi et al. 1999; Köhler et al. 1999; Köhler and Neuhaus 2000). The overlapping nature of the CaM- and CN-binding domains in plant CNGCs suggests an interaction between CaM and CN in regulating the channel (Hua et al. 2003; Li et al. 2005).
a Modular structure of AtCNGC10 protein. Membrane-spanning domains (S1–S6), pore (P), cyclic nucleotide binding domain (CNBD), calmodulin-binding domain (CaMBD), and epitope for antibody generation are denoted. The T-DNA insertion (SALK 071112) is found 60 amino acids downstream of the CN-binding domain. b T-DNA insertion sites (black circle) at the cngc10 locus, open arrow. The only insertion in the protein coding sequence, SALK 071112, is indicated with a blue circle at nucleotide 132536. The specific insertion sites from left to right are at nucleotides, 126920, 127414, 127549, 127748, 127796, 128581, 129085, 129263, 129349,129426, 130601, 130688, 131061/1, 131061/2, 131119, 131155/1, 131155/2, 131195, 131658, 131673/1, 131673/2, 131716, 131721, 131822, 135737, 136147, 137031, 137577, 137839, 138153, 138320, 138325, 138660, 140038, 140535, 140956, 141129, 141634, 141650, 142179
Recently, we confirmed the importance of this interaction by coexpressing CaM and Arabidopsis CNGC10 (AtCNGC10) in a potassium transport mutant of Escherichia coli. In these cells, Ca2+/CaM inhibited cell growth by 40%, while cGMP reversed the inhibition by Ca2+/CaM, in a AtCNGC10-dependent manner (Li et al. 2005). Because Ca2+/CaM and CN’s are key second messengers in several signal transduction pathways (Bush 1995; Schuurink et al. 1998; Zielinski 1998), the presence of their binding sites in this family implies a role for Ca2+/CaM and CN-crosstalk on channel-driven ionic fluxes in downstream signaling mechanisms (Talke et al. 2003). The signaling pathways connecting CNs and CaM with plant channel function have yet to be elucidated.
In Arabidopsis, there are 20 members of the CNGC family (Mäser et al. 2001). Homologues, HvCBT1 and NtCBP4, have been identified in barley and tobacco, respectively (Schuurink et al. 1998; Arazi et al. 1999) and in rice and bean (Talke et al. 2003). AtCNGC1 and AtCNGC2, partially rescue the yeast K+ uptake mutant CY162 (Köhler et al. 1999), suggesting weak K+ transport activity, whereas barley HvCBT1 does not rescue this mutant (Schuurink et al. 1998). CN’s have been shown to activate AtCNGC1, AtCNGC2 and AtCNGC4 (Leng et al. 2002; Balagué et al. 2003). AtCNGC2 is permeable to both K+ and other monovalent cations, Li+, Cs+, Rb+, but discriminates against Na+ (Leng et al. 1999; Leng et al. 2002). AtCNGC3 is involved in the non-selective uptake of Na+ and K+ and perhaps other monovalent cations (Gobert et al. 2006). AtCNGC4 preferably conducts Na+ and K+, but is blocked by Cs+ (Balagué et al. 2003). NtCBP4 is CN-gated and confers Ni2+ tolerance and Pb2+ sensitivity in transgenic plants (Arazi et al. 1999; Leng et al. 2002).
Phenotypes of six CNGC mutants have been described in Arabidopsis and disease response signaling is common to four of the CNGCs. A knockout mutation of AtCNGC1 results in improved tolerance to Pb2+ (Sunkar et al. 2000). AtCNGC3 mutants are susceptible to cation stress and have altered monovalent cation content (Gobert et al. 2006). Knockout AtCNGC2 plants are specifically hypersensitive to Ca2+, but not to Na+ or K+ (Chan et al. 2003). AtCNGC2 mutants suggests that it functions in disease-resistance, programmed cell death, and plant development mediated by Ca2+ (Clough et al. 2000). AtCNGC4, which clusters with AtCNGC2 (Talke et al. 2003), is also involved in hypersensitive disease resistance signaling (Balagué et al. 2003) as are AtCNGC11 and AtCNCG12 (Yoshioka et al. 2006).
Less is known about the function and regulation of the remaining 14 CNGC’s in plants and there is virtually no information on the high-resolution subcellular locations of any CNGCs. To expand our knowledge of CNGCs, we have analyzed AtCNGC10, a member of an entirely different phylogenetic cluster than any previously analyzed AtCNGC. Recently, we showed that AtCNGC10 rescues potassium channel mutants of E. coli (LB650) and yeast (CY162) and partially rescues Arabidopsis akt1 (Li et al. 2005). AtCNGC10 conferred tolerance to toxic levels of Na+ and Cs+ in a yeast K+-uptake mutant grown on low K+ medium. Antisense AtCNGC10 plants had >40% less potassium than wild type. Taken together, these and other results demonstrated a role for the CaM-binding channel, AtCNGC10, in mediating the uptake of K+ in plants.
In this report, we have further characterized the function of AtCNGC10 by determining its subcellular localization and the phenotypes resulting from the effects of antisense suppression of AtCNGC10 expression. Antisense AtCNGC10 plants were altered in flowering time, leaf thickness and surface area, palisade cell length, gravitropism and starch accumulation. Based on these results, we propose that AtCNGC10 is responsible for maintaining cellular K+ homeostasis across the plasma membrane, and that this balance helps regulate a variety of growth-related processes.
Materials and methods
Clones, strains and mutants
We have reported previously the AtCNGC10 sequence to Genbank (Accession number AF272002; Li et al. 2005). The Arabidopsis wild type Columbia, Landsberg and the phyB and SALK-071112 mutants were obtained from the Arabidopsis Biological Resource Center.
Construction of AtCNGC10 gene in the pBIN20 binary vector
The cauliflower mosaic virus 35S promoter was subcloned in the HindIII/BamHI sites of pBIN20 (Hennegan and Danna 1998). For the antisense lines, AtCNGC10 was inserted into the SalI/EcoRI sites through an intermediate vector, pET28b. The pBIN20 plasmids with AtCNGC10 inserts were transformed into Agrobacterium LBA4404 by electroporation.
Transformation of Arabidopsis plants
Transformation followed the floral dip method (Clough and Bent 1998). Plants were grown to the stage at which bolts were just emerging. The tip of the bolt was cut off to induce the growth of secondary inflorescence. The cutting was repeated several times to synchronize pots and to maximize the number of inflorescence meristems produced. Agrobacterium with the AtCNGC10 construct was inoculated into 5 ml LB medium with 50 ug/ml kanamycin and incubated at 37°C overnight. The culture was transferred to a 100 ml LB medium and incubated at 37°C till OD600 to 0.6. The cells were collected and resuspended in 10 mM MgCl2, 5% sucrose, 0.005% Silwet L-77 to OD600 0.8. The plants were dipped for 10 s with the Agrobacterium culture. Then the plants were covered with plastic dome overnight. Plants were grown until the seeds were mature. The T1 seeds were sown on selective medium (1/2 MS medium (Sigma), 0.8% Agar, Kanamycin 60 μg/ml) to identify transformants. After two weeks, the kanamycin-resistant seedlings were transferred to soil in pots and grown until seeds were mature. Seeds were prepared from two more generations of selfing and selection on kanamycin. Approximately 25% of the plants showed the mutant phenotype in the T2 generation and these were selected for selfing. The resulting T3 seeds were grown up on kanamycin and the presence of single copy transgenes was verified via PCR and Southern blot hybridization, using kanR gene specific primers and AtCNGC10 and kanR gene probes. The two antisense lines were used in all of the experiments.
Quantitative PCR
The PCR reactions were performed using a SYBR Green PCR Master Mix kit (Applied Biosystems) on a BioRad iCycler Thermal Cycler machine with iCycler Optical Module. Reverse Transcriptions were carried out in 25 μl using 1 μg total RNA and MMLV enzyme (Promega) according to users manual. The following primer sequences were checked against Arabidopsis database to ensure their specificity and designed to exon/exon boundaries (eliminating contaminating genomic DNA signals): CNGC10: 5′-cctcgatctcctcaagaaagtacct-3′ and 5′-tctccttcacggatcacgtaact-3′; CNGC13: 5′-cggaagaggcgagttggttgatgat-3′ and 5′-gacagctaagactacaagctgtggg-3′; actin2: 5′-cttccgctctttctttccaagctc-3′ and 5′-atcatctcctgcaaatccagccttc-3′; CBP20: 5′-tcgtgggttcttctccggtctcatgtc-3′ and 5′-cggaaacgacaaagagacgatgatcgc-3′. In general, amplicons were between 110 and 130 nt. The specificity of PCR amplifications was ensured by melting curve analysis and agarose gel electrophoresis (data not shown).
Immunoblot analysis
Total cellular proteins were isolated as described by Arazi et al. (1999). Fifty to one hundred microgram of each sample were electrophorized on 7% SDS-PAGE and blotted onto nitrocellulose membranes (Perkin-Elmer). The primary antibody was a polyclonal antibody (Zymed Co.) produced against a synthetic 20 aa oligopeptide Ser43-Asn62 that is unique to the AtCNGC10 protein. Blots were visualized with ECL Kit (Amersham). To test antibody specificity and binding to the CNGC polypeptide, the N-terminal half (300 aa) of AtCNGC10 was cloned into the pET25b vector and transformed into BL21 E. coli (Novagen) or used for in vitro translation (TNT, Promega Co. Madison, WI, USA). Induction of the recombinant cells were performed using the media of Zymo Co.
Plant growth and phenotypic measurements
Arabidopsis seeds were surface sterilized with 10% bleach, 0.2% Tween-20 and plated on 0.8% agar containing Murashige and Skoog medium and 2% sucrose. Plated seeds were refrigerated (4°C) for 15 h and then exposed to white fluorescent light (110–125 μmol m-2 s−1) at 22–24°C for 24 h to promote uniform germination. The germinating seeds were either kept in 14 h white light/10 h darkness for 18 days or were placed in light-tight controlled environment chambers (22–23°C) in a dark-room for 18 days (etiolated). Tissue was harvested by quick freezing in liquid nitrogen or fixation for microscopy. For immunoblot and potassium growth response analyses, Arabidopsis seeds were germinated and kept on artificial medium (Hirsch et al. 1998) supplemented with KCl in final concentrations of 1 mM, 0.1 mM or 0.001 mM.
For flowering and biomass measurements, seeds were planted in flats containing water-saturated potting mix (Lehle Seeds, Round Rock, TX, USA) and chilled (5°C) overnight (14 h) and exposed to a photoperiod of 13 h of darkness and 11 h of cool white light (fluorescent, 110–125 μmol m−2 s−1) at 22–24°C. They were watered with 0.5X-Miracle Gro complete soluble fertilizer (containing 1.6 mM KCl). For starch content measurements starch was isolated using HClO4 from 100 mg leaf tissue (protocol kindly provided by Steven M. Smith, University of Edinburgh, UK) at the end of the day and OD595 measured in 10% Lugol (potassium iodide) solution (Sigma). To measure gravity responses, seeds were germinated on MS plates turned to a vertical position in 12/12 DL. After 8 days, plates were rotated 180o or 90o. Root elongation was measured every 2 days (mm). To measure leaf area, the ten biggest leaves of pooled, 8-week-old COL antisense and T-DNA knockout plants were collected. The leaves were scanned between two glass plates and the surface area pixels were counted in Photoshop.
Light microscopy
Five leaves were selected from each of five wild type and mutant plants. Sample explants were removed from the mid-laminal region of each leaf. Explants were placed in an antifreeze/fixative that contained 4% (w/v) paraformaldehyde, 20% (v/v), dimethyl sulfoxide, 1% (v/v) Tween 20 and 0.05 M sodium cacodylate at pH 7.4. These were evacuated overnight to remove intercellular air and to assist penetration. Samples were stored at 5°C prior to cryosectioning at 30 μ in a Reichert–Jung Cryocut 1,800 freezing microtome at −20°C. Five widely separated transverse sections were photographed and measured for each explant. Twenty measurements of leaf thickness were made for each section with Sigma Scan Image (Jandel Scientific). Sections were photographed with a Nikon Coolpix 990 digital camera at a resolution of 2,048 × 1,536 pixels. The means of each leaf (n = 25) were used for an independent sample t test at the 0.01 probability level with Microsoft Excel 2000. Summary statistics, including the 95% confidence interval for each mean were also performed with Excel 2000.
Transmission- and immuno-electron microscopy
To observe the starch content of chloroplasts, specimens were fixed with 4% glutaraldehyde plus 5% paraformaldehyde and 0.1 M calcium chloride in 0.1 M sodium cacodylate buffer, pH 7.1 for 4 h, washed in 0.1 M cacodylate for 3 × 30 min, at RT, followed by postfixation with 2% OsO4 in 0.1 M cacodylate at 4°C overnight. Tissue was dehydrated in a graded ethanol series, substituted with propylene oxide, and embedded in Spurr’s resin. Ultrathin (60–80 nm) sections were obtained on a Reichert Ultracut E ultramicrotome, double stained with 5% uranyl acetate and 3% lead citrate, then viewed on a LEO 912 EFTEM at 100 kV.
For immunogold electron microscopy, Arabidopsis leaves (6 week old) were cryoprotected with 150 mM sucrose, placed into aluminum sample holders, and frozen in a Baltec 010 high pressure freezer (RMC Boeckeler Instruments, Tucson, AZ, USA). The samples were freeze substituted in 0.1% uranyl acetate/0.25% glutaraldehyde in anhydrous acetone for 5 days at −90°C. After slow warming to for −60°C 3 days, the samples were rinsed in acetone and infiltrated with 25, 50, 75% (24 h each), and 100% (4 days) LR-White (EMS, Fort Washington, PA, USA) in acetone, and UV polymerized at −60°C for 3 days. Processing of thin sections for immunolabeling was according to (Otegui and Staehelin 2000), with the anti-CNGC10 antibody being applied for 1.5 h at RT. Controls were performed by omitting the primary antibody and using pre-immune serum. The sections were viewed in a Philips CM10 microscope (Philips, Hillsboro, OR, USA).
Results
T-DNA knockout and antisense suppression data indicate that Atcngc10 is an essential gene
AtCNGC10 is predicted to have six membrane-spanning domains, a pore domain, and a CNBD that overlaps a CaMBD (Fig. 1a). A search for T-DNA mutants in the public and private genetic knockout resources produced only one mutant in this locus, which was at the extreme C-terminus (Fig. 1a) and clearly outside of any functional domain that could impact its activity. One kilobase-pair upstream in the promoter region is also absent of knockouts, despite the fact that T-DNAs have a predilection for inserting in the promoters of genes in the Arabidopsis genome (Pan et al. 2005). Clearly, the frequency of insertions in the AtCNGC10 locus is substantially lower than the upstream and downstream flanking regions (Fig. 1b). The scarcity of insertions is suggestive of an essential function for this gene. Therefore, we attempted to determine the impact of reduced levels of AtCNGC10 protein on the plant and assess its function by using antisense suppression of AtCNGC10 expression in the Arabidopsis Columbia background. Very few antisense AtCNGC10 lines survived to maturity. After verification of the presence of the kanR gene via PCR and Southern hybridization in the two surviving lines, (unpublished data), we conducted a molecular analysis of mRNA levels in these lines to verify the reduction in AtCNGC10 protein and mRNA levels.
We measured the specificity of the antisense expression of AtCNGC10 on the target mRNA (Fig. 2a). We quantitated the mRNA levels in leaves of AtCNGC10 and AtCNGC13 in both wild type, antisense AtCNGC10 and T-DNA mutant plants using RT-PCR and exon–exon junction-specific primers. AtCNGC13 was chosen for comparison with AtCNGC10 because it is the closest related member of this CNGC family in terms of conserved nt and amino acid sequences (Mäser et al. 2001). Antisense expression of AtCNGC10 had no significant effect on AtCNGC13 mRNA levels, but decreased AtCNGC10 mRNA levels by half. Therefore, the decreased levels of AtCNGC10 expression in the antisense lines was specific to this member of the channel family. In addition, we examined the levels of AtCNGC10 protein in wild type and the two antisense lines using an antiserum generated against a unique peptide within AtCNGC10- on immunoblots (Fig. 2b). The antiserum was generated against an amino-terminal epitope (Fig. 1a). As positive controls, recombinant AtCNGC10 protein produced in E. coli and via translation in vitro was used to verify antibody binding. A single AtCNGC10 protein of 75 kDa was detected in wild type plants, but was virtually undetected in the antisense lines, indicating that AtCNGC10 protein levels were decreased in these lines.
Protein and mRNA analysis. a Mean ( ± standard deviation, SD) of the relative levels of mRNAs for CNGC10 and CNGC13 of wild type and two antisense CNGC10 lines, and a T-DNA insertion line (Fig. 1). CNGC RNA levels were measured relative to actin2 and CBP20 gene mRNA levels, which served as an internal standard in both wild type and mutant lines. Antisense mRNA levels are shown as a percent of wild type levels. Statistically significant at the at 0.075 level (*) as determined with an independent sample t test from three independent replicated experiments. b AtCNGC10 protein is not detected on immunoblots of proteins from antisense plants. The antibody was produced against the unique AtCNGC10-specific epitope Ser43-Asn62 region. As positive controls, the first 185 residues containing the epitope were cloned in pET25b and expressed by in vitro translation (IVT) or in vivo in BL21 Escherichia coli. The AtCNGC10 antibody was used on protein samples from 6-week-old wild type (wt) and antisense (as1 and as2) Arabidopsis seedlings grown on artificial medium (Hirsch et al. 1998) containing low (0.1 mM) KCl. The protein produced by IVT is larger due to the pelB leader, which is cleaved in E. coli
AtCNGC10 localizes to the plasma membrane of leaf cells
In the next experiment, the AtCNGC10 antiserum was used in conjunction with immuno-electron microscopy to localize the AtCNGC10 protein on thin sections of leaf tissues. Immunogold particles were specifically detected along the plasma membrane inside the cell wall of mesophyll and parenchyma cells indicating that AtCNGC10 is a plasma membrane protein (Fig. 3). This is consistent with AtCNGC10 containing membrane spanning domains and having cation channel functions (Li et al. 2005). No gold particles were detected over chloroplasts, mitochondria, microbodies or vacuoles.
Immunolocalization of AtCNGC10 in Arabidopsis leaf cells by transmission electron microscopy. a, b Black arrowheads denote the 10 nm immunogold-anti-CNGC10 antibody conjugates along the plasma membrane (pm). Chloroplast (cp), cytoplasm (cy), and cell wall (cw) are labeled. b is an enlargement of the boxed region in a
Antisense AtCNGC10 plants exhibit multiple changes in growth and metabolism
In Table 1, the leaf surface area, lamina thickness, and palisade columnar cell length were reduced by 20% in the antisense lines relative to wild type, indicating that AtCNGC10 was required for normal leaf size, thickness and palisade columnar cell length. Antisense expression did not affect hypocotyl length when grown in continuous darkness. However, in the light, hypocotyls of antisense AtCNGC10 plants were twice as long relative to wild type.
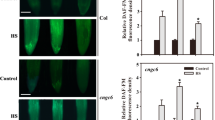
Leaves of antisense AtCNGC10 plants accumulated almost double the amount of starch than wild type on a fresh weight basis (Fig. 4a). As shown in the transmission electron micrographs, aberrantly large starch granules, identified according to the criteria of Gunning and Steer (1975) were present in the chloroplasts of antisense relative to wild type plants grown under identical conditions (Fig. 4b, c). Mitochondria were observed in close association with chloroplasts in both antisense and wild type plants (Fig. 4b, c). However, only in the antisense lines were peroxisomes frequently observed next to mitochondria and chloroplasts (Fig. 4c). Whether photorespiration is more active in the antisense lines will require further measurements.
Analysis of starch in leaf cells. a Quantitation of leaf starch content (St, mg/100 mg leaf tissue FW) in WT, two antisense (AS) and T-DNA (TD) insertion lines. Statistically significant at the 0.05 probability level (*) with an independent sample t test from three independent replicated experiments. Transmission electron micrographs of representative chloroplasts (Cp) from WT (b) and the AtCNGC10 AS plants (c). Both antisense lines had aberrantly large starch granules in chloroplasts relative to wild type. St, mitochondria (M), and peroxisomes (Px) are labeled
Flowering of the antisense lines occurred 10 days earlier than in wild type plants (Fig. 5), which was similar to the shortened time to flowering that was observed in the phyB mutant (Landsberg background). Flowering in the T-DNA mutant was unaffected. We emphasize that both wild type and antisense plants were grown under the same conditions together, watered adequately and were not environmentally stressed, which could cause plants to flower early, regardless of genotype.
Effect of antisense expression of AtCNGC10 on time to flowering. The days after germination (y-axis) leading up to the first appearance of the flower bud (Bud) and opening of the first flower are shown for two antisense AtCNGC10 lines, Columbia wild type (background for antisense lines), the phyB mutant, and Landsberg wild type (background for phyB). The mean ± standard error for two independent experiments done in duplicate are shown. In each replicate, 25 plants each of phyB, Columbia, Landsberg and two distinct antisense lines were measured. The error bars indicate low variability among the antisense lines
We examined the growth and elongation of roots from wild type and AtCNGC10 antisense lines. Root length of antisense plants was reduced by 38% compared to wild type plants (Fig. 6a, b). Upon gravistimulation by turning the plates 90o and 180o, a delay in the bending response was observed in the antisense lines by measuring the root angle relative to a straight line. Only 41% of the antisense plants exhibited visible bending after 2 days and 55% after 4 days relative to wild ype. Root elongation in antisense plants after bending remained approximately 50% lower than wild type, further indicating a slower growth rate of the roots specific to the antisense lines.
Antisense AtCNGC10 plants exhibit slower gravity response. a Picture of representative 7-day-old wild type (wt) and antisense (as) plants right before (0 day) and 4 days after turning the plate 90° (left panel) or 180° (right panel) The gravity response of the wild type is more pronounced. b Quantification of root length in millimeter (Mean ± standard deviation) and gravity response differences measured on wild type Columbia and two independent antisense lines (20 plants each) repeated in four independent experiments (two at 90° and 180°)
Because of the effect of the AtCNGC10 gene product on K+ transport in E. coli and yeast (Li et al. 2005), growth of the antisense lines was compared on media containing different concentrations of KCl (Fig. 7). Lower K+ decreased the growth of all plants. However, the decrease in growth, as measured by the dry weight, was much greater in the antisense than in the wild type plants, indicating that they were more sensitive to low K+ than the wild type. This suggested that they had a defect in K+ transport under K+-limiting conditions.
Effect of media containing low potassium on growth of WT plants and two AS lines. KCl concentrations in the media are indicated. The plants (25 plants per plate, 4 plates per line) were measured from WT and AS to determine the dry weight, mg DW, per plate (Mean ± standard deviation done in three independent experiments)
Discussion
Atcngc10 acts as an essential gene
No T-DNA inserts could be found inside any functional domain encoded by the Atcngc10 gene when the public and commercial T-DNA databanks were searched. However, numerous insertions were located upstream and downstream from the locus. We observed a high mortality rate when producing antisense AtCNGC10 plants on the wild type background, which led to a low frequency of antisense mutant recovery. We only recovered three antisense lines, one of which died in the T3 generation. The scarcity of knockouts and mutant recovery was suggestive that AtCNGC10 was an essential gene with limited genetic redundancy. A complete knock-out, and the resulting elimination of its unique cation channel activity in the plasma membrane, might be lethal or poorly viable. These observations prompted us to verify that the levels of AtCNGC10 mRNA and protein were indeed specifically decreased in the two main AtCNGC10 antisense lines that were recovered. The two antisense lines here had partial suppression of mRNA levels (Fig. 2a), virtually no detectable protein within the sensitivity of the immunoblots, and probably had a low level of gene expression required for survival. Together, these results suggest that Atcngc10 is an essential gene.
Interestingly, it would be expected that essential genes might be present in the T-DNA collections as heterozygotes, if their growth was adequate to yield seeds or that impaired growth did not escape detection by investigators. If the heterozygote is actually lethal (not simply impaired in growth), then this would be a case of haploinsufficiency (Fodde and Smits 2002; Veitia 2002), which has been well-described in animals. However, haploinsufficiency has been rarely observed in plants, such as in the porphyrin mutant of maize (Hu et al. 1998). Therefore, we conducted a more detailed analysis of chromosome−1 of Arabidopsis to determine the frequency of 2–5 to 3.0 kb gaps which lack T-DNA or transposon insertions. We analyzed 130 kb downstream from the Atcngc10 locus to the telomere and 300 kb upstream from the locus. We found the following seven gaps: 2.2 kb at At1g01030; 2.8 kb at At1g01040; 2.6 kb at At1g01260; 3.3 kb at At1g01790; 3.4 kb at At1g01960; 4.4 kb at At1g02010; and 4.5 kb at At1g02080. Of these, only two gaps completely spanned predicted genes, which were 2.6 kb at At1g01260 and 4.4 kb at At1g02010. When Atcngc10 (At1g01340) is included in the analysis, there were three uninterrupted genes in the 425 kb region out of 119 total predicted genes. This preliminary analysis showed that 2.5% of the predicted genes were uninterrupted, and 30 kb (7.1%) of the region existed as gaps greater than 2 kb in length. Therefore, 7.1% is the probability that 3 kb regions will be uninterrupted. The analysis further indicated that an uninterrupted gene is a rare occurrence.
AtCNGC10 is a plasma membrane channel
The AtCNGC10-specific antiserum cross-reacted with the truncated AtCNGC10 protein expressed in E. coli and in vitro, and the correct-sized 75 kDa protein from plants. Using this antiserum, immunolocalization electron microscopy was used to demonstrate that AtCNGC10 is located in the plasma membrane of leaf cells. To our knowledge, this study represents the first highly resolved localization of a member of the Arabidopsis CNGC family to a subcellular membrane in planta. The CNGC, NtCBP4, in tobacco appears to be associated with the plasma membrane fraction (Arazi et al. 1999). Recently, the AtCNGC3-GFP fusion appeared to be associated with the plasma membrane in leaf protoplasts (Gobert et al. 2006), suggesting other CNCGs may be in the plasma membrane. In turn, the localization of AtCNGC10 provides critical information on how CNGCs might exert their multiple effects on plant function. AtCNGC10 complemented potassium uptake mutants of E. coli, yeast and Arabidopsis on low potassium media, and modulated tissue levels of potassium by 40% (Li et al. 2005). Antisense lines had 1.98 mg K per 100 mg tissue DW, while wild type had 3.8 mg K per 100 mg tissue DW grown on 1 mM KCl (Li et al. 2005). Therefore, it is reasonable to propose that AtCNGC10 exerted its phenotypic effects via K+ transport across the plasma membrane and by altering potassium levels in the tissues. We recognize that there may be secondary effects of AtCNGC10 on the concentrations and transport of other cations that could also influence the phenotypes. Determining the specific cation transport specificity will require patch clamp experiments.
The orientation of the AtCNGC10 polypeptide in the membrane and the number of subunits comprising the channel are not yet known. In animals, cyclic nucleotide gated channels are tetrameric (Fnn et al. 1998; Trudeau and Zagotta 2002). The P-domains of each subunit cradle the pore that faces the extracellular side of the membrane, while the CN-binding domain of the c-terminus extends into the cytoplasm (Maathuis et al. 1997; Fox and Guerinot 1998). Electrophysiological studies show that some plant CNGCs are inward conducting channels (Leng et al. 2002;).
AtCNGC10 is required for light-modulated plant growth and development
We presented evidence that AtCNGC10 modulated leaf growth and enlargement, hypocotyl elongation, root growth rate in response to gravity, starch content and time to flowering. The AtCNGC2 knockout also significantly affected leaf development, which was interpreted to be in response to calcium stress (Clough et al. 2000; Chan et al 2003). In addition to AtCNGC10, mutant phenotypes for six other AtCNGCs have been reported previously. The majority of these phenotypes correspond to hypersensitive cell death, salt and to calcium and heavy metal responses (Sunkar et al. 2000; Chan et al. 2003; Talke et al. 2003; Balagué et al., 2003; Yoshioka et al. 2006). This study provides new information about the diverse developmental phenotypes caused by a unique member of the CNGC phylogenetic tree.
All of the phenotypes described in this paper could have been caused by general sub-optimal potassium levels in different tissues (Li et al. 2005) and may not have been caused by any single specific biochemical mechanism. Alternatively, K+ transport has been directly correlated with auxin-induced shoot growth and coleoptile expansion, light-stimulated leaf cell expansion, and gravity-induced growth and cell wall acidification (Maathuis et al. 1997; Philippar et al. 1999; Very and Sentenac 2003; Elumalai et al. 2002; Fuchs et al. 2003). Furthermore, auxin also stimulates expression of the K+ channels KAT1 and KAT2 (Philippar et al. 2004). It is possible that AtCNGC10 affected leaf cell expansion and the rate of root growth via the ‘acid-growth’ mechanism, as first proposed by Rayle and Cleland (1992), which requires the accumulation of solute (generally K+) for elevating turgor to drive cell expansion (Stahlberg and Van Volkenburgh 1999). Several K+ channels contribute to this process (Cao et al. 1995; Maathuis et al. 1997). We previously showed that the TEA insensitive nature of AtCNGC10 is consistent with the finding that a TEA-insensitive K+ channel is involved in the second stage of expansion of mature leaves via the acid-growth mechanism (Stiles et al. 2003). Inconsistent with this hypothesis, however, is our observation that antisense plants had longer hypocotyls (Table 1). Shoot cell expansion is more affected by KT2/KUP2 (Elumalai et al. 2002). To accurately elaborate further on these observations will require measuring H+ levels in sense and antisense lines to assess any influence of AtCNGC10 on cell wall acidification.
By exhibiting an alteration in the expected light responses, antisense AtCNGC10 lines behaved similar to weak photoreceptor mutants of phytochrome and cryptochrome (Quail 1997; Lin 2000; Weston et al. 2000; Chun et al. 2001). Light-regulated leaf lamina thickness and cell size are reduced in phytochrome mutants (Chory et al. 1989; Pyke and Lopez-Juez 1999; Weston et al. 2000) and involves a light-regulated K+ channel (Blum et al. 1992; Stiles and Van Volkenburgh 2002; Stiles et al. 2003). AtCNGC10 was also required for full inhibition of hypocotyl development, which is controlled by PhyA/PhyB and Cry1/Cry2. In addition, the shorter flowering time in antisense AtCNGC10 plants resembled the effect of the phyB mutant. The high levels of starch in antisense AtCNGC10 lines could be explained by defects in the allocation or export of carbohydrates from chloroplasts, which are decreased by K+ deficiency (Marschner 1995; Flügge 2000) and resemble high light-grown chloroplasts (Lichtenthaler et al. 1981). Other K+ channels regulated by light and the circadian clock include those affecting leaf motor pulvini (Moshelion et al. 2002) and guard cells (Schroeder et al. 2001). Regulation of the latter is mediated by the blue light photoreceptors, phot1 and phot2 (Kinoshita et al. 2001).
In mammals, CNGCs are essential downsteam targets of signaling pathways mediating olfactory and visual systems (Grunwald et al. 1999; Trudeau and Zagotta 2002). Based on the precedence for light-regulation of K+ channels in plants and animals, we speculate that AtCNGC10 modulates K+ fluxes across the plasma membrane involved in downstream steps in light signaling. This proposal is also consistent with the observations of Talke et al. (2003) and with studies in which the microinjection of cGMP and CaM into cells of the aurea phytochrome mutant of tomato showed that these signaling molecules modulated photomorphogenetic processes (Neuhaus et al. 1993; Bowler et al. 1994). Both CaM and cGMP affect AtCNGC10 function (Li et al. 2005), and are, therefore, candidates for modulating the effects of AtCNGC10 on plant development.
References
Anderson JA, Huprikar SS, Kochian LV, Lucas WJ, Gaber RF (1992) Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 89:3736–3740
Arazi T, Sunkar R, Kaplan B, Fromm H (1999) A tobacco plama membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J 20:171–182
Balagué C, Lin B, Alcon C, Flottes G, Malmström S, Köhler C, Neuhaus G, Pelletier G, Gaymard F, Roby D (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15:365–379
Blum DE, Elzenga TM, Linnemeyer PA, Van Volkenburgh E (1992) Stimulation of growth and ion uptake in bean leaves by red and blue light. Plant Physiol 100:1968–1975
Bowler C, Neuhaus G, Yamagata H, Chua NH (1994) Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77:73–81
Bush DS (1995) Calcium regulation in plant cells and its role in signaling. Annu Rev Plant Physiol Plant Mol Biol 46:95–122
Cao Y, Ward JM, Kelly WB, Ichida AM, Gaber RF, Anderson JA, Uozumi N, Schroeder JI, Crawford NM (1995) Multiple genes, tissue specificity, and expression-dependent modulation contribute to the functional diversity of potassium channels in Arabidopsis thaliana. Plant Physiol 109:1093–1106
Chan CW, Schorrak LM, Smith RK Jr, Bent AF, Sussman MR (2003) A cyclic nucleotide-gated ion channel, CNGC2, is crucial for plant development and adaptation to calcium stress. Plant Physiol 132:728–731
Chory J, Peto CA, Ashbaugh M, Saginich R, Pratt L, Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1:867–880
Chun L, Kawakami A, Christopher DA (2001) Phytochrome A mediates blue light and UV-A-dependent chloroplast gene transcription in green leaves. Plant Physiol 125:1957–1966
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–43
Clough SJ, Fengler KA, Lippok B, Smith RK, Yu IC, Bent AF (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97:9323–9328
Elumalai RP, Nagpal P, Reed JW (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14:119–131
Finn JT, Grunwald ME, Yau K-W (1996) Cyclic nucleotide-gated ion channels: an extended family with diverse functions. Annu Rev Physiol 58:395–426
Finn JT, Krautwurst D, Schroeder JE, Chen T-Y, Reed RR, Yau K-W (1998) Functional co-assembly among subunits of cyclic-nucleotide-activated nonselective cation channels, and across species from nematode to human. Biophys J 74:1333–1345
Flügge U-I (2000) Metabolite transport across the chloroplast envelope of C3-plants. In: Leegood RC, Sharkey TD, von Caemmerer S (eds) Photosynthesis: physiology and metabolism. Kluwar, Dordrecht, pp 137–152
Fodde R, Smits R (2002) Enhanced: a matter of dosage. Science 298:761–763
Fox TC, Guerinot ML (1998) Molecular biology of cation transport in plants. Annu Rev Plant Physiol Plant Mol Biol 49:669–696
Fuchs I, Philippar K, Ljung K, Sandberg G, Hedrich R (2003) Blue light regulates an auxin-induced K+-channel gene in the maize coleoptile. Proc Natl Acad Sci USA 100:11795–11800
Gobert A, Park G, Amtmann A, Sanders D, Maathuis FJM, (2006) Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J Exp Bot 57:791–800
Grunwald ME, Zhong H, Yau K-W (1999) Molecular determinants of the modulation of cyclic nucleotide-activated channels by calmodulin. Proc Natl Acad Sci USA 96:13444–13449
Gunning BES, Steer MW (1975) Ultrastructure and the biology of plant cells. Edward Arnold Publishers, London, pp 115–117
Hennegan KP, Danna KJ (1998) pBIN20: An improved vector for shape Agrobacterium-mediated transformation. Plant Mol Biol Rep 16:129–131
Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280:918–920
Hu G, Yalpani N, Briggs SP, Johal GS (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10:1095–1105
Hua BG, Mercier RW, Zielinski RE, Berkowitz GA (2003) Functional interaction of calmodulin with a plant cyclic nucleotide gated cation channel. Plant Physiol Biochem 41:945–954
Ketchum KA, Slayman CW (1996) Isolation of an ion channel gene from Arabidopsis thaliana using the H5 signature sequence from voltage-dependent K+ channels. FEBS Lett 378:19–26
Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414:656–660
Köhler C, Merkle T, Neuhaus G (1999) Characterization of a novel gene family of putatitive cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J 18:97–104
Köhler C, Neuhaus G (2000) Characterization of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett 4710:133–136
Leng Q, Mercier RW, Yao W, Berkowitz GA (1999) Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol 121:753–761
Leng Q, Mercier RW, Hua BG, Fromm H, Berkowitz GA (2002) Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol 128:400–410
Li X-L, Borsics T, Harrington HM, Christopher DA (2005) Arabidopsis AtCNGC10 rescues potassium channel mutants of E. coli, yeast and Arabidopsis and is regulated by calcium/calmodulin and cyclic GMP in E. coli. Funct Plant Biol 32:643–653
Lin C (2000) Plant blue-light receptors. Trend Plant Sci 5:337–342
Lichtenthaler HK, Buschmann C, Doll M, Fietz HJ, Bach T, Kozel U, Meier D, Rahmsdorf U (1981) Photosynthetic activity, chloroplast ultrastructure, and leaf characteristics of high-light and low-light plants of sun and shade leaves. Photosyn Res 2:115–141
Maathuis FJ, Ichida AM, Sanders D, Schroeder JI (1997) Roles of higher plant K+ channels. Plant Physiol 114:1141–1149
Marschner H (1995) Mineral nutrition of higher plants, 2nd edn. Academic, Boston
Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML (2001) Phylogenetic relationships with cation transporter families of Arabidopsis. Plant Physiol 126:1646–1667
Moshelion M, Becker D, Czempinski K, Mueller-Roeber B, Attali B, Hedrich R, Moran N (2002) Diurnal and circadian regulation of putative potassium channels in a leaf moving organ. Plant Physiol 128:634–642
Neuhaus G, Bowler C, Kern R, Chua NH (1993) Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell 73:937–952
Otegui M, Staehelin LA (2000) Syncytial type cell plates: a novel kind of cell plate involved in endosperm cellularization in Arabidopsis thaliana. Plant Cell 12:933–947
Pan X, Li Y, Stein L (2005) Site preferences of insertional mutagenesis agents in Arabidopsis. Plant Physiol 137:168–175
Philippar K, Fuchs I, Lüthen H, Hoth S, Bauer C, Haga K, Thiel G, Ljung K, Sandberg G, Böttger M, Becker D, Hedrich R (1999) Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc Natl Acad Sci, USA 96:2186–12191
Philippar K, Ivashikina N, Ache P, Christian M, Luthen H, Palme K, Hedrich R (2004) Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J 37:815–827
Pyke K, Lopez-Juez E (1999) Cellular differentiation and leaf morphogenesis in Arabidopsis. Crit Rev Plant Sci 18:527–546
Quail PH (1997) The phytochromes: a biochemical mechanism of signaling in sight? Bioessays 19:571–579
Rayle DL, Cleland RE (1992) The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol 99:1271–1274
Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52:627–658
Schuurink RC, Shartzer SF, Fath A, Jones RL (1998) Characterization of a calmodulin-binding transporter from plasma membrane of barley aleurone. Proc Natl Acad Sci USA 95:1944–1949
Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmon JM, Gaymard F, Grignon C (1992) Cloning and expression in yeast of a plant potassium ion transport system. Science 256:663–665
Stahlberg R, Van Volkenburgh E (1999) The effect of light on membrane potential, apoplastic pH and cell expansion in leaves of Pisum sativum L. var. Argentum. Planta 208:188–195
Stiles KA, Van Volkenburgh E (2002) Light-regulated leaf expansion in two Populus species: dependence on developmentally controlled ion transport. J Exp Bot 53:1651–1657
Stiles KA, McClintick A, Van Volkenburgh E (2003) A developmental gradient in the mechanism of K+ uptake during light-stimulated leaf growth in Nicotiana tabacum L. Planta 217:587–596
Sunkar R, Kaplan B, Bouche N, Arazi T, Dolev D, Talke IN, Maathuis FJ, Sanders D, Bouchez D, Fromm H (2000) Expression of a truncated tobacco NtCBP4 channel in transgenic plants and disruption of the homologous Arabidopsis CNGC1 gene confer Pb2+ tolerance. Plant J 24:533–542
Talke IN, Blaudez D, Maathuis FJ, Sanders D (2003) CNGCs: prime targets of plant cyclic nucleotide signalling? Trends Plant Sci 8:286–293
Trudeau MC, Zagotta WN (2002) Mechanism of calcium/calmodulin inhibition of rod cyclic nucleotide-gated channels. Proc Natl Acad Sci USA 99:8424–8429
Varnum MD, Zagotta WN (1997) Interdomain interactions underlying activation of cyclic nucleotide-gated channels. Science 278:110–113
Very AA, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54:575–603
Veitia RA (2002) Exploring the etiology of haploinsufficiency. Bioessays 24:175–84
Weston E, Thorogood K, Vinti G, Lopez-Juez E (2000) Light quantity controls leaf-cell and chloroplast development in Arabidopsis thaliana wild type and blue-light-perception mutants. Planta 211:807–815
Yoshioka K, Moeder W, Kang H-G, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF (2006) The chimeric Arabidopsis cyclic nucleotide-gated ion channel 11/12 activates multiple pathogen resistance responses. Plant Cell 18:747–763
Zielinski RE (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49:697–725
Acknowledgments
This research was supported by funds from the US Department of Energy grant no. DE-FG02-03ER15395 to DAC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Borsics, T., Webb, D., Andeme-Ondzighi, C. et al. The cyclic nucleotide-gated calmodulin-binding channel AtCNGC10 localizes to the plasma membrane and influences numerous growth responses and starch accumulation in Arabidopsis thaliana . Planta 225, 563–573 (2007). https://doi.org/10.1007/s00425-006-0372-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0372-3