Abstract
A pathogenesis related protein (AhPR10) is identified from a clone of 6-day old Arachis hypogaea L. (peanut) cDNA library. The clone expressed as a ∼20 kDa protein in E. coli. Nucleotide sequence derived amino acid sequence of the coding region shows its homology with PR10 proteins having Betv1 domain and P loop motif. Recombinant AhPR10 has ribonuclease activity, and antifungal activity against the peanut pathogens Fusarium oxysporum and Rhizoctonia solani. Mutant protein AhPR10-K54N where lys54 is mutated to asn54 loses its ribonuclease and antifungal activities. FITC labeled AhPR10 and AhPR10-K54N are internalized by hyphae of F. oxysporum and R. solani but the later protein does not inhibit the fungal growth. This suggests that the ribonuclease function of AhPR10 is essential for its antifungal activity. Energy and temperature dependent internalization of AhPR10 into sensitive fungal hyphae indicate that internalization of the protein occurs through active uptake.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are equipped with well-established defense machinery used to combat the war like situation during stress. This defense machinery acts by up regulating the expression of number of genes including microbe inducible phytoalexins (Darvill and Albersheim 1984), hydroxyproline rich glycoproteins (Hahn et al. 1989; Sommer-Knudsen et al. 1996, 1997), lytic enzymes, proteinase inhibitors and several low molecular weight proteins initially defined as pathogenesis-related (PR) proteins (Bowles 1990). A large number of PR proteins have been characterized in recent years and grouped into 17 families based on their primary structure, serological relationships, and biological activities (Christensen et al. 2002). Most PR-proteins are either extra cellular or intracellularly localized in the vacuole. In contrast, the PR-10 proteins are the only group that is intracellular and cytoplasmic. Intracellular PR10 proteins were first described in cultured parsley cells (Somssich et al. 1988). Members of PR 10 family are generally acidic, with a molecular mass of 16–19 kDa, lack signal peptide, resistant to proteases (Walter et al. 1990).
PR10 proteins have been reported in a number of species through out the plant kingdom, including pea (Fristensky et al 1988), bean (Walter et al. 1990, 1996), soybean (Crowell et al. 1992), alfalfa (Breda et al. 1996), potato (Matton and Brisson 1989), cotton (Zhou et al. 2002), pachyrrhizus (Wu et al. 2003), capsicum (Park et al. 2004) as well as several monocots including asparagus (Warner et al. 1992), lily (Huang et al. 1997), rice (Midoh and Iwata 1996) and sorghum (Lo et al. 1999). Apart from showing amino acid sequence similarity to the major food allergens of tree (Warner et al. 1994), PR10s also share homology to ribonuclease from phosphate-starved ginseng cells. (Moiseyev et al. 1994). Many PR10 proteins including Betv1 (Bufe et al. 1996), LaPR10 (Bantignies et al. 2000), GaPR10 (Zhou et al. 2002), SPE16 (Wu et al. 2003) have been reported to exhibit ribonuclease activity. Recently, a PR-10 protein from Capsicum annuum (CaPR10) has been shown to have both RNase and antiviral activity (Park et al. 2004). Lam and Ng (2001) have shown in vitro translation inhibitory activity for Panax RNase and predicted it to be responsible for its antifungal activity.
Although antifungal activity of PR10 proteins has been presumed to be mediated by its RNase activity, there has been no evidence for its internalization and the direct involvement of its ribonuclease activity in antifungal action. In this report, we have described the identification of a PR10 having RNase activity from the commercial crop peanut for the first time and explored the mode of its antifungal activity.
Materials and methods
Plant materials and growth conditions
Peanut seeds (Arachis hypogaea L. var. ICGS1) were obtained from National Research Center for groundnut (NRCG), Junagarh, Gujarat, India. The seeds were sterilized with 95% ethanol followed by 0.2% acidified mercuric chloride wash. The germinated seeds were grown for 6 days in agar medium and kept in aseptic conditions in a growth chamber programmed with alternate light and dark cycles of 12 h and 60–70% relative humidity at 28°C. Harvested and cleaned plant samples were stored in liquid nitrogen prior to use for RNA isolation.
Identification of cDNA clone and sequence analysis
Six-day old peanut roots were used for total RNA isolation using RNaesy plant mini kit followed by mRNA preparation using oligotex suspension (Qiagen). The cDNA library was constructed from polyA+ mRNA using SMART cDNA library construction kit (Clontech). Briefly, ds cDNAs synthesized were ligated to λTripleEx2 vector (Clontech) followed by packaging into lambda packaging extract (Gigapack, Stratagene). Unamplified library was titered to calculate pfu/ml. The average insert size of the library was calculated using PCR based insert screening of more than 200 cDNA clones. Primers used for the insert screening were provided by the manufacturer (Clontech). The cDNA clones were randomly picked up for expressed sequence tags analysis (ESTs) and sequenced using a dye terminator cycle sequencing kit (PE applied Biosystems) and an automated capillary DNA sequencer, ABI 3370, Prism (Applied Biosystems). Nucleotide sequences were analyzed for open reading frames using DNASTAR program (Lasergene, Madison, WI, USA) followed by homology search with BLAST (Altschul et al. 1997) against sequences in the NCBI database. Multiple sequence alignment of the nucleotide sequence deduced protein sequence was done using MegAlign program (Lasergene).
Construction of expression vector
The cDNA library vector carrying cDNA for putative AhPR10 was used as template to PCR amplify 453 bp coding region. Along with the wild type, three substitution mutants (K54N, F148S, and H150Q) were constructed by site directed mutagenesis using overlap extension PCR (Ho et al. 1989) with the following primer sets, Wild-F (5′CATGCCATGGGTGTTCACACTTTTGAAGA3′) and Wild-R (5′CGGAATTCCCGTGGACAAACATAACTCATA3′); K54N-F (5′CCTGGAACTGTCAACAAGGTTACCGCT3′) and K54N-R (5′AGCGGTAACCTTGTTGACAGTTCCAGG3′); F148S-R (5′CGGAATTCCCGTGGACAGACATAACCCTCATA3′) and F148S-F (5′CATGCCATGGGTGTTCACACTTTTGAAGA3′); H150Q-R (5′CGGAATTCCCCTGGACAAACATAACCCTCATA3′) and H150Q-F (5′CATGCCATGGGTGTTCACACTTTTGAAGA3′). NcoI and EcoRI restriction sites are underlined and mismatched nucleotides are bold faced. PCR were carried out at an annealing temperature of 56°C for 28 cycles. After digestion with NcoI and EcoRI, the PCR products were ligated into expression vector pET28a (Novagen) to express with 6× His affinity tag at C terminal end under the T7 promoter. Desired mutations were confirmed by DNA sequencing as described above.
Expression and purification of recombinant proteins
The pET28a vector containing wild type AhPR10 and its mutated coding regions were transformed into E. coli BL21 (DE3) strain and over expression of the cloned genes was induced with 1 mM IPTG at 37°C for 4 h.
For recombinant protein purification, bacterial cells were pelleted after induction, suspended in lysis buffer (50 mM sodium phosphate buffer, pH 7.0 containing 150 mM NaCl, 1 mM PMSF and 0.75 mg/ml of lysozyme) and sonicated on ice for 3 min (30 s pulse/min). Lysate was centrifuged at 14,000g, 4°C for 20 min and loaded onto Talon resin metal affinity column (Clontech), eluted with 100 mM of imidazole and desalted by centricon YM10 (Millipore). The recombinant proteins were further purified to apparent homogeneity by Sephadex G50 gel filtration chromatography and analyzed by 15% SDS PAGE (Laemmli 1970) followed by Western-blot analysis with monoclonal anti-His antibody (Qiagen) using the standard protocol (Towbin et al. 1979).
RNase assays
The RNA degradation assays by the method of Bantignies et al. (2000) was carried out using 10 μg of total RNA extracted from peanut root using TRIZOL reagent (Invitrogen) and 2 μg of purified FITC labeled or unlabeled AhPR10 or its mutant proteins at 37°C for 1 h. Reaction mixtures were deproteinized by extraction with phenol–chloroform followed by ethanol precipitation and analyzed by 1.2% agarose gel electrophoresis.
RNase activity units of the proteins were determined spectrophotometrically using acid soluble method (Wang and Ng 2000) using yeast tRNA as a substrate. Two hundred micrograms of yeast tRNA were incubated with AhPR10 or its mutant proteins in 150 μl of 0.1 M Tris–HCl buffer, pH 7.0 at 37°C for 15 min followed by addition of 350 μl of ice cold perchloric acid (3.4%) to terminate the reaction. The reaction mixture was kept on ice for 30 min before centrifugation (12,000g, 15 min at 4°C). The absorbance of the supernatant was read at 260 nm after suitable dilution. One unit of RNase activity was defined as the amount of enzyme that produces an absorbance increase of one in the acid soluble supernatant at A260 after 15 min incubation at 37°C.
Ribonuclease activities of wild and mutant proteins were also assayed by zymogram analysis method as described by Yen and Green (1991). Proteins exhibiting RNase activity were seen as bright bands against the blue background.
Antifungal activity assays
Antifungal activities of AhPR10 and its mutant proteins were determined by radial growth inhibition method as described by Schlumbaum et al. (1986) as well as micro spectrophotometry (Broekaert et al. 1990). Fungal pathogens, Fusarium oxysporum, Rhizoctonia solani, Sclerotium rolfsii, Aspergillus flavus, A. niger and Phytophthora infestans were used to check the antifungal activity of AhPR10s. For spectrophotometric assay, 10 μl of protein samples at different concentrations were mixed with 90 μl of Sabouraud dextrose broth (SDB, HiMedia) containing fungal spores that had been pre-germinated for 16 h in 96 well microtiter plates. After incubation for 12 h at 37°C without shaking, fungal growth was estimated by observing the absorbance at 595 nm in an ELISA plate reader (Molecular Devices, USA). IC50 values were calculated from twofold dilution steps (Terras et al. 1992). Growth inhibitions were also ascertained by visualizing under bright field phase contrast microscope (TS100, Nikon).
FITC labeling
Wild type AhPR10 and its mutant proteins were fluorescence labeled using fluorescein isothiocyanate (FITC, Sigma). Briefly, 0.5 mg of FITC powder was dissolved in 50 μl of anhydrous DMSO to give a reactive dye concentration of 10 mg/ml. One milligram of AhPR10 or mutant protein was dissolved in 200 μl of 50 mM carbonate–bicarbonate buffer, pH 9.0. FITC solution was added drop wise to the protein sample and mixed by stirring. Samples were incubated at 25°C in dark for 1 h. Unreacted FITC was removed by Sephadex G25 gel filtration and the protein concentration was determined by the Bradford method (Bradford 1977). An intense fluorescent band under UV light in SDS-PAGE gel verified the conjugation between FITC and protein. FITC labeled proteins were stored at 4°C until use.
Internalization assays by fluorescence microscopy
FITC labeled proteins were applied to fungi in a micro titer plate. To 90 μl of SDB (HiMedia) containing fungal spores (1 × 105 spores/ml) that had been pre-germinated for 16 h in 96 well microtiter plates, 12 μg/ml of FITC labeled AhPR10 proteins were added and incubated at 37°C and 4°C in presence or absence of 0.05% NAN3, for 1 h. For time course analysis of internalization, 100 μl hyphal suspension containing FITC labeled protein was incubated for different times starting from 15 min to 6 h. After incubation, hyphae were washed twice with 1× PBS and then observed under fluorescence microscope TS100 (Nikon) attached with FITC and propidium iodide (PI) filters. Images were captured using 40× objective under bright field as well as under fluorescent filters. Green and red images were merged using Image Pro software (Media cybernetics, USA) for co-localization of the FITC labeled protein and PI at different time points. For all the images a scale bar of 100 μm was set.
Propidium-iodide uptake assays
Fungal spores were pre germinated in SDB as described above, followed by addition of labeled or unlabeled AhPR10 or mutant AhPR10 along with 2 μM of PI. After different time intervals (15 min–6 h) hyphae were washed twice with 1× PBS to remove unincorporated dye and protein, and then monitored by fluorescence microscope.
Results
Identification of AhPR10 cDNA clone from peanut roots
Titer of unamplified peanut root cDNA library was found to 106 pfu/ml with an average insert size of 900 bp. Random sequencing of the library cDNA clones resulted in identification of a cDNA clone showing upto 70% homology with reported plant PR10 proteins in NCBI database. This clone represented the first PR10 identified from Arachis hypogaea L.
Sequence analysis
The cDNA clone identified as putative PR10 gene has the insert size of 847 bp with a single open reading frame (ORF) of 453 nucleotides to encode a protein of 150 amino acid residues. The nucleotide sequence also shows a 5′ untranslated region (5′ UTR) of 84 nucleotides and a 3′ UTR of 310 nucleotides along with a poly A signal AATAAA from 655–660 bp (Fig. 1a). The predicted translation product of the gene has the calculated molecular mass of 16.217 kDa and an isoelectric point of 5.3. The deduced amino acid sequence of AhPR10 shows the presence of conserved P loop motif GXGGXGXXK and a Betv1 domain as present in many PR-10 proteins (Fig. 1a). The sequence alignment using MegAlign (DNASTAR, Lasergene, Madison, WI, USA) of the predicted amino acid sequence of AhPR10 with different members of the PR10 family indicated that AhPR10 has the similarity with other PR10 proteins (Fig. 1b)
Nucleotide and nucleotide sequence derived amino acid sequences of putative AhPR10. a Nucleotide and deduced amino acid sequence of AhPR10 cDNA. P loop motif and Betv1 domain are underlined. Asterisk marks represent the translation start site and termination codon of the open reading frame. PolyA signal sequence is shown in the box. b Comparison of the predicted amino acid sequence of AhPR10 with Betv1 allergen (Betula pendula), [Z72431] CaPR10 (Capsicum annuum) [AF244121], GaPR10 (Gossypium arboreum) [AF416652], LaPR10 (Lupinus albus) [AJ000108], Ll PR10.1B (Lupinus luteus) [X79975], PANGI RNase2 (Panax ginseng) [P80890], SPE16 (Pachyrhizus erosus) [AY433943], and yPR10 (Malus domestica) [AJ417551]. The crucial amino acid residues implicated for RNase activity are shown in bold. The percentage amino acid homology of AhPR10 with other PR10s is indicated at the end of the sequence
Analysis of expressed recombinant AhPR10 proteins
Wild type AhPR10 and its mutants cloned in pET28a vector were confirmed by restriction enzyme digestion analysis (not shown). DNA sequencing confirmed the site directed mutagenesis at desired sites (S1). Protein molecular weights ( 20 kDa) of the putative wild type-recombinant AhPR10 and its three mutant proteins, K54N, F148S and H150Q expressed in E. coli are in agreement with their amino acid sequence derived molecular weights (S2). The apparent homogeneity of the protein preparations is evident from their SDS-PAGE analysis and identity of the expressed proteins was established by probing the Western-blot with monoclonal anti His-tag antibody (Fig. 2).
SDS-PAGE (15%) analysis of purified AhPR10 and its mutant proteins expressed in E. coli BL21 (DE3) cells. a Protein molecular weight markers (lane 1), purified AhPR10 (lane 2) and its mutant proteins K54N (lane 3), F148S (lane 4) and H150Q (lane 5). b Western-blot analysis with anti-His antibody of purified AhPR10 and its mutant proteins. Lanes 1–4, respectively, indicate wild type, K54N, F148S, and H150Q recombinant AhPR10 proteins. Lane M indicates the prestained protein molecular weight markers
Ribonuclease activity of AhPR10 and the mutant proteins
Differential RNase activities of wild and mutant AhPR10 proteins were observed in all three RNase assays as shown in the Fig. 3. Ribonuclease activity of AhPR10-K54N protein almost completely abolished whereas AhPR10-F148S and AhPR10-H150Q proteins lost their activities partially. The loss of activity in AhPR10-H150Q is more than AhPR10-F148S. Ribonuclease specific activity of wild type AhPR10 and mutants when calculated using acid soluble method was found to be 115 U/mg for wild type protein whereas for AhPR10-F148S and AhPR10-H150Q it reduced to 75 and 36 U/mg, respectively. Ribonuclease activity of AhPR10-K54N protein was almost completely lost with a value of 2.5 U/mg (Fig. 3b bottom). Ribonuclease activities of AhPR10 and its mutant proteins F148S and H150Q were not affected on FITC labeling (results not shown).
Activity of RNase of recombinant AhPR10 and its mutant proteins, F148S, H150Q and K54N as visualized in a 1.2% agarose gel using 2 μg purified recombinant proteins and 10 μg total RNA from peanut roots. b Comparison of RNase activities of wild type AhPR10 and its mutant proteins, by in gel (top) using 3 mg/ml of yeast tRNA in polyacrylamide gel and loading 2 μg of AhPR10 proteins in the well, and spectrophotometric assays (bottom) using 200 μg yeast tRNA per assay at pH 7.0 as described in Materials and methods. The RNase units plotted in (b, bottom) are the average of three independent determinations (P values ≤ 0.05)
Antifungal activity of AhPR10
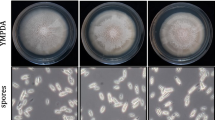
Preliminary screening of the antifungal activity of purified recombinant AhPR10 by halo formation indicated that it inhibits the growth of F. oxysporum and R. solani but not S. rolfsii, A. flavus, A. niger and P. infestans (results not shown).
In micro plate assays IC50 values of AhPR10 against F. oxysporum and R. solani were found to be 12.5 and 100 μg/ml (not shown) respectively. The effects on growth inhibition of F. oxysporum with increasing concentrations of AhPR10 and AhPR10-K54N are shown in the Fig. 4. AhPR10-K54N showed only 10% growth inhibition of F. oxysporum at 100 μg/ml concentration (Fig. 4). Inhibition of mycelial growth of F. oxysporum with increasing concentration of AhPR10 was also visible under microscope as shown in the insets of the Fig. 4.
Effect of AhPR10 and its mutant proteins, AhPR10-F148S, AhPR10-H150Q and AhPR10-K54N on growth of F. oxysporum and S. rolfsii in SDB medium at 28°C in 12 h. Each point of the plot were the average of four independent determinations. The inset shows the phase contrast view of F. oxysporum at three different concentrations of AhPR10
Internalization of AhPR10 in fungal hyphae
Internalization of FITC labeled AhPR10 occurred in F. oxysporum within 30 min of incubation at 37°C but not at 4°C. Uptake of the protein was inhibited by 0.05% sodium azide. The FITC labeled His tagged control protein (profilin from peanut) did not appear to internalize as no fluorescent signal was observed. In case of S. rolfsii fluorescent signal was not detected even after 4 h of incubation with labeled AhPR10 (Fig. 5).
AhPR10 interaction with fungal hyphae and its localization
Time course analysis showed increase in internalization of FITC labeled AhPR10 with time from 15 min to 4 h (Fig. 6b, c, d). FITC labeled AhPR10 was not localized in specific area in the hyphae, however, an intense green fluorescence at hyphal tip on 1 h incubation can be seen in F. oxysporum (Fig. 6g). Labeled AhPR10-K54N also internalized in F. oxysporum and appeared to be more concentrated in the vacuoles (Fig. 6i). AhPR10-F148S and AhPR10-H150Q were also internalized in F. oxysporum. Similar results were observed with R. solani (results not shown).
Fluorescence imaging of the interaction of FITC labeled AhPR10 and its mutant protein AhPR10-K54N with fungal hyphae. a and e Phase contrast images of F.oxysporum. b–d The internalization of FITC labeled AhPR10 (12.5 μg/ml) in F. oxysporum hyphae on incubation for 15 min, 1 h and 4 h, respectively, at 37°C. f Image of F. oxysporum after 4-h incubation with AhPR10-K54N protein. g Enlarged image of the hyphal tip shown by arrow in (c). Arrows in (h) and (i), respectively, indicate the vacuoles and vacuolar accumulation of internalized FITC labeled AhPR10-K54N in F. oxysporum
AhPR10 and membrane permeability
Incubation of FITC labeled AhPR10 or its mutant protein AhPR10-K54N at 37°C leads to internalization of the protein within 15 min. Internalization of PI occurred after 30 min of incubation with AhPR10 while that with mutant protein it did not occur even after 6 h of incubation (Fig. 7a, b). Heat killed hyphae showed positive staining of PI within 15 min (results not shown). The merged image in panel (c) of the Fig. 7 shows the co-localization of AhPR10 and PI in F. oxysporum hyphae.
Time course analysis of the effect of FITC labeled AhPR10 and AhPR10-K54N on membrane permeability of F. oxysporum hyphae. FITC labeled AhPR10 (a) and AhPR10-K54N (b) were used to check the effect of internalization on membrane integrity using propidium iodide uptake assay. Hyphae were observed under fluorescence microscope at different time points (15 min–6 h), and images were taken under phase contrast (PC), FITC filter and PI filter. c Co-localized FITC labeled AhPR10 protein and propidium iodide in F. oxysporum hyphae. Inset 4 of c shows the image obtained after merging green and red images of hyphae as observed under FITC (inset 2) and PI (inset 3) filters whereas inset 1 is the same image of F. oxysporum hyphae under phase contrast
Discussion
Pathogenesis related proteins of PR10 family are believed to have potential role in plant defense (Van Loon and Van Stein 1999). The putative AhPR10 has maximum amino acid sequence homology (70%) with drought induced protein RPR-10 from Retama raetam (Pnueli et al. 2002) followed by 58% homology with pea disease resistance response protein (Chiang and Hadwiger 1990). This constitutively expressed AhPR10 is inducible by drought and pathogen stress (manuscript in preparation). The presence of P loop motif (GXGGXGXXK) and 38–60% amino acid sequence homology of AhPR10 with PR10s having RNase activity indicated the possible RNase activity of AhPR10. The P loop has been considered to be the possible RNA phosphate binding site correlated with ribonucleotlytic activity (Bantignies et al. 2000; Hoffmann-Sommergruber et al. 1997). In other PR10s having RNase actvity tyr148 and glu150 are conserved but in AhPR10 these are replaced with phenylalanine and histidine respectively. However, glu96 and lys54 are also conserved in AhPR10.
The purified recombinant AhPR10 showed RNase activity in all three RNase assays (Fig. 3) as predicted by its amino acid sequence. Reduction of RNase activity of AhPR10-F148S and AhPR10-H150Q suggests the involvement of phe148 and his150 in its RNase activity. The lys54 appears to play a more crucial role in the RNase activity of AhPR10 as its mutation causes the almost complete loss of its RNase activity (Fig. 3). A similar role of lys55 in GaPR10 (Gossypium arboreum) and P loop in SPE16 (Pachyrrhizus erosus) in RNase activities were reported earlier (Wu et al. 2003; Zhou et al. 2002).
Growth inhibition of F. oxysporum and R. solani and not of S. rolfsii, A. niger, A. flavus and P. infestans by recombinant AhPR10 indicated the selectivity of inhibition by the protein. The non inhibition of the growth of F. oxysporum and R. solani by AhPR10-K54N protein lacking RNase activity suggests the possible role of its RNase function in fungal inhibition.
AhPR10 appears to exert its antifungal activity upon entering in the fungal hyphae of sensitive fungi as the protein is not internalized in S. roflsii. The temperature dependence and sodium azide inhibition of the entry of AhPR10 suggest the active transport of the protein in the fungal hyphae. Similar observations were also made earlier with antifungal histatins against C. albicans (Kim et al. 2001; Xu et al. 1999).
Internalization of AhPR10 mutant proteins implicates that RNase activity domain of AhPR10 is distinct from the site that interacts with the fungal membrane. It has been shown by Mogensen et al. (2002) that Bet v1 contains two ligand binding sites, one binds to 8-anilino-1-naphthalenesulfonic acid (ANS), a hydrophobic ligand and the other to kinetin, an adenine derivative. In AhPR10, lys54 adjacent to P loop may interact with RNA whereas the hydrophobic ligand binding site may interact with the receptor or target site on the fungal hyphae.
The uptake of PI (a commonly used cell viability marker which enters only through damaged cell membranes) after internalization of AhPR10 (Fig. 7a) suggests that membrane integrity is not disrupted by the protein unlike antifungal plant defensins, which directly permeabilize or disrupt the fungal membrane (Thevissen et al. 1999). Disruptions of the membrane integrity and PI uptake appear to be the post internalization phenomenon following the ribonuclease function of AhPR10 within the fungal hyphae. Plant defense proteins like ribosome inactivating proteins are also shown to act after internalization inside the fungi (Park et al. 2002). For some non plant antimicrobial peptides, such as buforin II from Bufo bufo garagriozans (Park et al. 1998), PR-39 from pig intestine (Boman et al. 1993) and tachyplesin I from Tachypleus tridentatus (Yonezawa et al. 1992) non-lytic internalization and nucleic acid binding properties have been reported. However, the intracellular nature of PR10 proteins suggests that such an interaction is possible only when the pathogen enters the host cells.
Thus, internalization appears to be essential for AhPR10 to display its antifungal activity through its RNase function. Energy and temperature dependence of AhPR10 for rapid internalization in the sensitive fungi without disrupting the membrane suggest that the internalization of the protein occurs through active uptake. Further studies on the type of putative receptor or target site on fungal membrane, and the trafficking pathway by which the wild and the mutant proteins are destined to different locations inside the hyphae will help to unravel the mechanism of interaction between the fungal membrane and AhPR10.
Abbreviations
- AhPR10:
-
Arachis hypogaea pathogenesis related 10 protein
- AhPR10-F148S:
-
AhPR10 mutant protein where phenylalanine148 is replaced by serine
- AhPR10-H150Q:
-
AhPR10 mutant protein where histidine150 is replaced by glutamine
- AhPR10-K54N:
-
AhPR10 mutant protein where lysine54 is replaced by asparagine
- FITC:
-
Fluorescein isothiocyanate
- IC50 :
-
Inhibitory concentration causing 50% growth inhibition
- IPTG:
-
Isopropyl thio-β-galactoside
- NaN3 :
-
Sodium azide
- PBS:
-
Phosphate buffered saline, pH 7.2
- PCR:
-
Polymerase chain reaction
- PI:
-
Propidium iodide
- PMSF:
-
Phenyl methyl sulfonyl fluoride
- PR:
-
Pathogenesis related
- SDS-PAGE:
-
Sodium dodecyl sulphate polyacrylamide gel electrophoresis
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bantignies B, Seguin J, Muzac I, Dedaldechamp F, Gulick P, Ibrahim R (2000) Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol Biol 42:871–881
Boman HG, Agerberth B, Boman A (1993) Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect Immun 61:2978–2984
Bowles DJ (1990) Defense-related proteins in higher plants. Annu Rev Biochem 59:873–907
Bradford MM (1977) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Breda C, Sallaud C, El-Turk J, Buffard D, de Kosak I, Esnault R, Kondorosi A (1996) Defense reaction in Medicago sativa: a gene encoding a class 10 PR protein is expressed in vascular bundles. Mol Plant Microbe Interact 9:713–719
Broekaert WF, Frankt RG, Terras Bruno PA, Cammue, Jos Vanderleyden (1990) An automated quantitative assay for fungal growth inhibition. FEMS Microbiol Lett 69:55–60
Bufe A, Spangfort MD, Kahlert H, Schlaak M, Becker WM (1996) The major birch pollen allergen, Bet V1, shows ribonuclease activity. Planta 199:413–415
Chiang CC, Hadwiger LA (1990) Cloning and characterization of a disease resistance response gene in pea inducible by Fusarium solani. Mol Plant Microbe Interact 3:78–85
Christensen AB, Cho BH, Naesby M, Gregersen PL, Brandt J, Madriz-Ordenana K, Collinge DB, Thordal-Christensen H (2002) The molecular characterisation of the two barley proteins establishes the novel PR-17 family of pathogenesis-related protein. Mol Plant Pathol 3:134–144
Crowell D, John ME, Russell D, Amasino RM (1992) Characterization of a stress-induced developmentally regulated gene family from soybean. Plant Mol Biol 18:459–466
Darvill AG, Albersheim P (1984) Phytoalexins and their elicitors-a defense against microbial infection in plants. Annu Rev Plant Physiol 35:243–275
Fristensky B, Horovitz D, Hadwiger LA (1988) cDNA sequences for pea disease response genes. Plant Mol Biol 11:713–715
Hahn MG, Buchell P, Cervone F, Doares SH, O’Neil RA, Darvill AG, Albersheim P (1989) Roles of cell wall constituents in plant–pathogen interactions. In: Kosuge T, Nester EW (eds) Plant–microbe interactions—molecular and genetic perspectives, 1st edn, vol 3. McGraw-Hill, New York, pp 131–181
Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59
Hoffmann-Sommergruber K, Vanek-Krebitz M, Radauer C, Wen J, Ferreira F, Scheiner O, Breiteneder H (1997) Genomic characterization of members of the Bet v 1 family: genes coding for allergens and pathogenesis-related proteins share intron positions. Gene 197:91–100
Huang JC, Chang FC, Wang CS (1997) Characterization of a lily tapetal transcript that shares similarity with a class of intracellular pathogenesis-related (IPR) proteins. Plant Mol Biol 34:681–686
Kim DH, Lee DG, Kim KL, Lee Y (2001) Internalization of tenecin 3 by a fungal cellular process is essential for its fungicidal effect on Candida albicans. Eur J Biochem 268:4449–4458
Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680
Lam SK, Ng TB (2001) Isolation of a novel thermolabile heterodimeric ribonuclease with antifungal and antiproliferative activities from roots of the sanchi ginseng Panax notoginseng. Biochem Biophys Res Commun 285:419–423
Lo SC, Hipskind JD, Nicholson RL (1999) cDNA cloning of a sorghum pathogenesis-related protein (PR-10) and differential expression of defense-related genes following inoculation with Cochliobolus heterostrophus or Colletotrichum sublineolum. Mol Plant Microbe Interact 12:479–489
Matton DP, Brisson N (1989) Cloning, expression and sequence conservation of pathogenesis related gene transcripts of potato. Mol Plant Microbe Interact 2:325–331
Midoh N, Iwata M (1996) Cloning and characterization of a probenaole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant Cell Physiol 37:9–18
Moiseyev GP, Beintema JJ, Fedoreyeva LI, Yakovlev GI (1994) High sequence similarity between a ribonuclease from ginseng calluses and fungus-elicited proteins from parsley indicates that intracellular pathogenesis-related proteins are ribonucleases. Planta 193:470–472
Mogensen JE, Wimmer R, Larsen JN, Spangfort MD, Otzen DE (2002) The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J Biol Chem 277:23684–23692
Park CB, Kim HS, Kim SC (1998) Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem Biophys Res Commun 244:253–257
Park SW, Stevens NM, Vivanco JM (2002) Enzymatic specificity of three ribosome inactivating proteins against fungal ribosome, and correlation with antifungal activity. Planta 216:227–234
Park CJ, Kim KJ, Shin R, Park JM, Shin YC, Paek KH (2004) Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J 37:186–198
Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R (2002) Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J 31:319–330
Schlumbaum A, Mooch F, Vögeli U, Boller T (1986) Plant chitinases are potent inhibitors of fungal growth. Nature 324:365–367
Sommer-Knudsen J, Clarke AE, Bacic A (1996) A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J 9:71–83
Sommer-Knudsen J, Clarke AE, Bacic A (1997) Proline- and hydroxyproline-rich gene products in the sexual tissues of flowers. Sex Plant Reprod 10:253–260
Somssich IE, Schmelzer E, Kawalleck P, Hahlbrock K (1988) Gene structure and in situ transcript localization of pathogenesis-related protein 1 in parsley. Mol Gen Genet 213:93–98
Terras FR, Schoofs HM, De Bolle MF, Van Leuven F, Rees SB, Vanderleyden J, Cammue BP, Broekaert WF (1992) Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem 267:15301–15309
Thevissen K, Terras FR, Broekaert WF (1999) Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl Environ Microbiol 65:5451–5458
Towbin H, Staehelin T,Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354
Van Loon LC, Van Stien EA (1999) The families of pathogenesis related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
Walter MH, Liu J-W, Grand C, Lamb CJ, Hess D (1990) Bean pathogenesis-related (PR) proteins deduced from elicitor-induced transcripts are members of a ubiquitous new class of conserved PR proteins including pollen allergens. Mol Gen Genet 222:353–360
Walter MH, Liu J-W, Wunn J, Hess D (1996) Bean ribonuclease-like pathogenesis-related protein genes (Ypr10) display complex patterns of developmental, dark-induced and exogenous-stimulus dependent expression. Eur J Biochem 239:281–293
Wang HX, Ng TB (2000) Quinqueginsin, a novel protein with anti-human immunodeficiency virus, antifungal, ribonuclease and cell-free translation—inhibitory activities from American ginseng roots. Biochem Biophys Res Commun 269:203–208
Warner SAJ, Scott R, Draper J (1992) Characterization of a wound-induced transcript from the monocot asparagus that shares similarity with a class of intracellular pathogenesis-related (PR) proteins. Plant Mol Biol 19:555–561
Warner SAJ, Gill A, Draper J (1994) The developmental expression of the asparagus intracellular PR protein (AoPR1) gene correlates with sites of phenylpropanoid biosynthesis. Plant J 6:31–34
Wu F, Yan M, Li Y, Chang S, Song X, Zhou Z, Gong W (2003) cDNA cloning, expression, and mutagenesis of a PR-10 protein SPE-16 from the seeds of Pachyrrhizus erosus. Biochem Biophys Res Commun 312:761–766
Xu Y, Ambudkar I, Yamagishi H, Swaim W, Walsh TJ, Oconnell BC (1999) Histatin 3-mediated killing of Candida albicans: effect of extracellular salt concentration on binding and internalization. Antimicrob Agents Chemother 43:2256–2262
Yen Y, Green PJ (1991) Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiol 97:1487–1493
Yonezawa A, Kuwahara J, Fujii N, Sugiura Y (1992) Binding of tachyplesin I to DNA revealed by footprinting analysis: significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 31:2998–3004
Zhou XJ, Shan Lu, Xu YH, Wang JW, Chen XY (2002) A cotton cDNA (GaPR-10) encoding a pathogenesis-related 10 protein with in vitro ribonuclease activity. Plant Sci 162:629–636
Acknowledgments
The authors gratefully acknowledge the Director of the Institute of Genomics and Integrative Biology for providing the infrastructural facilities to carry out this work. Authors also thank Dr. Pratibha Sharma of Mycology and Plant Pathology Division, IARI, New Delhi, India for providing the fungal pathogens. PC is the recipient of Research Fellowship from CSIR (India).
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence of AhPR10 reported in this paper is submitted to NCBI Nucleotide Sequence Database under the Accession number AY726607.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chadha, P., Das, R.H. A pathogenesis related protein, AhPR10 from peanut: an insight of its mode of antifungal activity. Planta 225, 213–222 (2006). https://doi.org/10.1007/s00425-006-0344-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0344-7