Abstract
Following the screening of a suppression subtractive library developed from durum wheat plants exposed to low temperature for 6 h, two early cold-regulated (e-cor) genes have been isolated. These genes, coding putatively for a ribokinase (7H8) and a C3H2C3 RING-finger protein (6G2), were characterized by the stress-induced retention of a subset of introns in the mature mRNA. This feature was dependent on cold for 7H8 and on cold and dehydration for 6G2. When other genes, such as the stress-related gene WCOR410c, coding for a dehydrin (one intron), or a gene coding for a putative ATP binding cassette transporter (16 introns) were analyzed, no cold-dependent intron retention was observed. Cold-induced intron retention was not observed in mutants defective in the chloroplast development; nevertheless treatment with cycloheximide in the absence of cold was able to promote intron retention for the 7H8 e-cor gene. These results suggest that the cold-induced intron retention reflects the response of the spliceosoma to specific environmental signals transduced to the splicing protein factors through a chloroplast-dependent pathway. Notably, when the 7H8 Arabidopsis orthologous gene was analyzed, no stress induction in terms of mRNA abundance and no cold-dependent intron retention was detected. Otherwise, 6G2 Arabidopsis homologous sequences sharing the same genomic structure of the durum wheat 6G2 showed a similar intron retention event although not strictly dependent on stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant growth and development are often limited due to extreme temperatures and osmotic stress, which comes from high salinity or drought conditions. Plants react to stress events through the synthesis and the accumulation of a number of proteins and small molecules which contribute to the stress tolerance as a whole. This is the result of a complex cascade of perception and transduction of the stress signal, in which few regulatory mechanisms direct the expression of many downstream genes, whose protein products have a structural role in stress tolerance, or catalyze the synthesis of small molecules with osmotic properties. The importance of regulatory mechanisms in determining stress tolerance has been clearly shown by means of studies on both mutants and transgenic plants (Kasuga et al. 1999; Saijo et al. 2000).
Genes encoding proteins having a key role in the regulation of the responsive mechanisms show, as a common feature, an early and transient increase in their mRNA amount. Examples are the genes coding for components of the signal transduction [e.g. Arabidopsis thaliana MAP kinase or ERK kinase kinase (ATMEKK) kinase], or for transcription factors [e.g. C-repeat binding factor/dehydration responsive element binding protein (CBF/DREB1)], known to be induced at the transcriptional level within 1–2 h by the onset of cold stress treatment in Arabidopsis (Mizoguchi et al. 1996; Jaglo et al. 2001) as well as in barley, wheat (Jaglo et al. 2001) and rice (Dubouzet et al. 2003).
Stress-related genes have been extensively analyzed for steady-state mRNA accumulation, but recent experimental data made evident the existence of complex pathways of post-transcriptional regulation. Finding discrepancies between the steady-state mRNA levels and relative transcription rates or protein amount demonstrates that the final protein amount is determined by multiple factors, depending on a number of intrinsic and environmental stimuli (Miller et al. 2001). An important role in sensing environmental stimuli and mediating the active response of plants is played by the chloroplast, which has been shown to control the accumulation of a number of stress-related proteins encoded by nuclear genes (Dal Bosco et al. 2003).
Post-transcriptional regulation mechanisms may act either quantitatively controlling the amount of mRNA and protein (synthesis/degradation rate), or qualitatively, modifying transcripts. Recent studies carried out with bioinformatics approaches on completely sequenced animal genomes describe the alternative splicing as one of the most significant components of genome complexity. A range of processes from sex determination to apoptosis are regulated by means of alternative splicing (Modrek and Lee 2002). The majority of changes in the protein product, also in plants, appear to be functionally relevant, such as replacement of the amino or carboxy terminus, or in-frame addition/removal of a functional unit. In this way, different polypeptides, with different functions or subcellular locations, are produced by a single gene. In some cases environmental stimuli can regulate the relative abundance of one form with respect to the other (Mano et al. 1999; Shi et al. 2002).
Many alternative splicing events, as described in the literature, are based on the retention of introns in mature mRNA following which shortened proteins may be formed because of early translational termination. In some cases introns within 3′UTR are also subjected to retention, although this has no influence on the protein sequence (Oh and Waxman 1994). Intron retention could reflect poor intron recognition (Brown and Simpson 1998) or represent the result of an active process inhibiting the splicing reaction. Examples have been described in both plants and animals, in which intron retention acts as an important regulatory mechanism of the biological protein activity. FCA, a gene coding for an RNA-binding protein that controls the photoperiod-independent flowering transition in Arabidopsis, is alternatively spliced into four transcripts, only one of them encoding a protein capable of complementing the flowering-time defective mutant fca-1 (Bailey-Serres et al. 1999). The Drosophila erect wing gene, which provides a vital neuronal function and is essential for the formation of certain muscles, is expressed in the head as well as in the body, but in a different manner, because a high level of the mature protein is lethal outside the nervous system. This differential regulation is ensured by the inefficient splicing of a subset of the nine introns of this gene in the body, where the unspliced transcripts are predominant with respect to the correctly spliced form (Koushika et al. 1999).
Furthermore, the shortened polypeptides formed following alternative splicing are not necessarily functionless forms of the full length protein, as shown for the N gene for resistance to tobacco mosaic virus where both full length and shortened alternative transcripts are needed for the complete resistance phenotype (Dinesh-Kumar and Baker 2000). Finally, many examples are available in the literature where alternatively spliced gene products are involved in signal transduction cascades or in regulation of gene expression. The FCA protein binds its own mRNA to negatively regulate its own expression (Quesada et al. 2003).
This work was aimed at identifying genes up-regulated during the early phases of durum wheat response to cold, and so putatively involved in resistance mechanisms, and to characterize their expression pattern at the transcriptional and post-transcriptional level.
Materials and methods
Plant material and growing conditions
The experiments were carried out with durum wheat (Triticum durum L., cultivar Ofanto), barley (Hordeum vulgare L., cultivars Nure and Tremois and albina mutant e-16—Henningsen et al. 1993), bread wheat (Triticum aestivum L., cultivars Cheyenne and Chinese Spring) and A. thaliana (ecotype Columbia). The seeds were grown in 50% sand and 50% soil in a daily regime of 9 h light (160 μmol m−2 s−1) at 20°C/15°C for 7 days (21 days for Arabidopsis), and cold acclimated in a daily regime of 9 h light at 3°C and 15 h dark at 2°C for the time indicated in each experiment. Leaves from 7-day-old durum wheat plants were cut and allowed to dehydrate for 0.5, 1.5 and 3 h in the growth chamber and relative water content (RWC) was determined. Average RWC values were 90% (control), 71% (0.5 h), 66% (1.5 h) and 63% (3 h).
The experiment with cycloheximide (CHX) was carried out with durum wheat plants grown on moist filter paper for 7 days at 20°C and then acclimated for 1 h onto a new filter paper completely saturated with 10 μg/ml CHX (SIGMA code n. C4859). Then the plants were maintained at 20°C or exposed to low temperature (3°C) for 4 h or 24 h.
Generation and screening of a subtracted library by suppression-subtractive hybridization
Poly(A) RNAs were isolated from plants of durum wheat variety Ofanto grown at 20°C for 7 days (driver) and then exposed to 3°C for 6 h (tester) by affinity chromatography on oligo(dT)-cellulose (Boehringer Mannheim) according to published methods (Sambrook et al. 1989).
Suppression-subtractive hybridization was performed using the PCR-Select cDNA Subtraction Kit (Clontech) according to the manufacturer’s recommendations. Approximately 100 ng PCR-amplified cDNA was ligated without further purification into 50 ng vector and cloned into pGEM-T Easy vector (Promega).
About 1,000 individual colonies were picked and the corresponding plasmid DNAs were isolated. An aliquot of each plasmid DNA sample was digested with EcoRI, loaded onto agarose gel and blotted onto nylon filters. Probes for hybridization were derived from the forward- and reverse-subtractive libraries as described in the PCR Select Kit manufacturer’s instructions (Clontech). About 200 ng for each probe was radiolabeled with [α−32P] dCTP by random-priming reaction. Hybridized filters were washed three times at 65°C with 2×, 1× and 0.5× SSC, 0.1% SDS for 20 min and exposed to film (Kodak).
Candidate clones were sequenced in both directions using the ABI Prism BigDye Terminator Cycle Sequencing kit on an ABI PRISM 310 Genetic Analyzer automated sequencing machine (PE Applied Biosystem). DNA sequences were compared with those in the nonredundant databases by using the BLAST-N and BLAST-X algorithms, available at the NCBI (http://www.ncbi.nlm.nih.gov) and TIGR (http://www.tigr.org) web sites. The Vector NTI software (Informax) was used to collect all sequences and to produce contigs, alignments, amino acidic translations, etc.
Northern blotting analysis
Equal amounts (2 μg) of poly(A) RNAs for each sample were separated on an 0.8% agarose formaldehyde gel and transferred to a positively charged nylon filter (Millipore). About 100 ng of DNA fragments was labeled with (α−32P) dCTP in a randomly primed reaction and used as probe. The hybridization was performed at 42°C in UltraHyb buffer (Ambion). The filters were washed three times at 65°C with 2×, 1× and 0.5× SSC, 0.1% SDS for 20 min. To control the integrity and the amount of loaded poly(A) RNAs, all filters were hybridized with an α−32P-labeled probe corresponding to the gene coding for ribosomal protein L12 (RPL12, Baldi et al. 2001).
RACE and RT-PCR
The clones 7H8 and 6G2 were subjected to 3′ and 5′ rapid amplification of cDNA ends (RACE) experiments performed using 3′ and 5′ RACE System kits (Invitrogen) according to the manufacturer’s instructions. The primers employed to extend the clones 7H8 and 6G2 are reported in the Electronic Supplementary Material, Table 1. PCR reactions were carried out by incubation in a thermal cycler at 94°C for 4 min, followed by 25 cycles at 94°C for 1 min, 55°C for 30 s and 72°C for 1 min.
Total RNA was isolated from leaves by means of the Trizol reagent (Invitrogen) following the manufacturer’s instructions. The single-stranded cDNA was synthesized using 200 U of SuperScript II RNase H− reverse transcriptase (Invitrogen) and a poly(T) primer on 1 μg of total RNA following the manufacturer’s recommendations (final volume of 20 μl).
The different fragments of 7H8, 6G2, wheat cold-regulated gene 410c (WCOR410c) and ATP binding cassette (ABC) transcripts were amplified by starting from 2 μl of the cDNA reaction as template and 5 U of platinum Taq DNA polymerase (Invitrogen) in a 28 cycles PCR program (94°C for 1 min, and 65°C for 1 min, 72°C for 1 min) followed by a final extension step at 72°C for 7 min. Twenty-eight cycles were chosen for all the amplifications because they allowed a semiquantitative evaluation of the abundance of analyzed mRNA sequences.
Normalization of the RT-PCR reactions was performed with primers designed on the gene coding for ribosomal protein RPL12 (Baldi et al. 2001) for reactions carried out onto wheat and barley RNAs, and on the actin2 gene (U41998) for the Arabidopsis RNAs. The utilized primers are listed in the Electronic Supplementary Material, Table 1. RT-PCR reactions were separated onto 2% high resolution agarose gel. The amplification products derived from the experiments described in Fig. 3 and 5 were eluted from the gel and sequenced in order to clarify their identity.
Results
Isolation of e-cor genes and expression analysis
Two early cold-regulated (e-cor) genes, 7H8 and 6G2, were isolated by means of a suppression subtractive library constructed with RNA from 7-day-old durum wheat plants grown at 20°C (RNA driver), or exposed to 3°C for 6 h (RNA tester). The selected fragments, characterized by a length of 421 bp and 525 bp (for 7H8 and 6G2, respectively), were employed as probes in Northern blotting experiments to detect the expression of the corresponding mRNAs. Plants of durum wheat were grown at 20°C and then exposed to 3°C for 4, 16, 24 h, 3, 7 and 10 days. Alternatively leaves were cut and allowed to dehydrate for 0.5, 1.5 and 3 h. Figure 1 shows the stress-dependent expression profile of 7H8 and 6G2 genes. We refer to our clones as early cold-regulated (e-cor) genes, since their mRNA level was induced or enhanced within the first 4 h of exposure to low temperature, in contrast to many “late responsive” cor genes, detectable, under the same conditions, after about 1 day of exposure to low temperature (Cattivelli and Bartels 1990). The isolated clones showed a common transient cold-dependent induction, being up-regulated by low temperature only for a short time after the cold exposure: they reached the maximum transcript amount between 16 h and 24 h. A different behavior was detected when dehydrated samples were tested: only 6G2 showed a clear up-regulation following water deficit.
Northern blots of the e-cor clones. Plants of durum wheat cv. Ofanto were grown for 7 days at 20°C (control, Ctr) and then transferred to 3°C for 4, 16 and 24 h, 3, 7 and 10 days. Otherwise leaves were cut and allowed to dehydrate for 0.5, 1.5 and 3 h. Normalization was assessed with the barley ribosomal probe RPL12
Sequence reconstruction and bioinformatic analysis
In vivo and in silico approaches were employed to reconstruct the gene sequences corresponding to the isolated e-cor fragments. The e-cor clone 7H8 was extended via RACE up to 1,307 bp. Homology search and analysis for known amino acid domains suggested that the e-cor clone 7H8 codes for a putative ribokinase: the 7H8 amino acid deduced sequence proved 78% and 66% identical to the rice and Arabidopsis putative ribokinases BAB16906 and NP_173390, respectively. Furthermore, the most similar domain found was the Pfam00294 consensus sequence for ribokinases. The ribokinases are components of the large family of carbohydrate kinases, which catalyze the phosphorylation of a number of substrates, like sugar and pyrimidines (Cheek et al. 2002).
The e-cor clone 6G2 was extended via RACE and linked to the wheat TC173020. The reconstructed sequence contains an ORF of 429 amino acids, showing a clear similarity with a rice hypothetical protein (BAA92399, 70% identity, modified following genomic vs cDNA comparison) and with four unknown proteins from Arabidopsis (best hit: NP_177673, 36% identity). When the 6G2 deduced protein plus the rice and Arabidopsis homologs were searched for conserved domains using Pfam and SMART databases, the presence of a RING-finger C3H2C3 type domain was identified from position 281–333. All the sequences contain the RING-finger pattern: C-x(2)-C-x(22)-C-x(1)-H-x(2)-H-x(2)-C-x(14)-C-x(2)-C, a sequence completely aligned with the highly stringent C3H2C3 consensus pattern described at the MIPS site (http://mips.gsf.de/proj/thal/ring/patterns.html). RING-finger is a protein–protein interaction domain implicated in protein ubiquitination (Matsuda et al. 2001). The three RING-finger containing sequences were also searched for the presence of putative small ubiquitin-related modifier (SUMO) consensus motifs, and a putative site has been found at the C-terminus in all three proteins (k346 in durum wheat, k348 in rice and k349 in Arabidopsis, with P values of 0.672, 0.672 and 0.957, respectively). SUMO consensus sequences promote the binding of the protein target to SUMO-1, a member of the ubiquitin superfamily. These data suggest that 6G2 gene could be a component of the ubiquitination system and that it could act in the post-translational control, via protein degradation, of gene expression.
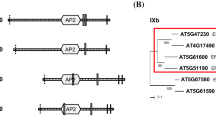
The durum wheat genomic sequences of 7H8 and 6G2 were completely reconstructed via PCR and compared with the genomic structures of 7H8 and 6G2 homologous genes of rice and Arabidopsis. A search in the EST databases of wheat, barley, rice and Arabidopsis always found a single 7H8 homologous sequence; the same results were also obtained at the genomic level in rice and Arabidopsis. The number and the position of all the introns of the e-cor gene 7H8 is highly conserved in the three species. The length of exons and introns is also conserved with the only exception being the first one (Fig. 2).
Genomic structure of 7H8 and 6G2 e-cor genes in durum wheat and their homologs in rice and Arabidopsis. Boxes represent the exons (length—bp base pair—is indicated); bars represent the introns with intron number above and length (bp) below. Each gene is aligned with the corresponding TCs or ESTs available in the databases (accession numbers are indicated). Dashed lines in the expressed sequences correspond to the genomic introns. The sources of the rice genomic sequences are AP002863.3 from 60,660 bp to 63,902 bp (reversed and complemented) and AP001366.1 from 21,710 bp to 27,365 bp. Reconstruction of the gene structures in durum wheat was experimentally determined, while in rice and Arabidopsis it was based on comparison between the genomic sequences and the indicated TCs or ESTs
Two wheat 6G2-related TCs (TC173931 and TC150727, 78% and 31% amino acid identity with 6G2, respectively) were found in the databases. A 6G2 homologous sequence was found in rice (68% of amino acid identity), while four different homologs were identified in Arabidopsis (At1g75400, At1g19860, At4g39140 and At2g21500), with an amino acid identity between 36% and 29%. The comparison among e-cor 6G2 homologous genes indicates a conserved position for all the introns except the last one. The fourth intron is positioned inside the ORF in the Arabidopsis genes At1g75400 and At1g19860 (only At1g75400 is reported in Fig. 2) and within the 3′ UTR in the Arabidopsis genes At4g39140 and At2g21500 (only At4g39140 is reported in Fig. 2), in the rice and durum wheat sequences. The full length genomic sequences of the e-cor 7H8 and 6G2 were submitted to the databank under the accession numbers AY466010 and AY465427, respectively.
The transient induction of 7H8 reflects the cold sensitivity of some splicing factors
Finding two early cold-regulated genes with many introns raises the possibility that these introns could have an influence on their expression in stress conditions. Furthermore, a putative alternative splicing event for the fourth 6G2 intron could be predicted in durum wheat, on the basis of the alignment between some cDNA sequences obtained via RACE and the wheat TC173020 sequence. For these reasons an extensive RT-PCR analysis was carried out in order to verify putative alternative splicing events following cold stress.
In Fig. 3a and c the amplification products are reported that were obtained after RT-PCR analysis for e-cor 7H8. All introns were tested with dedicated primer pairs designed on the flanking exons as well as on the intron sequence. Clear evidence of intron retention was observed for four introns. By using a primer upstream of the first intron and one downstream of the second intron, a 458 bp fragment was expected from the correctly spliced transcript. Nevertheless, another amplification product was present in samples exposed to low temperature for at least 4 h. This 542 bp product corresponded to a fragment containing the second 84 bp intron. With a primer pair upstream of the fifth and downstream of the sixth intron, four amplification products appeared after 4 h of cold treatment: a 238 bp band corresponding to the correctly spliced transcript, a 331 bp fragment containing only the fifth intron, a 413 bp band with the sixth intron, and a 506 bp fragment corresponding to the form containing both introns. Finally, primers upstream and downstream of the seventh intron yielded two bands beginning from 4 h of cold treatment, corresponding to the transcripts with or without the intron. The presence of transcripts containing introns following cold stress treatment has been further confirmed by using primer pairs designed on intron sequences (Fig. 3c, d).
RT-PCR analysis of 7H8 e-cor gene. Total RNA was extracted from leaves of durum wheat plants grown at 20°C (ctr) for 7 days and then exposed at low temperature for 2, 4, 8, 16 and 24 h. a Amplification products separated onto agarose gel obtained with exon-specific primers. b Description of the gel-detected fragments and position of utilized primers represented by arrows. c Amplification products separated onto agarose gel obtained with intron-specific primers. d Description of the gel-detected fragments and position of utilized primers represented by arrows. e Amplification products separated onto agarose gel obtained with primers designed inside the fifth and eighth exons. The figures of gel separation are presented in inverted colours
To resume, a cold treatment affects the spliceosoma involved in processing of the 7H8 mRNA promoting the retention of one or more introns. As a consequence, a population of 7H8 mRNAs, each of them characterized by the presence of a different intron pattern, is produced during the first day after the temperature drop. These features can be appreciated in particular when the RT-PCR is carried out using a primer pair encompassing the fifth, sixth and seventh introns (Fig. 3e). Notably, under these conditions the amount of the correctly spliced 7H8 transcript was lower in the 4-h-treated sample than in the control, and it increased to the initial level after 24 h of cold exposure. As a result the cold-induced intron retention of 7H8 led to a decreased amount of correctly spliced mRNA. All retained introns contain stop codons in frame in such a way that non-functional truncated polypeptides (without a complete ribokinase domain) are expected from all transcripts containing introns.
The comparison of RT-PCR gels with Northern blot (Fig. 1) indicates that the low temperature induction of the 7H8 mRNA steady-state amount, as seen by Northern blotting analysis, reflects the appearance of transcripts containing introns rather than an up-regulation of the fully spliced transcript. The presence of mRNA molecules containing one or more small introns might be responsible for the smeared signal detected in Fig. 1.
To test if the cold-dependent retention of the introns of the 7H8 e-cor gene was dependent on a neo-synthesized protein, an experiment was carried out using CHX, an inhibitor of the eukaryotic protein synthesis. Durum wheat 7-day-old plants were pre-treated with CHX (10 μg/ml) or water as a control for 1 h at 20°C, and then maintained at 20°C or transferred to 3°C for 4 h or 24 h. Total RNA was extracted, and RT-PCR was carried out with primers designed onto exons flanking the fifth and sixth introns (Fig. 4). Water-treated plants showed only one amplification product corresponding to the correctly spliced transcript at 20°C, and the expected four amplification products, on the basis of the presence/absence of the fifth and sixth introns, after exposure to low temperature for 4 h. The CHX treatment did not affect intron retention in cold-treated plants; nevertheless CHX promoted the intron retention yet in the absence of the cold stress, in fact a 331 bp fragment corresponding to the transcript containing the fifth intron was amplified in all samples kept at 20°C in the presence of the chemical inhibitor.
RT-PCR analysis of 7H8 e-cor gene (fifth and sixth introns) in the presence of cycloeximide (CHX). Total RNA was extracted from leaves of durum wheat plants grown at 20°C for 7 days and then treated for 4 h or 24 h with CHX (10 μg/ml) or water as a control at 20°C or 4°C. The figures of gel separation are presented in inverted colours
These results suggest that a protein factor(s) with a fast turnover, whose synthesis is inhibited by CHX, is involved in the correct splicing of 7H8 mRNA. On the basis of data shown in Figs. 3 and 4, we hypothesize that this splicing factor(s) is cold sensitive and its activity is impaired during exposure to low temperature.
The stress-induced expression of 6G2 is associated with partial retention of a 3′ UTR intron
Figure 5 shows the results of the RT-PCR experiment carried out on the e-cor gene 6G2. The amplification of the fragments encompassing introns one, two and three yielded intronless fragments only, while two amplicons were detected when primers flanking the fourth intron were used for RT-PCR. The two fragments differed because of the presence/absence of 102 bp corresponding to the fourth intron located in the 3′ UTR. The fragment containing the fourth intron was detected in samples exposed to low temperature for not less than 4 h. In contrast with the behaviour of 7H8 the amount of the correctly spliced 6G2 transcript arose in cold-treated plants.
RT-PCR analysis of 6G2 e-cor gene. Total RNA was extracted from leaves of durum wheat plants grown at 20°C (ctr) for 7 days and then exposed at low temperature for 2, 4, 8, 16 and 24 h, or allowed to dehydrate for 0.5, 1.5 and 3 h. a Amplification products separated onto agarose gel obtained with exon-specific primers. b Description of the gel-detected fragments and position of utilized primers represented by arrows. c Amplification products separated onto agarose gel obtained with intron-specific primers. d Description of the gel-detected fragments and position of utilized primers represented by arrows. e Analysis of retention of 6G2 fourth intron upon dehydration. The figures of gel separation are presented in inverted colours
The cold-dependent retention of the fourth intron was also confirmed with primers designed within the intron (Fig. 5c, d). Since 6G2 was shown to be also induced by dehydration, the intron retention was assayed on RNA samples extracted from durum wheat dehydrated leaves. The retention of the fourth intron was also associated with drought induction being detectable already after 0.5 h of dehydration (Fig. 5e).
Intron retention is not a general feature upon cold treatment
To assess whether intron retention is a general feature following cold stress in cereals we analyzed the influence of low temperature on intron splicing of two other genes with different genomic structure and expression profile. The wheat gene WCOR410c (TC104761) is a cold-regulated sequence 94% identical to the barley dehydrin 8 gene (dhn8, accession number AF181458). We inferred WCOR410c genomic structure on the basis of the barley dhn8 genomic sequence (AF043093). Dhn8 was found to contain a single intron not subjected to intron retention either at 20°C or during cold treatment (data not shown). We also investigated the effect of low temperature on the splicing process of a constitutively expressed gene characterized by a very complex genomic structure. The barley sequence AAG49002, coding for a putative ABC transporter, consists of 17 exons and 16 introns, ranging in size from 70 bp to 688 bp, all located into the ORF. The genomic sequence is available (accession number AY013246) and the predicted genomic structure has been confirmed by comparison with corresponding ESTs. We verified the homologous wheat gene using primer pairs designed to check for the presence of fragments containing from the 7th to the 16th intron. All primer pairs yielded a single amplification product either at 20°C or at low temperature, corresponding to the transcripts completely spliced and therefore without introns (data not shown).
The results obtained for WCOR410c and ABC genes clearly show that the effect of low temperature on splicing machinery is not a general effect, rather it involves a limited number of splicing factors.
7H8 and 6G2 homologous genes in wheat, barley and Arabidopsis show different intron retention behaviours
Other species have been investigated searching for cold-related intron retention in 7H8 and 6G2 homologous e-cor transcripts. When barley and bread wheat were analyzed, a behaviour identical (6G2—data not shown) or similar (7H8—Fig. 6a) to that of durum wheat was found. Intron retention of e-cor 7H8 was analyzed using primers upstream of the fifth and downstream of the sixth intron. No differences were detected between varieties within species; nevertheless some variations were present in the relative abundance of the different forms among species. In durum wheat all intron-containing amplicons remained visible until 24 h of cold treatment with the band containing the fifth intron being present in the highest amount after 24 h of cold exposure. In bread wheat after 8 h of cold exposure only the fragment containing the fifth intron was detectable, while no intron retention was visible after 24 h. In barley, the bands containing introns showed a higher size than in wheat, probably due to the presence of longer fifth and sixth introns; furthermore, only three amplification products were detected after 4 h of cold exposure, with the lacking of the fragment containing the sixth intron only. After 24 h of cold treatment no amplicons containing introns were detected. These results suggest that although the 7H8 cold-dependent intron retention is a general trait in cereals, the ability of the different species to normalize the splicing process is different, being faster and more efficient in bread wheat.
a RT-PCR analysis of 7H8 e-cor gene (fifth and sixth introns) in barley and wheat genotypes. Total RNA was extracted from leaves of plants grown at 20°C (ctr) for 7 days and then exposed at low temperature for 4, 8 and 24 h. Two barley genotypes (Nure—winter type and Tremois—spring type) and two bread wheat genotypes (Cheyenne—winter type and Chinese Spring—spring type) were compared with the durum wheat cultivar Ofanto. b RT-PCR analysis of 6G2 homologs in Arabidopsis (fourth intron). Total RNA was extracted from leaves of plants grown at 20°C (ctr) for 21 days and then exposed at low temperature for 2, 4, 8, 16 and 24 h. Normalization was assessed with primers designed on the Arabidopsis Actin2 gene. The figures of gel separation are presented in inverted colours
When the expression of the 7H8 homologous gene was analyzed in Arabidopsis, no amplification products corresponding to transcripts containing introns were detected within 24 h of cold treatment. This result was further confirmed by means of primers designed inside the introns (data not shown).
In contrast with 7H8, four 6G2-related sequences characterized by a different position of the fourth intron were identified in Arabidopsis (Fig. 2). When the cold-dependent expression of these genes was assessed by RT-PCR none of them was found to be induced by cold (Fig. 6b). On the contrary, a repression was observed after 24 h of cold treatment in At4g39140 ad At2g21500, the two genes with the fourth intron located into the 3′UTR. The same sequences showed the retention of the fourth intron with some enhancement by low temperature (Fig. 6b).
The analysis of the intron retention was also carried out on the mRNAs corresponding to two 6G2 related TC found in the wheat EST database (TC173931 and TC150727, 78% and 31% of amino acid identity with 6G2, respectively). The presence and the position of the fourth intron was inferred on the basis of alignment with the 6G2 nucleotide sequence and assessed by amplification on cDNA and genomic DNA. Based on RT-PCR analysis, the expression of these sequences was not induced by cold or drought stress. Furthermore, any 3′ UTR located intron was found for TC150727, while the TC173931 showed an intron about 170 bp long within 3′ UTR; nevertheless no intron retention was observed.
Barley albina mutants fail to retain introns following cold stress
Barley albina mutants are characterized by a recessive nuclear mutation resulting in a block of the chloroplast development. At the molecular level, they are also impaired in a signal transduction pathway controlling the chloroplastic-mediated expression of nuclear cold-related genes (Dal Bosco et al. 2003). In order to test if this way could affect the splicing efficiency in stress conditions, barley plants carrying a mutation at the e-16 locus were grown for 7 days at 20°C and then exposed to 3°C for 4 h. The RT-PCR reactions were carried out from the leaf-extracted total RNA to analyze the cold-dependent retention of the fifth and sixth introns of 7H8, and of the fourth intron of 6G2. Wild-type plants showed a similar cold-dependent intron retention pattern with respect to that found in the other barley cultivars shown in Fig. 6 and three amplification products were detectable after 4 h of cold treatment for e-cor 7H8. By contrast, the cold-dependent intron retention in the mutated genotype was strongly reduced for both 7H8 and 6G2 (Fig. 7).
RT-PCR analysis of 7H8 and 6G2 e-cor genes in the barley e-16 albina mutant. Total RNA was extracted from green (wild type—wt) and white (alb) leaves of the barley mutant e-16 grown at 20°C (ctr) and then exposed at low temperature for 4 h. The figures of gel separation are presented in inverted colours
Discussion
In this work we describe two e-cor genes, a putative ribokinase (7H8) and a RING-finger domain containing protein (6G2), isolated after screening of a suppression subtractive library from durum wheat plants exposed to cold (3°C). The expression pattern, analyzed via Northern blot, revealed a transient induction for both these genes driven specifically by cold stress for 7H8, and by cold and dehydration at the same extent for 6G2. When gene expression was studied via RT-PCR a more complex frame was revealed, with 7H8 and 6G2 showing different features in terms of mRNA accumulation. The regulation of both 7H8 and 6G2 genes in durum wheat was associated with the stress-induced retention of a subset of introns in the mature mRNA. Four out of the seven 7H8 introns were found to be independently retained in mature mRNA and their presence resulted in masking, in the Northern blot, a transient down-regulation of the correctly spliced 7H8 transcript. In contrast, only a single 6G2 intron, located outside of the ORF, was retained during the up-regulation of the properly processed transcript.
When homologous genes in Arabidopsis were taken in consideration none of them was induced by cold at the level of steady-state mRNA abundance, as shown by our data as well as by data from array analysis (Chinnusamy et al. 2003). Furthermore, the stress-dependent intron retention was not observed in the 7H8 orthologous gene of Arabidopsis. A search in maize EST database identified two 7H8 homologous TCs (TC193781 and TC193779) differing only by the presence in TC193779 of two insertions corresponding to the fifth and sixth introns, a finding suggesting that the maize 7H8 homologous gene is also subjected to intron retention. These data suggest a different cold sensitivity of the spliceosome machinery of Arabidopsis compared with that of cereal species. In contrast, analysis of the four 6G2-related genes in Arabidospis indicated that the retention was associated more with the intron position than with stress response. The genes carrying the fourth intron in the 3′UTR partially retained the intron although not induced by cold.
The experimental data presented in this work demonstrate that cold and dehydration can act on the operating capacity of the spliceosome modifying the population of some gene transcripts. This effect is specific for some genes and for some introns inside each gene, depending on the fact that different introns need different splicing factors to be correctly processed.
The intron retention in mature mRNA could reflect the effect of the stress in impairing the splicing efficiency of some sensible factors. The influence of the temperature on splicing efficiency has been studied in several works. In many cases the splicing efficiency has been reported to decrease, for cryptic splicing junctions, following a high-temperature treatment, due to a less stable interaction between mRNA and splicing factors. A single-nucleotide polymorphism at the leader intron 5′ splice site of the rice waxy gene reduces the splicing efficiency and promotes the retention of the intron particularly at high temperatures, in cultivars with the AG TTATA allele compared to the ones carrying the AG GTATA allele (Larkin and Park 1999). The ap3-1 allele of APETALA3 gene, required for stamen and petal development in Arabidopsis, is characterized by a missense mutation near a 5′ splice site, which determines a high temperature-dependent splicing defect. For this reason, ap3-1 plants behave as a conditional mutant showing a normal phenotype when grown at low temperature, but abnormal flower structure when grown at non-permissive temperatures (Sablowski and Meyerowitz 1998). Nevertheless, in a recent work, Iida et al. (2004) analyzed the correlation between alternative splicing events and environmental conditions in a database of about 280,000 RIKEN Arabidopsis full length cDNAs. They found that environmental stresses, cold above all, affected significantly splicing profiles, in particular by inducing intron retention. According to the these findings we report the retention of introns in response to cold treatment, a condition that is expected to increase the stability of the interactions between splice junctions and protein factors. A treatment with the protein synthesis inhibitor CHX promotes 7H8 intron retention at 20°C suggesting that low temperature intron retention is due to the absence/degradation of a specific protein factor. The progressive normalization of 7H8 expression after 24 h of cold treatment can be interpreted as a recovery of the splicing efficiency after the initial cold damage to the splicing apparatus. Furthermore the cold-dependent intron retention for 7H8 and 6G2 was suppressed in the absence of a fully developed chloroplast, as shown by the barley albina mutant e16. Identical results have been obtained with other non-allelic albina mutants (data not shown). It is known that chloroplast has a key role in the perception of the environmental changes (Huner et al. 1998) and in the control of the cold molecular response (Ndong et al. 2001). As a consequence of the mutation, albina plants show great alterations in their cold response: some Cor genes are not expressed in response to cold (Dal Bosco et al. 2003), while others are constitutively up-regulated also in the absence of stress (Baldi et al. 2001).
These data strongly suggest that the cold-dependent intron retention described for the e-cor genes 7H8 and 6G2 is not due to cryptic splicing junction sequences (identical between albina mutants and wild type), but rather to a cold-sensitive protein factor(s) characterized by a fast turnover, reflecting the response of the spliceosoma to specific environmental signals.
Finally, the absence of intron retention in Arabidopsis 7H8 orthologous gene would reflect a lower cold sensitivity of some components of the splicing machinery in this species compared to Triticeae.
Abbreviations
- ATMEKK:
-
Arabidopsis thaliana MAP kinase or ERK kinase kinase
- CBF/DREB1:
-
C-repeat binding factor/dehydration responsive element binding protein
- RWC:
-
Relative water content
- RPL12:
-
Ribosomal protein L12
- e-cor :
-
Early cold-regulated
- Pfam:
-
Protein families database
- SMART:
-
Simple modular architecture research tool
- RING:
-
Really interesting gene
- SUMO:
-
Small ubiquitin-related modifier
- CHX:
-
Cycloheximide
- dhn8 :
-
Dehydrin 8
- WCOR410c:
-
Wheat cold-regulated gene 410c
- ABC:
-
ATP binding cassette
References
Bailey-Serres J, Rochaix JD, Wassenegger M, Filipowicz W (1999) Plants, their organelles, viruses and transgenes reveal the mechanisms and relevance of post-transcriptional processes (EMBO Workshop Report). EMBO J 18:5153–5158
Baldi P, Valè G, Mazzucotelli E, Govoni C, Faccioli P, Stanca AM, Cattivelli L (2001) The transcript of several components of the protein synthesis machinery are cold-regulated in a chloroplast-dependent manner in barley and wheat. J Plant Physiol 158:1541–1546
Brown JWS, Simpson CG (1998) Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol 49:77–95
Cattivelli L, Bartels D (1990) Molecular cloning and characterization of cold-regulated genes in barley. Plant Physiol 93:1504–1510
Cheek S, Zhang H, Grishin NV (2002) Sequence and structure classification of kinases. J Mol Biol 320:855–881
Chinnusamy V, Ohta M, Kanrar S, Lee B, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Dal Bosco C, Busconi M, Govoni C, Baldi P, Stanca AM, Crosatti C, Bassi R, Cattivelli L (2003) Cor gene expression in barley mutants affected in chloroplast development and photosynthetic electron transport. Plant Physiol 131:793–802
Dinesh-Kumar SP, Baker BJ (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Nat Acad Sci USA 97:1908–1913
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Henningsen KW, Boyton JE, von Wettstein D (1993) Mutants at xantha and albino loci in relation to chloroplast biogenesis in barley (Hordeum vulgare L.). The Royal Danish Academy of Sciences and Letters, Copenhagen
Huner NPA, Oquist G, Sarhan F (1998) Energy balance and acclimation to light and cold. Trends Plant Sci 3:224–230
Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K (2004) Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res 32:5096–5103
Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127:910–917
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Koushika SP, Soller M, De Simone SM, Daub DM, White K (1999) Differential and inefficient splicing of a broadly expressed Drosophila erect wing transcript results in tissue-specific enrichment of the vital EWG protein isoform. Mol Cell Biol 19:3998–4007
Larkin PD, Park WD (1999) Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol Biol 40:719–727
Mano S, Hayashi M, Nishimura M (1999) Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J 17:309–320
Matsuda N, Suzuki T, Tanaka K, Nakano A (2001) Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J Cell Sci 114:1949–1957
Miller WA, Waterhouse PM, Brown JWS, Browning KS (2001) The RNA world in plants: post-transcriptional control III. Plant Cell 13:1710–1717
Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K (1996) A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 93:765–769
Modrek B, Lee C (2002) A genomic view of alternative splicing. Nat Genet 30:13–19
Ndong C, Danyluk J, Huner NPA, Sarhan F (2001) Survey of gene expression in winter rye during changes in growth temperature, irradiance or excitation pressure. Plant Mol Biol 45:691–703
Oh Y, Waxman SG (1994) The β1 subunit mRNA of the rat brain Na+ channel is expressed in glial cells. Proc Natl Acad Sci USA 91:9985–9989
Quesada V, Macknight R, Dean C, Simpson GG (2003) Autoregulation of FCA pre-mRNA processing controls Arabidopsis Flowering time. EMBO J 22:3142–3152
Sablowski RWM, Meyerowitz EM (1998) Temperature-sensitive splicing in the floral homeotic mutant apetala3-1. Plant Cell 10:1453–1463
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Shi H, Xiong L, Stevenson B, Lu T, Zhu JK (2002) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role forvitamin B6 in plant salt tolerance. Plant Cell 14:575–588
Acknowledgements
This work was supported by the Ministry of Science (Progetto FIRB-plant stress) and Ministry of Agriculture (Progetto FRUMISIS) of Italy. The authors thank Dr. Matteo Busconi (Institute for Plant Genetics, Catholic University of Sacro Cuore, Piacenza, Italy) for his technical assistance during sequencing.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Mastrangelo, A.M., Belloni, S., Barilli, S. et al. Low temperature promotes intron retention in two e-cor genes of durum wheat. Planta 221, 705–715 (2005). https://doi.org/10.1007/s00425-004-1475-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1475-3