Abstract
Glandular trichomes are a major site of plant natural product synthesis and accumulation for protection against insect predation. However, to date few studies have attempted to obtain a global view of trichome gene expression. Two contrasting approaches have been adopted to investigate genes expressed in glandular trichomes from alfalfa (Medicago sativa L.). In the first approach, 5,674 clones from an alfalfa glandular trichome cDNA library were sequenced. The most highly abundant expressed sequence tag (EST) corresponded to a lipid transfer protein. The presence of ESTs corresponding to enzymes for all steps in the biosynthesis of flavonoids suggests that these are important metabolites in alfalfa trichome biology, as confirmed by histochemistry and metabolite profiling. No ESTs corresponded to enzymes of cyclized terpenoid biosynthesis. In a second approach, microarray analysis was used to compare levels of alfalfa transcripts corresponding to 16,086 Medicago truncatula A17 genes in stems with and without trichomes. This revealed over 1,000 genes with strong preferential expression in the trichome fraction of the stem, 70% of which are of unknown function. These define a class of genes that are not trichome-specific, since M. truncatula A17 does not itself have glandular trichomes, but has potential importance for trichome function within the stem.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trichomes are epidermal appendages found on the stems and leaves of many plants. Although their morphology varies considerably, two major classes can be distinguished, the glandular and non-glandular trichomes. Glandular trichomes have received considerable attention in view of their capacity to synthesize, store and secrete secondary metabolites that help protect the plant against insect predation and other biotic challenges (Wagner 1991; Ranger and Hower 2001; Wagner et al. 2004). For example, the peltate glandular trichomes of mint produce a suite of defensive monoterpenes that are the major components of, and give the characteristic smell and flavor to, mint oil (McCaskill et al. 1992; Voirin and Bayet 1996); trichomes from tomato species collectively produce a number of insecticidal sesquiterpenes, acyl sugars and methyl ketones (Li et al. 1999; Antonious 2001; Maluf et al. 2001); and tobacco trichomes produce diterpenes and acyl sugars (Kandra et al. 1990; Guo and Wagner 1995). Some plants accumulate phenylpropanoids (Gang et al. 2001, 2002) or flavonoids and related compounds (Wollenweber 1984; Hirosawa et al. 1995; Stevens et al. 1998) in their trichomes. The potentially hydrophilic flavonoids are often substituted by methylation or prenylation to increase their hydrophobicity (Wollenweber 1984; Stevens et al. 1998), a property beneficial for secretion. Trichomes may also be important for sequestration of toxic metals (Choi et al. 2001). Because of these functions, trichomes are natural targets for genetic manipulation strategies to improve plant disease or pest resistance by introduction of constitutively produced antimicrobial or insecticidal compounds, or heavy metal tolerance by introduction of metal chelating proteins or peptides.
Because trichome density and morphology are easily scored, significant progress has been made in understanding the molecular genetic basis of trichome development, particularly in the model plant Arabidopsis thaliana (Walker et al. 1999; Szymanski et al. 2000). In contrast, less is known about the molecular aspects of trichome metabolism and secretion. Because of the low level of trichome biomass relative to that of the organs on which the trichomes are located, it is not clear how well trichome-expressed genes are represented in the whole organ cDNA libraries, which constitute the basis of most of the existing plant expressed sequence tag (EST) databases. cDNA libraries have been constructed from trichomes of mint (Lange et al. 2000), sweet basil (Gang et al. 2001), and wild and cultivated tomatoes (http://www.tigr.org/tdb/tgi/). The mint and tomato trichomes show a strong preponderance of transcripts (represented by ESTs) encoding enzymes of terpene metabolism.
Recently, varieties of alfalfa (Medicago sativa), the world’s major forage legume, have been introduced which have a high density of glandular trichomes on the stems and increased resistance against the potato leafhopper (Ranger and Hower 2001). Although the morphology of the erect and procumbent glandular Medicago trichomes has been described (Kreitner and Sorensen 1979a, b; Ranger and Hower 2001), virtually nothing is known about the genes that are expressed in these trichomes in relation to their development, metabolic activity, or protective functions. To begin to address these issues, over 5,000 ESTs have been sequenced from an alfalfa glandular trichome cDNA library. In addition, use has been made of the close genetic relationship between alfalfa and the model legume Medicago truncatula to apply DNA array technology with M. truncatula arrays for the examination of transcripts that are preferentially expressed in alfalfa stem trichomes. Nearly 200,000 EST sequences are currently available for M. truncatula (Bell et al. 2001), although A17, the line for which the majority of genomics resources are available, does not have glandular trichomes. Thus, the microarray analysis addresses genes that are important for trichome function within stem tissue, but that are also expressed elsewhere in the plant. The EST and microarray analyses support preliminary metabolite profiling data indicating that Medicago trichomes primarily produce flavonoid and lipid compounds.
Materials and methods
Plant material
Alfalfa plants (provided by Forage Genetics International, West Salem, WI, USA) were of an elite clone (6-30-5) from a potato leafhopper selection nursery established at West Salem in May 2000. The clone was selected on the basis of plant vigor and high potato-leafhopper tolerance, and had a high density of glandular hairs. Plants were grown in a Conviron growth chamber [16-h days, full lights (250 μmol/m2/s), 24°C] in 40 one-gallon pots in Metromix 350 (GraceSierra, Milpita, CA, USA) and fertilized with MiracleGro (Scotts Company, Marysville, OH, USA) as needed. They were cut back closely to encourage the emergence of vigorous shoots. Plants of M. truncatula cv Jemalong A17 were grown under the same conditions.
Isolation of trichomes and trichome RNA
Trichomes were isolated from stems, approximately 8–12 in. long, clipped from plants 2–3 in. above the crown. With minimal handling of the stems, the apical buds, leaves, and nodes were excised and discarded, while the internode segments were frozen by immersion in liquid nitrogen. Small batches of frozen stem segments (approximately 20–30 pieces) were placed in 50 ml plastic polypropylene screw-cap tubes containing liquid nitrogen with the lids loosened. After the nitrogen had evaporated, the tube was vortexed for 45 s to shear off the trichomes. The tube was immediately placed back in liquid nitrogen, and the stem segments removed. Approximately 5 ml of TriReagent (MRC Inc, Cincinnati, OH, USA) was added to coat the inside of the tube. As the TriReagent thawed, the tube was vortexed. Additional batches of trichomes were isolated as above, but instead of adding fresh TriReagent to each tube, the solution from the previous isolation was transferred to the next tube to increase the trichome RNA concentration. Total RNA was isolated according to the manufacturer’s standard protocol. Quality and quantity of RNA were assessed using both an Eppendorf Biophotometer (Eppendorf, Hamburg, Germany) and formamide gel electrophoresis according to standard protocols.
cDNA library construction and DNA sequence analysis
The cDNA library was constructed using the Creator Smart cDNA library construction kit (Clontech, Palo Alto, CA, USA) following the manufacturer’s protocol for small amounts of RNA. Approximately 1 μg of total RNA was used. Plasmid preparations were made using a Biomek 2000 robot with standard protocols. Average insert size was evaluated by agarose gel electrophoresis to be 1.5 kb. cDNA clones were sequenced (single pass, 5′-end sequencing) using an ABI3700 sequencer and submitted to ESTAP (Mao et al. 2003) for analysis of EST data.
Microarray analysis
The Medicago Genome Oligo Set Version 1.0 (Qiagen Operon, Alameda, CA, USA) was used for microarray studies (see Electronic Supplementary Material).
Total RNA was isolated from 0.5 g of alfalfa stems (with and without trichomes, prepared as in Fig. 1d, e) using 5 ml of Tri-Reagent following the manufacturer’s protocol. The SuperScript Indirect cDNA Labeling System (Invitrogen Life Technologies, Carlsbad, CA, USA) was used to label 20 μg of RNA following the manufacturer’s protocol. The product was resuspended in 50 μl of SlideHyb no. 1 hybridization buffer (Ambion Inc, Austin, TX, USA), denatured at 95–100°C for 2 min, and then pipetted onto the slides and coverslip (Corning, Corning, NY, USA) before sealing into a hybridization chamber (Corning). The sealed chambers were wrapped in aluminum foil and incubated at 42°C for 16–24 h. The arrays were subsequently washed with 1x SSC, 0.1% (wt/vol) SDS, followed by a wash in 0.5x SSC, 0.01% SDS, and a third wash with 0.05x SSC at room temperature for 5 min each. The slides were dried by centrifugation.
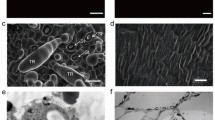
Trichome phenotypes on alfalfa and Medicago truncatula stems. a Trichomes on alfalfa line 6-30-5. b Magnified view showing glandular (2) and non-glandular (1) trichomes. c M. truncatula stem with non-glandular trichomes. d, e Alfalfa stems before (d) and after (e) physical removal of trichomes. f Micrograph showing alfalfa trichome preparation. H head, S stalk. g Alfalfa trichomes stained with DMACA reagent. Yellow bar = 1 mm, red bar = 100 μ
Arrays were read with a ScanArray 4000 scanner (Packard, Palo Alto, CA, USA) at 10-μm resolution and variable photomultiplier tube voltage settings to obtain the maximal signal intensities with <1% probe saturation. The fluorescence intensity for each fluor and each element on the array was captured using GenePix Pro 4.1 (Axon, Union City, CA, USA). The local background was subtracted from the values of each spot on the array. A pixel intensity cut-off of 300 was established based on pixel intensity values obtained from negative control features (e.g., nptII, gus) within the array. Normalization of Cy3 and Cy5 signal was performed by adjusting the signal intensities of the two images (Global Normalization). Within-array normalization of Cy3/Cy5 channel intensities was performed using a LOWESS procedure, incorporating signal intensities of all four elements for each gene (three replicate hybridizations and one dye-swap), that reduces differential dye effects (Cleveland 1979; Yang et al. 2002). The significance of results was determined using an SAM scatter plot analysis (Goss et al. 2001).
RT–PCR analysis
RT–PCR analysis of selected genes was performed using the SuperScript One-Step RT–PCR System with Platinum Taq DNA Polymerase (Invitrogen) following the manufacturer’s protocol. PCR was performed for 40 cycles of denaturation for 1 min at 95°C, annealing for 2 min at 60°C and extension for 2 min at 72°C, followed by a final extension of 5 min. Primers for each of the selected genes were 20–22 bases long, optimized for Tm and GC content. The primer sequences used were; TC49455 (Forward 5′-TCTCCCAAATTCTCAACATCTC-3′, Reverse 5′-AAATTTGCCCCAGCATTTCT-3′), TC52596 (Forward 5′-TGCAAGCTCATTGCAAAAAC-3′, Reverse 5′-TTGATCATTGGCATCCTTCA-3′), TC44811 (Forward 5′-GGGATATTCCTGAAGCAGCA-3′, Reverse 5′-GGGTTGGTTTGGTTTTGAGA-3′), and TC56805 (Forward 5′-CCTTGCTGTCTCCTCCAAAG-3′, Reverse 5′-GTCATCACTTCGCCAGTCAA-3′).
Staining of trichomes for proanthocyanidins
Plant tissues were submerged in p-dimethylaminocinnamaldehyde reagent [DMACA; 0.1% w/v, freshly prepared in ethanol: 6 M HCl (1:1, v/v)] for 5–10 min and then de-colorized by replacing the DMACA with ethanol: acetic acid (75:25, v/v) until complete removal of chlorophyll had occurred. Under these conditions, proanthocyanidins stain deep blue (Li et al. 1996).
Metabolite profile analysis
Trichomes from 40 g fresh weight of stem internodes were extracted in acetone for 24 h and the extracts concentrated under a nitrogen stream. A portion of the sample was hydrolyzed in 1 N HCl at 90°C for 2 h, extracted twice with ethyl acetate, and the organic phases pooled and evaporated. Samples were dissolved in methanol for HPLC analysis of an aliquot (corresponding to trichomes from 16.7 g of fresh stem material) on an Agilent HP1100 HPLC equipped with an autosampler, quaternary pump, and diode array detector. The eluant from the reverse-phase column was monitored at 254, 270, 280, 315, and 524 nm. Solvent A was 1% phosphoric acid in pure water and solvent B was HPLC grade acetonitrile. The flow rate was 1 ml/min, and the gradient was isocratic at 5% B for 5 min, followed by a multi-step gradient from 5% to 10% B in 5 min, 10–17% B in 15 min, 17–23% B in 5 min, 23–50% B in 35 min; and 50–100% B in 4 min.
For gas chromatography/mass spectrometry (GC/MS) analysis, trichomes were extracted in 5 ml of chloroform, filtered through glass microfiber to remove debris, the solvent removed by evaporation, and the sample taken up in 40 μl pyridine plus 40 μl MSTFA [N-methyl-N-(trimethylsilyl)-trifluoroacetamide]. A 1-μl sample was injected at a 1:1 split ratio onto an Agilent 6890 GC equipped with a 60 m DB-5 MS column coupled to a 6973 mass selective detector. The oven was ramped from 80°C (2 min) to 315°C (15 min) at 5°C/min. Column flow was held constant at 1.0 ml/min. The inlet was held at 250°C, the transfer line at 250°C, and the ionization source at 230°C. The quadrupole scanned m/z 50–650.
Results and discussion
Trichome isolation and chemical composition
Stems of alfalfa clone 6-30-5 possess a very high density of capitate glandular trichomes, of which approximately 96% are of the erect glandular type, the remainder being erect non-glandular hairs, which probably have little, if any, secretory function (Fig. 1a, b). In contrast, stems of M. truncatula Jemalong A17 contain a low density of non-glandular trichome hairs (Fig. 1c). Simply freezing alfalfa stem sections in liquid nitrogen followed by physical agitation in plastic tubes resulted in the shearing off of the vast majority of the trichomes (Fig. 1d, e). The resulting trichome preparations consisted primarily of sheared heads and stalks (Fig. 1f).
It has been suggested that trichomes of potato leafhopper-resistant alfalfa lines may contain proanthocyanidins (oligomeric flavan-3-ols) (M. McCaslin, personal communication). Treatment of stems with DMACA reagent resulted in deep blue staining only in the heads of the trichomes, suggesting the presence of proanthocyanidins in the secretory space beneath the epidermis (Li et al. 1996) (Fig. 1g). Analysis of organic extracts of trichome preparations by HPLC with diode array detection resulted in the detection of a series of conjugated flavones and flavonols (note that the extraction and HPLC methods do not detect oligomeric flavonoids). There was also a major peak representing a conjugated hydroxycinnamic acid (Fig. 2a). Hydrolysis in weak acid resulted in a major change of profile (Fig. 2b), and appearance of new peaks identified, on the basis of retention times and UV spectra as the probable aglycones of the conjugates seen in Fig. 2a; these were free hydroxycinnamic (4-coumaric, caffeic, and ferulic) acids, as well as the flavone luteolin and the flavonol kaempferol. Trichomes were also extracted in chloroform to yield the lipid fraction, which was analyzed by GC/MS. The lipids identified included wax components (eicosanoic, 3-hydroxy-eicosanoic, tetracosanoic, hexacosanoic, and octacosanoic acids; heptacosane, nonacosane, heneicosane, dotriacontane, and tritriacontane; hexacosanol, octacosanol, and nonacosanol), nonanoic acid, octadecadienoic acid, and spinasterol (a triterpene) (data not shown). Ferulic and sinapic acids were also detected, in addition to minor peaks confirming the presence of flavonoids. Mono-, sesqui-, and di-terpenes were not detected.
Metabolite profiles from alfalfa trichomes. a HPLC/UV analysis of trichome extracts before acid hydrolysis. UV absorption was measured at 254 nm. Compounds were tentatively identified on the basis of retention times and UV spectra compared to authentic standards. 1 hydroxycinnamic acid conjugate, 2 flavone conjugate, 3 luteolin (flavone) conjugate, 4–9 flavone or flavonol conjugates, 10 putative methoxyflavone. b HPLC/UV analysis of trichome extracts after acid hydrolysis. 1 caffeic acid, 2 p-coumaric acid, 3 ferulic acid, 4 flavonol conjugate, 5 luteolin (flavone), 6 kaempferol (flavonol)
Analysis of alfalfa trichome ESTs
Trichomes were sheared from frozen alfalfa stems as described in Experimental procedures, collected directly in Tri-Reagent for isolation of RNA, and a cDNA library prepared. After plating and random selection, the inserts in 7,629 cDNA clones were sequenced, and the sequences entered into the ESTAP database (Mao et al. 2003). The high-quality sequences (5,657) were clustered into tentative consensus sequences (TCs), and TCs and singletons annotated by BLAST (Altschul et al. 1990) analysis against the nonredundant GenBank database. The ESTs clustered into 555 TCs and 2,245 singletons. Of these 2,800 apparent unigenes, 788 (28.1%) had no database matches based on the ESTAP BLAST search.
Functional annotation (gene ontology) was assigned to 1,000 randomly chosen ESTs (Fig. 3). Of these ESTs, 27.2% had no predicted function (no database hit, or a match to a gene with unknown function), with the second largest group representing proteins of lipid metabolism or transport. In this latter group, most of the ESTs corresponded to one lipid transfer protein (LTP) gene that was the most highly expressed gene represented in the trichome library (8.3% of the total ESTs) (Table 1). LTPs are also highly expressed in mint and basil trichomes (Lange et al. 2000; Gang et al. 2001). They may be involved in the formation of the epicuticular waxes that coat the trichome, and their high abundance may simply reflect the greater proportion of epidermis to total cellular mass in a trichome preparation than in a whole stem or leaf. However, LTPs are less strongly represented in tomato trichome EST libraries (http://www.tigr.org/tdb/tgi/).
EST analysis of alfalfa glandular trichome cDNA library. a Functional ontology of 1,000 transcripts. Gene classes are: 1 primary metabolism (3.7%), 2 secondary metabolism (4.6%), 3 cytoskeleton (0.6%), 4 protein synthesis, processing, trafficking and organization (5.6%), 5 protein degradation (2.2%), 6 lipid synthesis and transport (16.4%), 7 stress response proteins (6.8%), 8 microbial defense response proteins (3.3%), 9 photosynthesis (5.9%), 10 cell wall metabolism (3.1%), 11 signal transduction (includes transcription factors) (5.9%), 12 transport processes (2.2%), 13 respiratory processes (1.1%), 14 redox reactions and protection (1.2%), 15 RNA synthesis and metabolism (3.6%), 16 chromosomal proteins (1.8%), 17 unknown (27.2%), 18 others (4.8%). The remaining panels list natural product pathway gene ESTs (within gene class 2) in the trichome library (numbers represent EST counts per 5,647 ESTs)
Among the 20 most abundant of the 5,657 alfalfa trichome ESTs (Table 1), five had no predicted function and four (GI: 13359451, 729185, 1076525 and 1781279) corresponded to genes potentially involved in abiotic or biotic stress resistance. This may reflect the exposed position of the trichome on the exterior of the plant. The LTP might also be involved in protection against biotic stress, either through signaling or direct antimicrobial activity (Molina et al. 1993; Maldonado et al. 2002; Ge et al. 2003). One putative pathogen resistance protein, an ortholog of M. truncatula MtN13, has sequence identity to the PR10 family of pathogenesis-related proteins. MtN13 has previously been classified as a nodulin, and described as a specific marker for the root nodule outer cortex (Gamas et al. 1998).
Nearly 1% of the 5,657 trichome ESTs represent a protein with similarity to a rice intron-encoded homing endonuclease (IEHE). Such enzymes induce the transposition of mobile intervening sequences containing their open reading frame by making a double strand break in an intronless allele (Jurica and Stoddard 1999). M. truncatula contains at least three IEHE-like genes, two of which are represented by only two ESTs each in the nearly 200,000 ESTs currently available (http://www.tigr.org/tdb/mtgi/). The potential function of the IEHE in the trichome is intriguing, and can now be addressed by gene-knockout technology. A similar approach may yield functional information for the second most highly expressed gene in the trichome library; its sequence identity to the yeast transcript antisense to ribosomal RNA is quite low, and may not provide an accurate functional annotation.
To interrogate trichome secondary metabolism, the BLAST annotations of the 5,647 ESTs were searched for enzymes involved in phenylpropanoid, (iso)flavonoid, terpenoid, and alkaloid biosynthesis. The most highly abundant classes of enzymes potentially involved in secondary metabolism were peroxidases (72 ESTs, clustering into 22 TCs) and cytochrome P450s (68 ESTs falling into 20 TCs). Cyclized mono-, sesqui-, di-, or tri-terpenes are often produced in trichomes as defensive compounds, but there were no significant hits to any of the families of terpene cyclases, including β-amyrin synthase (the cyclase for production of the well-characterized Medicago triterpene saponins) (Suzuki et al. 2002). ESTs annotated as 3-hydroxymethylglutaryl CoA reductase, isopentenyl diphosphate isomerase, farnesyl diphosphate synthase, and squalene epoxidase were present at low abundance, with only one EST annotated as a “terpene synthase.” The presence of ESTs corresponding to these early enzymes of terpene metabolism most likely reflects the biosynthesis of membrane sterols and photosystem-protective carotenoids; trichomes are photosynthetically active (Peterson and Vermeer 1984) and transcripts encoding two photosynthesis-related proteins (chlorophyll a/b binding protein, ribulose bisphosphate carboxylase small subunit) are among the top 20 most highly expressed transcripts. The photosynthetic activity is doubtlessly associated with the trichome stalk tissue present in the trichome preparations.
In contrast to the above, ESTs annotated as corresponding to every enzyme required for the biosynthesis of flavones, flavonols, anthocyanins, isoflavone, monolignols (coniferyl and sinapyl alcohols), and lignans (secoisolariciresinol) were found, with the exception of the hydroxycinnamoyl CoA: quinate/shikimate hydroxycinnamoyl transferase (Hoffmann et al. 2003) of the core phenylpropanoid pathway (Fig. 3). Putative acyl transferases annotated as anthranilate: N-hydroxycinnamoyl/benzoyl transferase and benzoyl CoA: benzyl alcohol benzoyl transferase were, however, highly represented. Chalcone synthase and leucoanthocyanidin dioxygenase (LDOX) were represented by five and six ESTs, respectively. This suggests a significant production of flavonoids and anthocyanin derivatives in the trichomes, consistent with the results in Fig. 2a, b and the detection of proanthocyanidins in Fig. 1g. However, no ESTs corresponding to the key anthocyanidin or leucoanthocyanidin reductases (ANR, LAR) of proanthocyanidin biosynthesis (Tanner et al. 2003; Xie et al. 2003) were found by additional BLAST searches, although members of the dihydroflavonol reductase (DFR)- and isoflavone reductase (IFR)-like families to which these enzymes belong were represented. The high expression of LDOX transcripts, coupled with the lack of apparent accumulation of anthocyanins, suggests that the pathway to proanthocyanidins via anthocyanidins may be operative in the trichomes. However, DMACA-staining can also detect monomeric flavan 3-ol (Li et al. 1996), which was not, however, detected in the HPLC analyses. The presence of proanthocyanidins requires further chemical confirmation.
The level of representation of natural product pathway gene transcripts in alfalfa trichomes is much lower than that of terpene pathway transcripts in EST libraries from mint or tomato trichomes (Lange et al. 2000) (http://www.tigr.org/tdb/tgi/). For example, in trichomes from leaves of Lycopersicum hirsutum, a TC corresponding to one sesquiterpene cyclase accounts for 2.8% of 2,463 ESTs, whereas genes with strong similarity to flavonol synthase and cinnamyl alcohol dehydrogenase, the most strongly expressed secondary metabolism genes in the alfalfa trichomes (apart from peroxidase), each account for only 0.22% of the total alfalfa trichome ESTs. This also contrasts with the very high percentage of phenylpropanoid pathway gene transcripts observed among 1,215 ESTs obtained from a basil peltate trichome cDNA library, where caffeoyl CoA 3-O-methyltransferase transcripts accounted for nearly 2.4% of the total ESTs (Gang et al. 2001). Overall, the functional ontology analysis of the M. sativa trichome ESTs suggests that the gene-expression pattern within these organs is not predominantly associated with a particular metabolic specialization. In fact, the relative proportion of genes involved in metabolism is not dissimilar to that found overall in the Arabidopsis genome (Initiative 2000).
Subtractive microarray analysis of trichome-expressed genes from alfalfa stems
Of the 5,647 alfalfa trichome ESTs, BLAST analysis against all known M. truncatula EST sequences indicated that 4,804 had M. truncatula orthologs with E values of −20 and below. Remarkably, 2,622 of these had an E value of zero (100% nucleotide sequence identity to the M. truncatula ortholog). Among genes represented in the M. sativa trichome ESTs as TCs that have orthologs in M. truncatula, 66.5% have 100% sequence identity to the corresponding M. truncatula ortholog. This analysis validates the use of available M. truncatula DNA arrays for the study of gene expression in alfalfa. No hits were obtained for 598 of the ESTs, which might therefore represent genes present in M. sativa but not M. truncatula (including trichome-specific genes), or genes present in both but yet to be sequenced in M. truncatula.
RNA microarray analysis was performed to determine the relative transcript levels of potential alfalfa orthologs of 16,086 M. truncatula genes in intact alfalfa stems compared to stems from which trichomes had been physically removed. The 16,086 genes were represented by 70-mer oligonucleotides, designed to correspond to unique M. truncatula sequences, printed on glass slides. Due to the very high degree of sequence similarity between alfalfa genes and their M. truncatula orthologs, a significant signal was observed for most of the 16,086 genes represented on the arrays. The overall number of features with a signal more than 300 pixels above the background was the same as observed when the same arrays were hybridized with labeled RNA from M. truncatula stems.
Transcripts encoding 1,088 genes were more than threefold lower in the minus trichome samples (the highest relative decrease being 48.4-fold). We chose threefold as a conservative cutoff value because no genes were more than threefold lower in the plus trichome sample (Fig. 4a, b). A threefold lower expression level in the minus trichome sample equates to an approximately 60-fold lower value on an RNA basis, calculated from direct measurement of the RNA level in a trichome preparation from mature stem tissue. Predicted functional classes for the top 1,000 most “trichome preferentially expressed” sequences are shown in Fig. 4c. Remarkably, over 70% of these genes were of unknown function (compared to 33% of the genes represented on the array), the second most abundant class being genes potentially involved in signal transduction and transcriptional regulation. Clearly, additional genes in the twofold to threefold lower expression class in Fig. 4b may also be preferentially expressed in trichomes. However, as all genes on the microarray come from a species (M. truncatula) that lacks glandular trichomes (see below), we have not further analyzed this class of genes since their function in processes critical for trichome biology may be less apparent than that for genes with stronger differential expression.
Detection of genes with enhanced expression in M. sativa glandular trichomes by differential microarray analysis. a Scatter plot of normalized log 2 (ratios) of Cy3 (stems with trichomes) versus log 2 (ratios) of Cy5 (stems without trichomes). b Distribution of genes in various fold-change categories based on ratio of expression level in intact stems compared to stems minus trichomes. c Functional ontology of the 1,000 genes with highest relative reduction in transcript level following removal of trichomes from alfalfa stems. The percentage of the 1,000 genes in each class was: 1 5.1%, 2 3.2%, 3 0.4%, 4 1.7%, 5 1.3%, 6 1.4%, 7 0.6%, 8 1.2%, 9 0.2%, 10 0.4%, 11 5.8%, 12 2.0%, 13 0.5%, 14 0.8%, 15 1.1%, 16 0.8%, 17 70.1%, 18 3.6%. See legend to Fig. 3 for description of gene classes
To confirm trichome preferential expression in alfalfa stem tissues, four genes (with 43.7-, 10.2-, 5.1- and 2.0-fold lower expression in stems without, compared to with, trichomes, and therefore covering the full range of differential expression), were selected for analysis by RT–PCR with gene-specific primers (Fig. 5). Even the gene with the twofold difference by array analysis was expressed with a much greater than twofold preference in the trichome-containing stems (Fig. 5). The low signal in the minus trichome samples could result from the presence of a few residual trichomes (Fig. 1e).
RT–PCR analysis of transcript levels in alfalfa stem and trichome samples. The material analyzed was stems plus trichomes 1, stems minus trichomes 2 and trichomes alone 3. Also listed is the fold-difference in transcript level in stems with (T+) compared to without trichomes based on microarray analysis, the presence or absence of the transcript in the alfalfa trichome EST library (SGT) (with E value for the alfalfa ortholog), the abundance of the transcript in the M. truncatula EST library database (number of ESTs per total number of ESTs in all libraries in which the transcript occurred), and the tissue source of the libraries in which ESTs were found. The genes were: TC49455, similar to ACC synthase; TC52596, similar to nitrate reductase; TC44811, similar to NAC domain protein; TC56805, unknown. The cDNA libraries were: Sd drought-stressed seedlings, Ra root (all libraries), R root, Lps phosphate-starved leaf, S stem, Ce elicited cell culture, F flower, Lih leaf subjected to insect herbivory
The 20 alfalfa genes (possessing sequenced orthologs in M. truncatula) with the highest relative trichome expression in the stem are listed in Table 2, along with information as to their presence or absence in the 5,647 alfalfa trichome ESTs and the E values for orthologous matches between M. truncatula and M. sativa; data for all 16,000+ genes represented on the microarray are available in the Electronic Supplementary Material. Twelve of the 20 genes have no predicted function (no hit, or hits to a protein of unknown function), and only one appears to encode an enzyme of secondary metabolism (a putative LDOX). Those genes with an annotated function show high sequence identity between M. truncatula and M. sativa.
Although only one of the 20 genes listed in Table 2 is annotated as encoding an enzyme of secondary metabolism (an LDOX), the microarray analysis indicated that additional flavonoid pathway genes were, within stem material, preferentially expressed in trichomes. These include TCs annotated as encoding chalcone synthase (4.1-fold reduced expression in stems lacking trichomes), chalcone isomerase (2.7-fold), flavanone 3-hydroxylase (3.6-fold), flavone synthase II (2.4-fold), flavonol synthase (2.5-fold), and flavonoid 3′,5′-hydroxylase (2.5-fold). Thus, on the basis of transcript levels, alfalfa trichomes appear to have an increased flavonoid biosynthetic capacity as compared to the stem alone. The alfalfa flavanone 3-hydroxylase promoter is expressed in the stalks of leaf trichomes of tobacco (Charrier et al. 1996), suggesting that some of the flavonoid synthesis inferred from the present experiment may as likely take place in the trichome stalks as in the heads.
The tissue-specific expression patterns of the M. truncatula genes orthologous to the alfalfa genes with high trichome expression in the stem were estimated by EST counts in the nearly 40 different cDNA libraries available in the TIGR Medicago gene index (http://www.tigr.org/tdb/mtgi/). Among the 20 most “trichome preferentially expressed” transcripts in the alfalfa stem, only one (TC57602) met the criteria for potential trichome specificity in M. truncatula (expressed in stem, or leaf, but not in roots, nodules, seeds, or cell cultures which do not contain trichomes) (Table 2). Many of these genes were expressed in root tissues, particularly roots associated with symbiotic bacteria (nodules) or fungi (mycorrhizal roots). The reason for this is not clear.
The stem trichomes of M. truncatula Jemalong A17 are non-glandular, and, furthermore, trichome-specific transcripts may, because of their overall low abundance, be underrepresented in most cDNA libraries from which EST databases have been derived. This accounts for the differences in the identities of the highly expressed genes in trichomes revealed by EST sequencing and the group of genes with trichome preferential expression in stems revealed by subtractive microarray analysis; indeed, the two approaches as used here target very different groups of genes. The results highlight the need for trichome EST projects in other species, or from trichomes from different tissues of the same species (note that most molecular analyses to date have targeted leaf trichomes). Such projects will provide valuable information on trichome biochemistry for those species which have yet to receive detailed phytochemical analysis.
In conclusion, combined transcript and metabolite profiling indicate that alfalfa glandular trichomes express a wide range of genes with functions in defense and production of (primarily flavonoid) secondary metabolites. The results also highlight the value of the genomics tools developed in the model legume M. truncatula for studies in alfalfa.
Abbreviations
- EST:
-
Expressed sequence tag
- LTP:
-
Lipid transfer protein
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Antonious GF (2001) Production and quantification of methyl ketones in wild tomato accessions. J Environ Sci Health B36:835–848
Bell C, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW, May GD (2001) The Medicago genome initiative: a model legume database. Nucleic Acids Res 29:114–117
Charrier B, Leroux C, Kondorosi A, Ratet P (1996) The expression pattern of alfalfa flavanone 3-hydroxylase promoter-GUS fusion in Nicotiana benthamiana correlates with the presence of flavonoids detected in situ. Plant Mol Biol 30:1153–1168
Choi YE, Harada E, Wada M, Tsuboi H, Morita Y, Kusano T, Sano H (2001) Detoxification of cadmium in tobacco plants: formation and active excretion of crystals containing cadmium and calcium through trichomes. Planta 213:45–50
Cleveland W (1979) Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc 74:829–836
Gamas P, De Billy F, Truchet G (1998) Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding proteins homologous to plant defense proteins. Mol Plant-Microbe Interact 11:393–403
Gang DR, Wang J, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E (2001) An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol 125:539–555
Gang DR, Beuerle T, Ullmann P, Werck-Reichhart D, Pichersky E (2002) Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol 130:1536–1544
Ge X, Chen J, Sun C, Cao K (2003) Preliminary study on the structural basis of the antifungal activity of a rice lipid transfer protein. Protein Eng 16:387–390
Goss V, Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarray applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121
Guo Z, Wagner GJ (1995) Biosynthesis of cembratrienols in cell-free extracts from trichomes of Nicotiana tabacum. Plant Sci 110:1–10
Hirosawa T, Saito T, Tanaka T, Matsushima H. (1995) SEM observation and HPLC analysis of the accumulation of alpha- and beta-acids in the fresh developing hop (Humulus lupulus L.) peltate glandular trichomes. J Electron Microsc 44:145–147
Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278:95–103
Initiative The Arabidopsis Genome (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Jurica MS, Stoddard BL (1999) Homing endonucleases: structure, function and evolution. Cell Mol Life Sci 55:1304–1326
Kandra G, Severson R, Wagner GJ (1990) Modified branched-chain amino acid pathways give rise to acyl acids of sucrose esters exuded from tobacco leaf trichomes. Eur J Biochem 188:385–391
Kreitner GL, Sorensen EL (1979a) Glandular trichomes on Medicago species. Crop Sci 19:380–384
Kreitner GL, Sorensen EL (1979b) Glandular secretory system of alfalfa species. Crop Sci 19:499–502
Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97:2934–2939
Li YG, Tanner G, Larkin P (1996) The DMACA-HCl protocol and the threshold proanthocyanidin contant for bloat safety in forage legumes. J Sci Food Agric 70:89–101
Li AX, Eannetta N, Ghangas GS, Steffens JC (1999) Glucose polyester biosynthesis. Purification and characterization of a glucose acyltransferase. Plant Physiol 121:453–460
Maldonado AM, Dixon RA, Lamb C, Doerner P, Cameron RK (2002) A putative lipid transfer protein is involved in systemic signaling to establish acquired resistance in Arabidopsis thaliana. Nature 419:399–403
Maluf WR, Campos GA, das Gracas CM (2001) Relationships between trichome types and spider mite (Tetranychus evansi) repellence in tomatoes with respect to foliar zingiberene contents. Euphytica 121:73–80
Mao C, Cushman JC, May GD, Weller JW (2003) ESTAP—an automated system for the analysis of EST data. Bioinformatics 19:1720–1722
McCaskill D, Gershenzon J, Croteau R. (1992) Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha piperita L.). Planta 187:445–454
Molina A, Segura A, García-Olmedo F (1993) Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett 316:119–122
Peterson RL, Vermeer J (1984) Histochemistry of trichomes. In: Rodriguez E, Healey PL, Mehta I (eds) Biology and chemistry of plant trichomes. Plenum Press, New York, pp 71–94
Ranger CM, Hower AA (2001) Glandular morphology from a perennial alfalfa clone resistant to the potato leafhopper. Crop Sci 41:1427–1434
Stevens JF, Miranda CL, Buhler DR, Deinzer ML (1998) Chemistry and biology of hop flavonoids. J Am Soc Brew Chem 56:136–145
Suzuki H, Achnine L, Xu R, Matsuda SPT, Dixon RA (2002) A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J 32:1033–1048
Szymanski DB, Lloyd AM, Marks MD (2000) Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci 5:214–219
Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278:31647–31656
Voirin B, Bayet C (1996) Developmental changes in the monoterpene composition of Mentha X piperita leaves from individual peltate trichomes. Phytochemistry 43:573–580
Wagner GJ (1991) Secreting glandular trichomes: more than just hairs. Plant Physiol 96:675–679
Wagner GJ, Wang E, Shepherd RW (2004) New approaches for studying and exploiting an old protuberance, the plant trichome. Ann Bot 93:3–11
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasdan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA 1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11:1377–1349
Wollenweber E (1984) The systematic implication of flavonoids secreted by plants. In: Rodriguez E, Healey PL, Mehta I (eds) Biology and chemistry of plant trichomes. Plenum Press, New York, pp 53–69
Xie D, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299:396–399
Yang Y, Dudoit S, Lin D, Peng V, Ngai J, Speed T (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15.1–e15.10
Acknowledgements
We thank Dr Stephen Temple, Forage Genetics International, for provision of glandular-haired alfalfa lines and helpful discussions, Jack Blount and Corey Broeckling for assistance with HPLC and GC analysis, respectively, Dr Yuanji Zhang for assistance with bioinformatics, Dr David Galbraith (University of Arizona) for printing of microarrays, and Drs Li Tian and Mary Sledge for critical reading of the manuscript. This work was supported by the Samuel Roberts Noble Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Data deposition
The alfalfa trichome EST sequences reported in this paper have been deposited in the GenBank database (accession nos. CO511688 to CO517334).
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Aziz, N., Paiva, N.L., May, G.D. et al. Transcriptome analysis of alfalfa glandular trichomes. Planta 221, 28–38 (2005). https://doi.org/10.1007/s00425-004-1424-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1424-1