Abstract
The MtTrHb1 and MtTrHb2 genes of the model legume Medicago truncatula Gaertn. encode proteins homologous to truncated hemoglobins (TrHb) from plants and a range of different microorganisms. Induction of MtTrHb1 in root nodules and expression of MtTrHb2 in root nodules, as well as in mycorrhizal roots, were shown by quantitative real-time reverse transcription–polymerase chain reaction (RT–PCR). The promoters of both genes were PCR-amplified and fused to the gusAint coding region. By analysing these gusAint-fusions in transgenic root tissues, we were able to localize their activity in root nodules and in roots colonized by arbuscular mycorrhizal (AM) fungi. Whereas the promoter of MtTrHb1 was activated in the infected cells of the nitrogen-fixing zone of root nodules, the MtTrHb2 promoter was predominantly active in the nodule vascular tissue. This expression pattern correlates with the presence of an ‘organ-specific element’ (OSE)-like sequence in the MtTrHb1 promoter, which is not present in the MtTrHb2 regulatory unit. Concerning the AM symbiosis, only the MtTrHb2 promoter mediated an expression in arbuscule-containing cells and in the root vascular tissue of mycorrhizal root segments colonized by the fungus Glomus intraradices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Legumes can establish symbiotic associations with bacteria of the genus Rhizobium and with arbuscular mycorrhizal (AM) fungi of the order Glomeromycota (Schüssler et al. 2001). The Rhizobium symbiosis leads to the formation of a specialized organ, the root nodule, in which the bacteria convert atmospheric nitrogen to ammonium, which is supplied to the plant in exchange for carbohydrates. AM symbiosis offers several benefits to the host plant, including improved nutrition (mainly phosphorus), enhanced drought resistance, and protection from pathogens. Thus, both symbioses have to be looked upon as ecological and economical factors of great importance (Smith and Read 1997).
In the case of the N2-fixing symbiosis, the exchange between the two symbiotic partners occurs in the rhizobia-infected cells of the nitrogen-fixing zone III of root nodules (Provorov et al. 2002). For the AM symbiosis, the main site of exchange between plant and fungus is the arbuscule, a highly branched structure that is formed by the fungus within inner cortical cells of the root (Harrison 1999). These complex symbiotic systems require sophisticated processes of defence suppression, structural changes in the root tissue, and an altered metabolism. Due to obvious similarities between these two interactions, and the existence of mutations resulting both in nod− and myc− phenotypes, a common genetic basis for the two symbioses was supposed (Duc et al. 1989; Gianinazzi-Pearson 1997). In fact, both microsymbionts share steps in their symbiotic signalling pathways during the initiation of root nodules and arbuscular mycorrhizas (Cullimore and Denarie 2003), and several nodulin genes have already been shown to be induced not only during nodulation but also in legume root tissues colonized by mycorrhizal fungi (Lum and Hirsch 2002).
The most prominent nodule-specific proteins are the leghemoglobins (Lbs), which are expressed in the infected cells just prior to the onset of nitrogen fixation. These oxygen-binding heme proteins are supposed to be responsible for supporting the flux of oxygen to the nitrogen-fixing bacteroids (Appleby 1984). One member of this family, the Vicia faba Lb gene Vflb29 was shown to be expressed in both symbiotic interactions (Frühling et al. 1997), and recently the promoter of this gene was found to be specifically active not only in the infected cells of the root nodule but also in the arbuscule-containing cells of mycorrhizal roots (Vieweg et al. 2004).
The discovery of hemoglobins (Hbs) in all kingdoms of organisms suggests that the corresponding genes are derived from a very early common ancestor. Alignments of Hb sequences from various species reveal a high structural variability, but their common characteristic is the ability to reversibly bind oxygen. The variability among Hb proteins indicates different functional features, and in fact many recent studies have revealed that, apart from the transport of oxygen between tissues, Hbs can fulfil additional functions ranging from intracellular oxygen transport to the detoxification of nitric oxide (NO) and the catalysis of redox reactions (Kundu et al. 2003).
In the case of plants, apart from symbiotic Lb genes, several genes encoding Hbs have been found in many legume and non-legume species (Seregelyes et al. 2000). For these non-symbiotic Hbs, it was previously shown that they seem to have a function different from oxygen transport. This function is associated with detoxification of the NO that is produced as a response to altered metabolism under anaerobic conditions (Seregelyes et al. 2003; Dordas et al. 2003).
Standard Hb proteins, including the animal blood Hbs as well as the Lbs and non-symbiotic Hbs, occur ubiquitously in eukaryotes and exhibit a characteristically conserved structure of the globin fold, which consists of a 3-on-3 sandwich of α-helices (Kundu et al. 2003). In bacteria, Hb proteins can be found that exhibit an aberrant tertiary structure based on a 2-on-2 α-helical sandwich. Due to this structural property, these proteins were named 2-on-2 or truncated hemoglobins (TrHb), and were categorized as a separate family within the Hb super-family. It has recently been discovered that the TrHbs are not restricted to prokaryotes since, although they are not present in animals or yeast, they have been discovered in several plant species (Wittenberg et al. 2002). So far, little is known about the role of these genes in plants, since the only TrHb from plants described in more detail is the glb3 gene from Arabidopsis thaliana. This gene is expressed throughout the plant with an enhanced expression in roots. Its induction is not altered by hypoxia or treatment with hormones, which have already been proven to induce some of the non-symbiotic plant Hb genes (Watts et al. 2001). In the case of the model legume M. truncatula, a TrHb gene was mentioned amongst more than 300 nodule-specific genes investigated by in silico profiling. The induction of this TrHb gene was confirmed by macroarray and RNA-blot hybridizations but was not analysed or discussed further (Fedorova et al. 2002). This TrHb gene is one of the two TrHb genes investigated in more detail within this study.
For bacteria, several studies indicate a function for TrHbs in the detoxification of NO that is induced under hypoxic conditions (Thorsteinsson et al. 1999) or produced by the host as a defence reaction in the case of pathogenic bacteria (Ouellet et al. 2002). Nitric oxide turned out to be an important messenger molecule in plants, where it is known to be involved in a broad range of pathogenic and developmental processes (Lamattina et al. 2003). Thus, a function for plant TrHbs in connection with NO is a point of speculation.
In this study we report on the characterization of two M. truncatula genes encoding TrHbs that are expressed in root endosymbioses. A possible role for the encoded proteins in association with NO is discussed.
Materials and methods
Databases and tools for sequence analyses
The TIGR M. truncatula Gene Index was used for in silico screening (http://www.tigr.org/tigr-scripts/tgi/T_reports.cgi?species=medicago). A description of the libraries based on nodule and mycorrhizal tissues can be found at http://www.tigr.org/tdb/tgi/mtgi/searching/xpress_search.html. Multiple sequence alignments were performed using MultAlin (http://prodes.toulouse.inra.fr/multalin/multalin.html), and a phylogenetic tree was constructed using TreeTop (http://www.genebee.msu.su/services/phtree_reduced.html).
Isolation of nucleic acids, recombinant DNA techniques and hybridizations
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Plasmid DNA was prepared from Escherichia coli, grown on Pennassay agar plates containing appropriate antibiotics, using the Plasmid Mini Kit (Qiagen). All in vitro manipulations of DNA and the hybridization of bacterial artificial chromosome (BAC) filters representing the M. truncatula genome (Clemson University, SC, USA) with MtTrHb2 probes were performed using standard protocols.
Quantitative real-time RT–PCR
Quantitative real-time reverse transcription–polymerase chain reaction (RT–PCR) was designed according to Bustin (2000), and carried out using the QuantiTect SYBR Green RT–PCR kit (Qiagen). For RT–PCR, 50 ng of total RNA was used in a total volume of 25 μl. Reactions were run on an Opticon real-time PCR cycler (MJ Research, Waltham, MA, USA), and quantification was done with the Opticon Monitor analysis software version 1.05. The program used was as follows: reverse transcription (50°C for 30 min), polymerase activation (95°C for 15 min), amplification and quantification cycles repeated 44 times (94°C for 15 s, 45°C for 30 s, 72°C for 30 s with a single fluorescence measurement), and melting curve generation (40°C to 95°C with one fluorescence read every 1°C). A relative expression ratio was calculated as the ratio of normalized gene expression of the gene of interest against a constitutively expressed M. truncatula gene that encodes an elongation factor 1-alpha (MtTefa, TC67846, TIGR M. truncatula Gene Index, http://www.tigr.org/tdb/mtgi). The following formula was used: relative expression ratio of the gene of interest is 2−ΔCT with ΔCT=CTgene−CTTefa and CT denoting the threshold cycle. All primers had a calculated Tm of 53°C (±0.1°C) and the primer sequences (Table 1) were unique in the TIGR M. truncatula Gene Index.
Construction of the MtTrHb1 and MtTrHb2 promoter–gusAint fusions
The MtTrHb1 (−1405/−15, in relation to the ATG) and the MtTrHb2 (−2620/−80, in relation to the ATG) promoters were inserted as XhoI/EcoRI and XhoI/SmaI fragments, respectively, into the plasmid pGUS-INT (Küster et al. 1995). Both MtTrHb–gusAint fusions were subsequently cloned as XbaI/StuI fragments (filled in using Klenow polymerase) into the SmaI site of the binary Vector pRedRoot (Limpens et al. 2004), and the resulting binary vectors were transformed into Agrobacterium rhizogenes Arqua1 (Quandt et al. 1993).
Induction of transgenic hairy roots and conditions of plant growth
Transgenic roots were induced in open pots as described by Vieweg et al. (2004). Root nodules were harvested at 21–25 days post inoculation (dpi) and grown in Petri dishes containing nitrogen-free nodulation buffer according to Quandt et al. (1993), and mycorrhizal (myc+) roots were harvested 21–25 dpi with Glomus intraradices (Premier Tech Biotechnologies, Rivière-de-Loup, Québec, Canada). For the induction of hairy roots, Medicago truncatula Gaertn. cv. Jemalong A17 (T. Huguet, INRA Toulouse, France) was used. A. rhizogenes strain Arqua1 (Quandt et al. 1993) was used for the induction of hairy roots. Nodules were induced on 12- to 14-day-old transgenic hairy roots of M. truncatula by Sinorhizobium meliloti strain 1021 (Casse et al. 1979). Inoculations with G. intraradices were carried out with a commercially available inoculum based on aseptic liquid medium containing spores (Premier Tech Biotechnologies, Rivière-de-Loup, Québec, Canada). For inoculation, roots were dipped in the liquid medium and subsequently planted in pots.
Myc+ and sterile roots were fertilized weekly with half-strength Hoagland’s solution (Arnon and Hoagland 1940) containing 20 μM phosphate and grown in pots with sterilized clay granulate (Seramis, Masterfoods, Verden, Germany). Nodulated plants were fertilized without the nitrogen-containing component. All plants were grown in growth chambers under a photoperiod of 16 h at 22°C with a decrease to 18°C during the dark phase. The relative humidity was set to 70%.
Histochemical analysis of transgenic tissues
β-Glucuronidase (GUS) activity was assayed histochemically as described by Jefferson et al. (1987). The GUS substrate solution contained 2 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide cyclohexylammonium salt; Biosynth, Switzerland), 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, 100 mM Tris (pH 7.0), 50 mM sodium chloride (pH 7.0) and 0.1% Tween-20. The incubation time for semi-thin sections was 4 h and roots were stained overnight. Roots were first vacuum-infiltrated prior to incubation in the dark at 37°C. GUS assays of mycorrhizal roots and nodules expressing promoter–gusAint fusions were performed 30 dpi. In order to verify the presence of the fungus, GUS-stained roots were treated with black ink as described by Vierheilig et al. (1998). Before the GUS staining of mycorrhizal roots and root nodules, semi-thin sections of 80–120 μm were prepared with a microtome (VT/000S; Leica, Wetzlar, Germany). Examination of tissues was carried out by light microscopy (BH-2; Olympus, Hamburg, Germany), and documentation was done using a digital camera (Olympus C-2000Z).
Results
The M. truncatula proteins MtTrHb1 and MtTrHb2 are highly homologous to TrHbs from various plants and bacteria
In the TIGR M. truncatula Gene Index, two tentative consensus sequences (TCs) can be found representing different TrHb genes. First, TC80635 is composed of expressed sequence tags (ESTs) from libraries based on nodule tissues. This TC represents the TrHb gene that was first mentioned by Fedorova et al. (2002) and is referred to as MtTrHb1 in this study. Second, TC76399 is assembled from ESTs predominantly derived from libraries constructed from nodule tissues, as well as roots colonized by AM fungi, and represents a gene designated MtTrHb2. The results of these TIGR Gene Index searches tentatively allowed us to assume that expression of these two TrHb genes was related to symbiotic associations, and formed the starting point of this study.
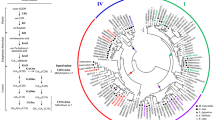
Phylogenetic analyses of amino acid sequences from different members of the known plant Hb families in combination with some bacterial Hbs generate two main branches with two subgroups each (Fig. 1). A branching that reflects the origin and functional classification in plant non-symbiotic Hbs, Lbs, TrHbs, and bacterial TrHbs becomes obvious. It is evident that the TrHbs from plants exhibit a closer structural relationship to the bacterial TrHbs than to the other plant Hbs. Plant TrHbs contain a central region flanked by N- and C-terminal sequences containing amino acid residues characteristic for all TrHbs (Watts et al. 2001). Both truncated hemoglobins, MtTrHb1 and MtTrH2, presented in this study, clearly fit into the group of other plant TrHbs identified so far. MtTrHb1 and MtTrH2 are 60% identical to each other, with MtTrHb1 showing 75% and MtTrH2 77% identity to the A. thaliana GLb3, which is the only plant TrHb described in more detail so far (Watts et al. 2001). Interestingly, the closest relationship can be found to the TrHb of Datisca glomerata with an identity of 76% for MtTrHb1 and 83% for MtTrH2. Although D. glomerata does not belong to the legume family, this species is able to form N2-fixing nodules in symbiosis with actinomycetes (Pawlowski and Bisseling 1996), and the TrHb transcript was, in fact, found in Datisca nodules, as stated in the GenBank entry AJ489324.
Phylogenetic tree showing the relationships among leghemoglobins (Lb), hemoglobins (Hb) and truncated hemoglobins (TrHb) from plants and bacteria. Bootstrap values are shown at branching sites. The Medicago truncatula MtTrHb1 and MtTrHb2 proteins are highlighted, as is the Vicia faba symbiotic leghemoglobin VfLb29 that is expressed in arbuscule-containing cells in addition to the infected cells of root nodules. References are indicated by GenBank accession numbers
Quantitative real-time RT–PCR reveals induction of MtTrHb1 and MtTrHb2 in root nodules and additional expression of MtTrHb2 in mycorrhizal roots
The sequences of the TCs representing MtTrHb1 and MtTrHb2 were used to design gene-specific primers which were used in quantitative real-time RT-PCR experiments, in order to investigate the expression profiles of both genes in different plant tissues. To determine the abundance of both TrHb transcripts in non-symbiotic tissues, RNA probes derived from uninfected roots were used. For the transcriptional analysis of symbiotic tissues, we used several biological replicates of probes representing root nodules (30 dpi) and roots colonized with the AM fungus G. intraradices (30 dpi) based on different inoculation approaches. The parallel amplification of transcripts encoding a constitutive translation elongation factor 1-alpha (MtTefa; Wulf et al. 2003) served for normalization of gene expression. As a control, transcripts of the early nodulin gene ENOD18 were amplified as a marker for nodulation, as described in Hohnjec et al. (2003). For quantification of the mycorrhization status we used the detection of transcripts of the M. truncatula phosphate transporter MtPT4, which is induced in arbuscule-containing cells of mycorrhizal roots (Harrison et al. 2002). Quantitative real-time RT–PCR experiments resulted in specific products and their identities were subsequently verified.
The experiments demonstrated that ENOD18 was induced exclusively in nodules, whereas MtPT4 expression was confined to mycorrhizal roots (data not shown). The results presented in Fig. 2 represent the mean of five technical and three biological replicates based on different inoculation approaches. The quantitative real-time RT–PCR experiments clearly indicated induction of MtTrHb1 in root nodules and expression of MtTrHb2 in nodules as well as in mycorrhizal roots (Fig. 2). In root nodules, MtTrHb1 was 132-fold and MtTrHb2 13.6-fold induced. In the case of MtTrHb2 we additionally detected a 2.6-fold enhanced expression in mycorrhizal roots, whereas no induction was found for MtTrHb1 in this tissue. These induction factors are calculated relative to the expression in sterile roots, which is set to 1.
The MtTrHb1 and MtTrH2 promoters are active in M. truncatula root nodules
A 1,390-bp MtTrHb1 promoter fragment was isolated by PCR amplification using the BAC sequence Mth2-16N19 (GenBank accession AC135413) which contained the MtTrHb1 coding and promoter sequences. This BAC sequence was generated in the M. truncatula genome-sequencing project (University of Oklahoma; http://www.genome.ou.edu/medicago.html).
In the case of MtTrHb2, the three BACs, Mth2-175E1, Mth2-183F15, and Mth2-160C2, carrying coding sequences and promoter regions were identified by hybridization of BAC-filters representing the M. truncatula genome (Clemson University, SC, USA). Starting from the known TC sequence, a “primer walking” was done using BAC DNA as a template, in order to sequence the MtTrHb2 promoter region. In this way, a 2,554-bp MtTrHb2 promoter fragment (GenBank accession AY673965) was isolated by PCR amplification from BAC Mth2-175E1.In order to study the expression properties of both MtTrHb promoters during symbiotic nitrogen fixation, chimeric pMtTrHb1–gusAint and pMtTrHb2–gusAint constructs were prepared. After 4 weeks, transgenic hairy roots of M. truncatula plants transformed with either construct were histochemically analyzed for GUS activity in roots and different stages of nodule development. The pMtTrHb1–gusAint fusions showed GUS activity in the infected cells of nitrogen-fixing zone III, as well as in the interzone II–III, and only a low level of GUS activity in the infection zone of mature nodules (Fig. 3a). GUS activity in nodule primordia or during other stages of nodule development was not observed (Fig. 3c). Apart from root nodules, the MtTrHb1 promoter was additionally found to be active in the root tip (Fig. 3b). The expression pattern of pMtTrHb1–gusAint in the root nodule and root tips can be looked upon as an activity with striking similarities to that of known Lb gene promoters (Franche et al.1998).
Histochemical localization of GUS activity in tissues of transgenic M. truncatula hairy roots expressing pMtTrHb–gusAint constructs. a Activity of pMtTrHb1–gusAint as shown in a longitudinal semi-thin section (80 μm) of a root nodule. GUS activity is located in the infected cells of the nitrogen-fixing zone III as well as in the interzone II–III and only at a low level in the infection zone. b Root tips of pMtTrHb1–gusAint transgenic roots showing GUS activity. c Activity of pMtTrHb1–gusAint as shown by a longitudinal semi-thin section (80 μm) of a nodulated root. The arrow indicates a nodule primordium showing no GUS activity in the vicinity of a GUS-positive adult nodule (asterisk). d Activity of pMtTrHb2–gusAint in a root nodule. Enhanced GUS activity is located in the base and vascular tissue of the nodule. e Activity of pMtTrHb2–gusAint as shown by a longitudinal semi-thin section (80 μm) of a root nodule. GUS activity is located in the vascular tissue as well as the nodule base, with low GUS expression in the non-infected cells of the nitrogen-fixing zone III, the interzone II–III, and in the infection zone. f Mycorrhized root segment of M. truncatula showing GUS activity in the vascular tissue and in arbuscule-containing cells. The arrow indicates cortical cells containing arbuscules. g Close-up of two arbuscule-containing cells and the root vascular tissue showing GUS activity. Bars = 0.1 mm
The activation pattern of pMtTrHb2 in root nodules was found to be completely different from that of pMtTrHb1, since the strongest activation of pMtTrHb2 was confined to the vascular tissue of the root nodule and the nodule base (Fig. 3d,e). Lower levels of GUS expression were additionally detected in the non-infected cells and the interzone II–III. Remarkably, no GUS activity was observed in the infected cells of the nitrogen-fixing zone III (Fig. 3e).
The MtTrHb2 promoter is active in root segments colonized by the AM fungus G. intraradices
Since quantitative RT–PCR results demonstrated an abundance of MtTrHb2 transcripts not only in root nodules but also in root tissues colonized by the AM fungus G. intraradices, the activity of pMtTrHb2 during this symbiotic interaction was investigated and compared with that of the MtTrHb1 promoter. Again, transgenic hairy roots of M. truncatula carrying the pMtTrHb1–gusAint and pMtTrHb2–gusAint constructs were used. After root transformation, the plants were inoculated with the AM fungus G. intraradices. At 30 dpi, colonized roots were harvested for histological GUS staining, and were examined by light microscopy. As a control, wild-type roots were also examined, but no gus expression was observed in mycorrhizal root segments or in cells containing arbuscules (data not shown). In the case of roots transformed with the pMtTrHb1–gusAint construct, no GUS staining above an occasional unspecific background staining (Hänsch et al. 1995) was observed. By contrast, an examination of transgenic hairy roots carrying the pMtTrHb2–gusAint fusion revealed GUS staining in the vascular tissue of mycorrhizal root segments and inner cortical cells with enhanced gus expression (Fig. 3f,g). To confirm the MtTrHb2 promoter activity in root segments colonized by the AM fungus, GUS-stained roots were treated with black ink (Vierheilig et al. 1998) to visualize fungal structures within the plant root tissue. This double staining confirmed that gus expression was restricted to mycorrhizal root segments and that arbuscules were present in the GUS-stained inner cortical cells (data not shown). There was little variability in the intensity of GUS staining among the different AM colonizations in large numbers of independent hairy roots. Thus, MtTrHb2 transcript abundance in mycorrhizal roots of M. truncatula was extended to the cellular level by the localization of promoter activity in mycorrhizal root segments.
The MtTrHb1 promoter shows similarities to Lb gene promoters
Since the expression pattern of pTrHb1–gusAint resembled that typical of a plant Lb gene promoter, it appeared possible that similarities might also be present on the nucleotide level. To address this question, the MtTrHb1 and MtTrHb2 promoter fragments were compared by multiple alignments to Lb promoter sequences from various species. In accordance with the absence of MtTrHb2 promoter activity in the inner nodule tissue, no significant homology of the MtTrHb2 promoter sequence to the MtTrHb1 promoter or to various Lb gene promoters was found (data not shown). In the case of pMtTrHb1, however, the alignments revealed similarities to Lb promoters (Fig. 4). The most characteristic common motifs of Lb promoters are two consensus sequences AAAGAT and CTCTT (Fig. 4). Both motifs are typical elements of Lb and other nodulin gene promoters, and are important regulatory elements with respect to the expression in the infected cells of root nodules. These two motifs are part of the ‘organ-specific element’ (OSE) sequences originally identified in the soybean Lb lbc3 promoter reported by Stougaard et al. (1987). Although the motifs are not exactly conserved in the MtTrHb1 promoter, a noticeable similarity can be found in the area of Lb promoters that comprise the OSE. Here, pMtTrHb1 shows a sequel of ten thymidine nucleotides that allows pMtTrHb1 to be aligned with Lb promoters in the region of the OSE. Furthermore, the alignment shows that the position of this OSE-like element is conserved in its distance from the TATA-box. It should be mentioned that the V. faba Lb3 promoter also lacks the typical CTCTT motif, but shows a CTTT instead, with no loss of promoter activity (Vieweg et al. 2004). Thus, the activity of MtTrHb1 in the infected cells of root nodules can be related to a promoter region that is at least similar to classical Lb promoter motifs.
Alignment of Lb-promoter sequences from various species in comparison to pMtTrHb1. The consensus sequences of the organ-specific element characteristic of promoters activated in infected cells of root nodules and the TATA-box sequences are highlighted, the positions of these motifs are marked by boxes, and conserved nucleotides are marked in bold. Corresponding GenBank entries are indicated by superscript letters: a, M32883; b, X57733; c, X57732; d, P93848; e, AB042718; f, X73283; g, AJ001752
Discussion
In this report, we present the characterization of two TrHb genes, MtTrHb1 and MtTrH2, of M. truncatula. Comparative analyses of amino acid sequences revealed that both Hbs clearly fit into the group of already- known plant TrHbs, including their characteristic close relationship to bacterial TrHbs. In this context it is remarkable that the closest relationship for both TrHbs presented in this study is to a TrHb expressed in root nodules of D. glomerata. This finding allows us to conclude that the symbiotic induction of TrHbs is not restricted to M. truncatula.
Expression analyses by quantitative real-time RT–PCR revealed that both MtTrHb genes are induced in response to symbiosis: MtTrHb1 was found to be expressed in root nodules, and MtTrH2 was active in root nodules as well as in mycorrhizal roots.
The activity of promoter–gusAint fusions in transgenic hairy roots revealed that MtTrHb1 and MtTrH2 had completely different cellular expression patterns. pMtTrHb1 was active in the infected cells of the nitrogen-fixing zone of root nodules, thus exhibiting a cellular expression pattern typical of an Lb gene promoter (Franche et al. 1998). Interestingly, this finding is in accordance with the structural characteristics of the MtTrHb1 promoter sequence, which shows significant similarities to Lb promoters of various species at the position where the consensus sequence motifs of the OSEs are usually located. These motifs are required for expression in the infected cells of root nodules (Stougaard et al. 1987). By contrast, the MtTrH2 promoter does not exhibit such similarities to Lb promoters, as reflected by its activity in the base and vascular tissue of nodules, and its lack of activity in the infected cells.
The up-regulation of MtTrH2 in mycorrhizal roots was verified by the observation of MtTrH2 promoter activity in arbuscule-containing cells and the vascular tissue of root segments colonized by the AM fungus G. intraradices. The activity in vascular tissue appears to be specific for the MtTrH2 promoter, since neither the incubation time used nor previous results obtained for other legume gene promoters only active in arbuscule-containing cells (Vieweg et al. 2004) showed an unspecific staining in the vascular tissue. A similar activation during both AM- and nodule-type symbiotic interactions was also shown in the case of the early nodulin gene MtEnod11 (Journet et al. 2001). Using promoter–gus fusions, the strongest expression was observed in the very early stages of nodule development, whereas in mature nodules GUS activity was restricted to the distal region. The observations concerning the activity of pMtEnod11 during AM symbiosis resemble those for pMtTrHb2, since both promoters show activation in mycorrhizal root segments, and here in cells containing arbuscules.
The most striking correlation in the case of MtTrHb2, however, can be found with the Lb gene VfLb29, which was also shown to be induced in the root nodule and in mycorrhizal roots (Frühling et al. 1997). However, the cellular expression pattern of VfLb29 is different from that of MtTrHb2. The VfLb29 promoter was shown to be specifically active in the infected cells of the root nodule (Vieweg et al. 2004), in contrast to pMtTrHb2, which was found to be active only in the nodule base and vascular tissue. The observations concerning the activity of pVfLb29 during the mycorrhiza symbiosis resemble to some extent those for pMtTrHb2, since both promoters show activation in cells containing arbuscules. In the case of pVfLb29, however, this activation was shown to be restricted to these cells (Vieweg et al. 2004), whereas pMtTrHb2 is additionally active in the vascular tissue of the whole mycorrhizal root segment. Since the phylogenetic analyses clearly indicate that VfLb29 and MtTrHb2 belong to different Hb subfamilies, it has to be emphasized that VfLb29 and MtTrHb2 cannot be regarded as orthologous genes, although it might be possible that they serve similar functions concerning their induction in symbiotic interactions.
In recent years, several studies have indicated a role for Hbs in the detoxification of NO. In animals, NO is detoxified to nitrate by a reaction with blood Hb or myoglobin (Flögel et al. 2000). For plants, the overproduction of two non-symbiotic Hbs was recently shown to prevent cell death caused by NO (Dordas et al. 2003; Seregelyes et al. 2003), and in the case of VfLb29 we previously speculated about a possible function in the suppression of NO, to avoid cell death, and in defence gene induction in symbiotic interactions (Vieweg et al. 2004).
NO is a highly diffusible, bioactive molecule produced in plant tissues in response to various factors. It is supposed to be a key signalling molecule with diverse functions in a broad spectrum of pathophysiological as well as developmental processes (Lamattina et al. 2003). In addition, it is assumed that NO is involved in symbiotic interactions, either as a messenger molecule, or as a by-product of the altered metabolism in root nodules (Mathieu et al. 1998; Herouart et al. 2002), and this might also be the case for AM symbiosis. For instance, the synthesis of NO in root nodules as a by-product of nitrogen fixation and its inhibitory effect on nitrogenase activity has been demonstrated (Cueto et al. 1996; Trinchant and Rigaud 1982). Furthermore, NO is known to be involved in plant responses against pathogens and, here, NO scavengers were able to suppress plant defense (Delledonne et al. 1998; Durner et al. 1998). In accordance with that, transgenic Nicotiana tabacum over-expressing an alfalfa non-symbiotic Hb showed reduced necrosis after inoculation with pathogens or application of exogenous NO (Seregelyes et al. 2003). Finally, Ouellet et al. (2002) were able to show for bacterial TrHbs a strong detoxification effect on NO that was 20-fold more rapid than myoglobin in the case of a TrHb of Mycobacterium bovis. Considering the fact that the structural properties of TrHbs are highly conserved among different species, the authors proposed that possibly all TrHbs fulfil a function in the detoxification of NO. Against this background it appears plausible that MtTrHb1 and MtTrHb2 might serve in parallel to suppress NO accumulation during nodule and mycorrhizal symbioses. MtTrHb1 might be involved in the detoxification of NO produced in the infected cells of root nodules due to their high metabolic activity, in the occurrence of defense processes, or in the release of NO by nitrogen-fixing bacteroids, whereas MtTrH2 might serve as a scavenger of NO in the base and vascular tissues of nodules, as well as in arbuscule-containing cells. It should be noted that defense suppression takes place in cells containing arbuscules, since an induction of genes related to defense suppression was shown to exclusively occur in these cells (Salzer et al. 1999; Bonanomi et al. 2001). Since NO detoxification can be mediated by either Lb or TrHb proteins, different legume species might have recruited different Hb genes for this purpose.
It should be mentioned that in the TIGR M. truncatula Gene Index, the TC corresponding to MtTrHb2 also contains ESTs from plant tissues colonized by both the fungus Colletotrichum trifolii and the root knot nematode Meloidogyne incognita, suggesting that MtTrH2 can be induced during these pathogenic interactions, which were not investigated in this study. It is most notable that although C. trifolii and M. incognita are pathogens, both organisms exhibit a biotrophic lifestyle, which is characteristic also for symbiotic organisms. Interestingly, no MtTrHb2 EST can be found in libraries that are derived from tissues colonized with non-biotrophic pathogens. Biotrophic parasitism shows interesting correlations with symbiotic interactions, and there is evidence for conserved signalling pathways, common gene expression, and the induction of defence suppression processes, which obviously play a key role in the establishment of all biotrophic interactions (Mathesius 2003). Thus, if it is assumed that MtTrHb2 has a role in the suppression of defence processes based on NO detoxification or scavenging, induction of this gene during biotrophic interactions in general would not be surprising and should be the subject of further investigation.
Abbreviations
- AM :
-
Arbuscular mycorrhizal
- BAC :
-
Bacterial artificial chromosome
- dpi :
-
Days post inoculation
- EST :
-
Expressed sequence tag
- Hb :
-
Hemoglobin
- Lb :
-
Leghemoglobin
- NO :
-
Nitric oxide
- OSE :
-
Organ-specific element
- TC :
-
Tentative consensus sequence
- TrHb :
-
Truncated hemoglobin
References
Appleby CA (1984) Leghemoglobin and Rhizobium respiration. Annu Rev Plant Physiol 35:443–476
Arnon DI, Hoagland DR (1940) Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci 50:463–483
Bonanomi A, Wiemken A, Boller T, Salzer P (2001) Local induction of a mycorrhiza-specific class III chitinase gene in cortical root cells of Medicago truncatula containing developing or mature arbuscules. Plant Biol 3:194–199
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193
Casse F, Boucher D, Julliot JS, Michel M, Denarie J (1979) Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J Gen Microbiol 113:229–242
Cueto M, Hernandez-Perera O, Martin R, Bentura ML, Rodrigo J, Lamas S, Golvano MP (1996) Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett 398:159–164
Cullimore J, Denarie J (2003) Plant sciences. How legumes select their sweet talking symbionts. Science 302:630-633
Delledonne M, Xia YJ, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394:585–588
Dordas C, Hasinoff BB, Igamberdiev AU, Manac’h N, Rivoal J, Hill RD (2003) Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. Plant J 35:763–770
Duc G, Trouvelot A, Gianinazzi-Pearson V, Gianinazzi S (1989) First report of non-mycorrhizal plant mutants (myc−) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.). Plant Sci 60:215–222
Durner J, Wendehenne D, Klessig DF (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95:10328–10333
Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130:519–537
Flogel U, Merx MW, Godecke A, Decking UKM, Schrader J (2001) Myoglobin: a scavenger of bioactive NO. Proc Natl Acad Sci USA 98:735–740
Franche C, Diouf D, Laplaze L, Auguy F, Frutz T, Rio M, Duhoux E, Bogusz D (1998) Soybean (lbc3), Parasponia, and Trema hemoglobin gene promoters retain symbiotic and nonsymbiotic specificity in transgenic Casuarinaceae: implications for hemoglobin gene evolution and root nodule symbioses. Mol Plant Microbe Interact 11:887–894
Frühling M, Roussel H, Gianinazzi-Pearson V, Pühler A, Perlick AM (1997) The Vicia faba leghemoglobin gene VfLb29 is induced in root nodules and in roots colonized by the arbuscular mycorrhizal fungus Glomus fasciculatum. Mol Plant Microbe Interact 10:124–131
Gianinazzi-Pearson V (1997) Have common plant systems co-evolved in fungal and bacterial root symbioses? In: Legocki A, Bothe H, Pühler A (eds) Biological fixation of nitrogen for ecology and sustainable agriculture: NATO ASI Series, vol G39. Springer, Berlin Heidelberg New York, pp 321–324
Hänsch R, Koprek T, Mendel RR, Schulze J (1995) An improved protocol for eliminating endogenous beta-glucuronidase background in barley. Plant Sci 105:63–69
Harrison MJ (1999) Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol 50:361–389
Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14:2413–2429
Herouart D, Baudouin E, Frendo P, Harrison J, Santos R, Jamet A, Van de Sype G, Touati D, Puppo A (2002) Reactive oxygen species, nitric oxide and glutathione: a key role in the establishment of the legume–Rhizobium symbiosis? Plant Physiol Biochem 40:619–624
Hohnjec N, Perlick AM, Puhler A, Kuster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16:903–915
Jefferson RA, Kavanagh TA, Bevan MV (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14:737–748
Kundu S, Trent JT, Hargrove M (2003) Plants, humans and hemoglobins. Trends Plant Sci 8:387–393
Küster H, Quandt HJ, Broer I, Perlick AM, Pühler A (1995) The promoter of the Vicia faba L. VfENOD-GRP3 gene encoding a glycine-rich early nodulin mediates a predominant gene expression in the interzone II–III region of transgenic Vicia hirsuta root nodules. Plant Mol Biol 29:759–772
Lamattina L, Garcia-Mata C, Graziano M, Pagnussat G (2003) Nitric oxide: the versatility of an extensive signal molecule. Annu Rev Plant Biol 54:109–136
Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55:983–992
Lum MR, Hirsch AM (2002) Roots and their symbiotic microbes: strategies to obtain nitrogen and phosphorus in a nutrient-limiting environment. J Plant Growth Regul 21:368–382
Mathesius U (2003) Conservation and divergence of signalling pathways between roots and soil microbes—the Rhizobium–legume symbiosis compared to the development of lateral roots, mycorrhizal interactions and nematode-induced galls. Plant Soil 255:105–119
Mathieu C, Moreau S, Frendo P, Puppo A, Davies MJ (1998) Direct detection of radicals in intact soybean nodules: presence of nitric oxide–leghemoglobin complexes. Free Rad Biol Med 24:1242–1249
Ouellet H, Ouellet Y, Richard C, Labarre M, Wittenberg B, Wittenberg J, Guertin M (2002) Truncated hemoglobin HbN protects Mycobacterium bovis from nitric oxide. Proc Natl Acad Sci USA 99:5902–5907
Pawlowski K, Bisseling T (1996) Rhizobial and actinorrhizal symbioses: what are the shared features? Plant Cell 8:1899-1913
Provorov NA, Borisov AY, Tikhonovich IA (2002) Developmental genetics and evolution of symbiotic structures in nitrogen-fixing nodules and arbuscular mycorrhiza. J Theor Biol 214:215–232
Quandt HJ, Pühler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6:699–706
Salzer P, Corbiere H, Boller T (1999) Hydrogen peroxide accumulation in Medicago truncatula roots colonized by the arbuscular mycorrhiza-forming fungus Glomus intraradices. Planta 208:319–325
Schüssler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421
Seregelyes C, Mustardy L, Ayaydin F, Sass L, Kovacs L, Endre G, Lukacs N, Kovacs I, Vass I, Kiss GB, Horvath GV, Dudits D (2000) Nuclear localization of a hypoxia-inducible novel non-symbiotic hemoglobin in cultured alfalfa cells. FEBS Lett 485:127
Seregelyes C, Barna B, Hennig J, Konopka D, Pasternak TP, Lukacs N, Feher A, Horvath GV, Dudits D (2003) Phytoglobins can interfere with nitric oxide functions during plant growth and pathogenic responses: a transgenic approach. Plant Science 165:541–550
Smith SE, Read DJ (1997) Mycorrhizal symbiosis. Academic Press, London
Stougaard J, Sandal N, Gron A, Kuhle A, Marcker K (1987) 5′ analysis of the soybean leghaemoglobin lbc3 gene: regulatory elements required for promoter activity and organ specificity. EMBO J 6:3565–3569
Thorsteinsson MV, Bevan DR, Potts M, Dou Y, Eich RF, Hargrove MS, Gibson QH, Olson JS (1999) A cyanobacterial hemoglobin with unusual ligand binding kinetics and stability properties. Biochemistry 38:2117–2126
Trinchant JG, Rigaud J (1982) Nitrite and nitric oxide as inhibitors of nitrogenase from soybean bacteroid. Appl Environ Microbiol 44:1385–1388
Vierheilig H, Coughlan AP, Wyss U, Piche Y (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64:5004–5007
Vieweg MF, Frühling M, Quandt HJ, Heim U, Bäumlein H, Pühler A, Küster H, Perlick AM (2004) The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and non-legume plants. Mol Plant Microbe Interact 17:62–69
Watts RA, Hunt PW, Hvitved AN, Hargrove MS, Peacock WJ, Dennis ES (2001) A hemoglobin from plants homologous to truncated hemoglobins of microorganisms. Proc Natl Acad Sci USA 98:10119–10124
Wittenberg JB, Bolognesi M, Wittenberg BA, Guertin M (2002) Truncated hemoglobins: a new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. J Biol Chem 277:871–874
Wulf A, Manthey K, Doll J, Perlick AM, Linke B, Bekel T, Meyer F, Franken P, Kuster H, Krajinski F (2003) Transcriptional changes in response to arbuscular mycorrhiza development in the model plant Medicago truncatula. Mol Plant Microbe Interact 16:306–314
Acknowledgements
We thank the Deutsche Forschungsgemeinschaft for supporting this work in the frame of the SPP1084 (MolMyk) project Pe814/1-2 and Prof. Alfred Pühler for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vieweg, M.F., Hohnjec, N. & Küster, H. Two genes encoding different truncated hemoglobins are regulated during root nodule and arbuscular mycorrhiza symbioses of Medicago truncatula. Planta 220, 757–766 (2005). https://doi.org/10.1007/s00425-004-1397-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1397-0