Abstract
The Wilms' tumour gene, Wt1, encodes a zinc finger protein, which is mutated in a subset of paediatric renal carcinomas known as Wilms' tumours (nephroblastomas). Recent findings indicate that Wt1, beside its role in genitourinary development, is also necessary for normal vascularisation of the embryonic heart, and may even be involved in tumour angiogenesis. The original purpose of this study was to decipher potential downstream signalling pathways of Wt1 for blood vessel formation. We found that the Wt1(−KTS) protein, which functions as a transcription factor, stimulated the expression of cadherin 5 (CDH5, vascular endothelial (VE) cadherin) and other vascular genes, i.e. those encoding vascular endothelial growth factor receptors 1 and 2, and angiopoietin-2. Furthermore, an enhancer element was identified in the first intron of the CDH5 gene, which bound to the Wt1(−KTS) protein and was necessary for reporter gene activation by Wt1(−KTS) in transiently transfected cell lines. Wt1 and VE-cadherin proteins could be co-localised by double immunofluorescence staining in maturating glomeruli of embryonic murine kidneys. VE-cadherin transcripts were reduced in some but not all tissues of Wt1-deficient mouse embryos. These results indicate that Wt1 can stimulate vascular gene transcription. By demonstrating that Wt1(−KTS) protein trans-activates an enhancer element in the first intron we identified CDH5 as a novel target gene of Wt1. It is suggested that transcriptional activation of CDH5 by Wt1 fulfils regulatory functions during vascular development and kidney formation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wt1 was originally identified as a tumour suppressor gene by reason of loss-of-function mutations in a childhood malignancy of the kidneys known as nephroblastoma or Wilms' tumour [32]. Wilms' tumours can arise when pluripotent progenitor cells in the developing kidney continue to proliferate instead of differentiating to glomeruli and tubules [28, 32]. It is assumed that the Wt1 gene product enables cells to switch between an epithelial and mesenchymal state [2, 25, 26]. Failure of epithelial cell differentiation in the kidneys due to Wt1 inactivation is responsible for approximately 10% of Wilms' tumours. The Wt1 gene encodes a zinc finger protein that can function as transcription factor [33]. Alternative RNA splicing gives rise to the insertion of three additional amino acids (lysine, threonine, serine (KTS)) in the C-terminal zinc finger domain [13]. Molecules with the KTS tripeptide exhibit increased RNA-binding affinity and are presumably involved in post-transcriptional processes [21, 27].
Besides its role in tumourigenesis, Wt1 is also necessary for the development of certain organs. Mice with homozygous disruption of Wt1 (Wt1 −/−) are embryonic lethal due to heart failure resulting from myocardial growth arrest [20, 26]. Other abnormalities of Wt1-null mice include defects of the kidneys and gonads, mesothelial tissues, spleen, adrenal glands, retina and olfactory epithelium [14, 20, 26, 39, 41]. We have previously found that Wt1 is also required for blood vessel formation in the developing heart [40]. Coronary vascular cells are derived from the epicardium, which forms an epithelial sheet on the outer heart surface [31]. During cardiogenesis Wt1-positive epicardial cells undergo epithelial-to-mesenchymal transition (EMT), which permits them to become cardiovascular progenitor cells [11]. It has been shown recently that EMT does not occur in embryoid bodies of Wt1-knockout mice [25]. Furthermore, Wt1-deficient embryonic bodies expressed endothelial genes, i.e. Flk1/Kdr and cadherin 5 (Cdh5, vascular endothelial (VE)-cadherin), at a rather low level [25]. These findings raise the interesting possibility that Wt1 facilitates the differentiation of progenitor cells along the endothelial lineage through up-regulation of vascular growth factor receptors and endothelial cell adhesion molecules. Consistently, Wt1 was expressed de novo in resident vascular smooth muscle and endothelial cells after myocardial infarction in rats suggesting a role in neovascularisation of the ischemic heart [38]. Wt1 was recently also detected in the vascular endothelium of various tumours [34, 37]. While these observations provided first evidence for a link between Wt1 and tumour angiogenesis, little is known about the function of Wt1 in the vasculature of normal and neoplastic tissues. Important hints hereunto can be obtained from the knowledge of candidate target genes that are regulated by Wt1. As a step towards this issue, we explored vascular gene regulation by Wt1 in cells derived from osteosarcoma, a highly vascularised bone tumour with strong metastatic potential and the capacity of vascular mimicry [3, 7]. We report here that Wt1 stimulates indeed the expression of several vascular genes including CDH5, VEGF receptor 1 and 2 (Flt1 and Flk1), and angiopoietin-2 (angpt2). Furthermore, we identified an enhancer element in the first intron of the CDH5 gene, which is trans-activated by the Wt1(−KTS). It is proposed that activation of CDH5 and other vascular genes by Wt1 supports blood vessel assembly in the heart and may also promote tumour angiogenesis.
Materials and methods
Cell culture
The UB27 and UD28 lines, which express the Wt1(−KTS) and Wt1(+KTS) proteins under control of a tetracycline-regulated promoter, were the gift of Dr. Christoph Englert [8]. These cells are derived from the parental U-2OS osteosarcoma cell line (ATCC no. HTB-96) and were grown in DMEM nutrient (PAA Laboratories, Pasching, Austria) supplemented with 10% FCS (Biochrom KG, Berlin, Germany), 100 IU/ml penicillin (Invitrogen GmbH, Karlsruhe, Germany), 100 μg/ml streptomycin (Invitrogen), 1 μg/ml tetracycline, and 1 μg/ml puromycine. Stimulation of Wt1 expression was achieved by incubation of UB27 and UD28 cells in tetracycline-free DMEM as described [8, 40]. Human embryonic kidney (HEK) 293 cells (catalogue no. ACC 305) were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) and kept in supplemented DMEM.
Cell transfections and reporter assays
UB27 and UD28 cells were expanded to approximately 50% confluence in 24-well tissue culture plates. Tetracycline was removed from the medium 24 h before the transfections in the moiety of cells to stimulate Wt1(−KTS) and Wt1(+KTS) expression, respectively [8]. Nine hundred nanograms of firefly luciferase constructs harbouring regulatory sequences of the VE-cadherin gene and 100 ng of a renilla luciferase plasmid were transiently co-transfected with the Fugene6® reagent (3 μl per well) according to the manufacturer's protocol (Roche Diagnostics, Mannheim, Germany). Transfection of the empty pGL3-basic reporter vector served as negative control. The transfected cells were incubated for 48 h with and without tetracycline (1 μg/ml) prior to lysis in Reporter Lysis Buffer (Promega, Mannheim, Germany). Luciferase activities were measured in a luminometer (Microlite TLX1, MGM Instruments, Hamden, CT) as described [5, 17]. Data are presented as relative light units normalised to renilla luciferase activities. Transfection of HEK 293 cells was performed as described [17].
Reverse transcription real-time PCR
Total RNA was isolated from cultured cells and tissues with the Trizol reagent (Invitrogen) according to the manufacturer's protocol. First-strand cDNA synthesis was performed with 2 μg of total RNA using oligo(dT) primers and superscript II reverse transcriptase (Invitrogen). Forty nanograms of cDNA were taken for real-time PCR amplification with SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, USA). The PCR reactions were carried out on a StepOnePlus thermocycler (Applied Biosystems, Darmstadt, Germany) as follows (45 cycles): DNA denaturation (15 s) at 94°C, primer annealing (15 s) and extension (60 s) of double-stranded DNA at 60°C, detection of SYBR® Green fluorescence at 77°C (30 s). The PCR primers used for the amplification reactions are listed in Table 1. The Ct values for the genes of interest were subtracted by the Ct values for β-actin to obtain delta Ct values. Differences in transcript levels were calculated according to the equation 2deltadeltaCt.
Plasmids
A DNA sequence extending from −2,790 to +99 base pairs (bp) relative to the transcription start site in the human cadherin 5 (CDH5, VE-cadherin) gene (National Center for Biotechnology Information (NCBI) accession no. NT_010498) was amplified by PCR using a bacterial artificial chromosome (imaGenes, Berlin, Germany, clone RP11-93H5) as template. The PCR product was ligated into the KpnI restriction site of the pGL3-basic reporter plasmid (Promega). This construct was designated pGL3bVEcadprom. Likewise, various regions of the first intron of the CDH5 gene were cloned and ligated downstream of the promoter into the SacI/HindIII sites of pGL3bVEcadprom. Site-directed base-pair mutations were introduced in the first intron of the VE-cadherin gene by PCR as described elsewhere [5, 17]. All constructs were analysed by automated DNA sequencing.
Immunoprecipitation and SDS-PAGE
UB27 osteosarcoma cells were kept for 72 h either in the absence or presence of tetracycline (1 μg/ml) to stimulate or inhibit Wt1 expression. Cell lysates were prepared in RIPA buffer (1× PBS, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 0.1 mg/ml PMSF, 0.1 IU/ml aprotinin, 1 mM sodium orthovanadate) as described elsewhere [5, 17]. The lysates (250 μg protein each) were pre-cleared for 1 h at 4°C with Protein A Sepharose™ CL-4B (Amersham Bioscience, Uppsala, Sweden), and the beads were pelleted by centrifugation at 1,000×g (5 min, 4°C). Prior to immunoprecipitation, aliquots of the supernatants were removed for immunoblotting with anti-actin antibody (see below). One microgram of mouse monoclonal anti-VE-cadherin antibody (C-19, catalogue no. sc-9989, Santa Cruz Biotechnology) was added to the supernatants (1 ml), which were then incubated for 2 h at 4°C. Control experiments were performed with mouse IgG instead of primary antibody. After addition of pre-washed Protein A Sepharose beads (50 μl each) the tubes were shaken overnight at 4°C on a rocker platform. The next morning, the beads were centrifuged at 1,000×g for 5 min (4°C), washed four times with RIPA buffer, and after the final wash resuspended in Laemmli buffer and heated to 95°C for 3 min. Following centrifugation (1,000×g for 5 min) the supernatants were loaded and separated on a 7.5% polyacrylamide gel. The proteins were transferred onto polyvinylidene difluoride membranes (Amersham Pharmacia Biotech, Freiburg, Germany) with the use of a semidry blotting apparatus (BioRad, München, Germany). Non-specific binding activity was reduced by incubating the membranes for 60 min at room temperature in PBS, 5% non-fat milk (Roth, Karlsruhe, Germany), 0.05% Tween-20 (Serva, Heidelberg, Germany). Incubation with anti-VE-cadherin antibody from goat (catalogue no. sc-6458, diluted 1:200 in PBS, 2.5% non-fat milk, 0.05% Tween-20) was performed overnight at 4°C. The primary antibody was detected with a peroxidase-coupled donkey anti-goat-IgG (catalogue no. sc-2020, Santa Cruz Biotechnology, 1:1,000 dilution), and the reaction products were visualised with the enhanced chemiluminescence system (Amersham Pharmacia Biotech.). A goat polyclonal anti-β-actin antibody (catalogue no. sc-1615, Santa Cruz Biotechnology, diluted 1:500) was applied for immunoblotting of aliquots that had been collected before immunoprecipitation to assess equal protein content of the samples.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described in detail elsewhere [24]. In brief, UB27 cells were grown for 48 h either in the presence or absence of tetracycline (1 μg/ml) to inhibit or stimulate expression of Wt1(−KTS). Cross-linking of DNA-bound protein was accomplished by incubating the cells for 10 min in a 1% formaldehyde solution. The cell lysates were ultrasonicated to obtain an average DNA fragment size of 200–1,000 bp. Immunoprecipitations were performed with the following antibodies (1 μg each) overnight at 4°C: rabbit polyclonal anti-Wt1 antibody (sc-192, Santa Cruz Biotechnology), rabbit polyclonal anti-acetylated histone 3 antibody (cat. no. 06–599, Upstate-Millipore, Darmstadt, Germany). Incubation with normal rabbit IgG (Cell Signaling, Boston, USA) served as a negative control. The antibody-bound proteins were precipitated for 1 h at 4°C with DNA-blocked protein G-agarose (Millipore, Darmstadt, Germany). After several washes in low- and high-salt buffer, the DNA was eluted from the agarose beads, extracted by phenol:chloroform treatment, and precipitated in 100% ethanol. The DNA pellet was resuspended in 30 μl of ddH2O and 2 μl of a 1:40 dilution was taken for quantitative PCR amplification of the first intron of the human CDH5 gene using the following primers: 5′-GAGCTGAGCCGATCTCAC-3′ (forward), 5′-GCCGGTCATGTGGTGAAA-3′ (reverse) on a StepOnePlus thermocycler (Applied Biosystems). Amplification of genomic β-actin DNA with the following primers 5′-GTGAGTGGCCCGCTACCT-3′ (forward) and 5′-CCTTGTCACACGAGCCAG-3′ (reverse) served as a control.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were performed with GST-purified recombinant Wt1 protein as described in detail elsewhere [5, 17, 40]. In brief, a double-stranded oligonucleotide (5′-GTCTGTATGCTCCCACAGCCTCCTCGAT-3′) was selected on the basis of a predicted Wt1 binding site in the first intron of the CDH5 gene and end-labelled with [γ-32P]ATP (7,000 Ci/mmol, catalogue no. 35020, ICN Biochemicals, Eschwege, Germany). Likewise, a second double-stranded oligonucleotide with mutation of the core Wt1 binding motif (underlined) was used for the binding reactions: 5′-GTCTGTATGCGAATTCAGCCTCCTCGAT-3′. Competition experiments were performed with an unlabelled oligonucleotide carrying the previously identified Wt1 binding site [40] from the human NTRK2 gene (NCBI accession no. AL390777): 5′-TGTGAACTCCCACATGCTGCTG-3′. The binding reactions were separated on a non-denaturing 4% polyacrylamide gel. The dried gels (gel dryer, model 583, BioRad) were exposed on X-ray film for 6 h and developed.
Immunohistochemistry
Morphological studies were performed as described previously [5, 17, 19, 38, 40]. Staged embryos were fixed overnight in paraformaldehyde (3% in PBS) at 4°C and frozen in tissue-Tek O.C.T. compound (Sakura Finetek, Zoeterwoude, Netherlands). Ten-micrometre cryostat sections were permeabilised with 0.1% Triton X-100 in PBS and blocked for 5 min at room temperature in serum-free DakoCytomation protein block (catalogue no. X0909, Dako, Hamburg, Germany). The following primary antibodies were diluted 1:50 in ready-to-use antibody diluent (Zymed Laboratories Inc., Berlin, Germany): rabbit polyclonal anti-Wt1 antibody (C-19, catalogue no. sc-846, Santa Cruz Biotechnology) and goat polyclonal anti-VE-cadherin antibody (C-19, catalogue no. sc-6458, Santa Cruz Biotechnology). The reaction products were visualised by incubation (1.5 h at room temperature) with Cy3 (Wt1) and Cy2 (VE-cadherin) conjugates. Nuclei were sometimes counterstained with 4′,6-diamidino-2-phenylindole [5, 17]. Tissue sections were viewed under an epifluorescence microscope (Axiovert S100, Zeiss, Jena, Germany), which was connected to a digital camera (Spot RT Slider, Diagnostic Instruments, Sterling Heights, USA) equipped with the Metamorph V4.1.2 software (Molecular Devices Inc. Downington, USA).
Chemicals
If not otherwise indicated, all chemicals were obtained from Sigma–Aldrich, Hamburg, Germany.
Statistics
Values are presented as means ± SD. ANOVA with Bonferroni test as post hoc calculation and Student's t test were performed as indicated to reveal statistical significances. Values of P < 0.05 were considered statistically significant.
Results
Wt1 activates expression of vascular genes in osteosarcoma cells
Genetically modified cells are valuable tools to identify transcription factor target genes. We took advantage of a previously engineered osteosarcoma cell line (U-2OS) harbouring a Wt1 expression cassette under control of the tetracycline repressor [8] to identify putative downstream effectors of the Wt1 protein. Osteosarcoma is a highly vascularised bone tumour whose cells are capable of vascular mimicry, e.g. forming blood vessel-like tubular networks with concomitant expression of vascular cell markers [3, 7]. Thus, one can assume that osteosarcoma-derived cell lines are endowed with a pro-angiogenic programme, which makes them suitable for studying vascular gene regulation. The two clonal osteosarcoma cell lines that we used differ in their expression either of the Wt1(−KTS) (UB27 cells) or the Wt1(+KTS) (UD28 cells) protein [8]. Wt1 isoforms without the lysine–threonine–serine (KTS) splice insertion in the zinc finger domain can function as transcription factors, whereas Wt1(+KTS) molecules have a presumed role in mRNA processing [21, 27]. Total RNA was obtained from four independent experiments, and a quantitative reverse transcription (RT)-PCR technique was applied to determine relative transcript levels of genes involved in blood vessel formation. Remarkably, the mRNAs of CDH5, VEGFR1 (Flt1), VEGFR2 (Flk1) and angiopoietin-2 (angpt2) were enhanced significantly upon induction of the Wt1(−KTS) protein in UB27 cells (Fig. 1). In contrast, no significant differences in angiopoietin-1 (angpt1) and Tie2 mRNAs were observed between cells with high and low Wt1(−KTS) expression (Fig. 1). No changes in vascular gene transcripts were detected upon induction of Wt1(+KTS) protein in UD28 cells (Fig. 1).
Messenger RNA levels of vascular genes in osteosarcoma cells. Transcripts of VE-cadherin (VE-cadh.), vascular endothelial growth factor receptors (VEGFR) 1 and 2, angiopoietins (Angpt) 1 and 2, and Tie2 were measured by real-time RT-PCR in stable osteosarcoma cell lines with inducible expression of Wt1(−KTS; clone UB27) and Wt1(+KTS) proteins (clone UD28), respectively [8]. Relative mRNA levels were determined on the basis of deltaCt values. Shown are the fold differences of gene-specific mRNAs normalised to Gapdh mRNA in cells with stimulated vs. repressed Wt1(−KTS) (grey bars) and Wt1(+KTS) (black bars), respectively. Values are means ± S.D. of four independent experiments, each. Statistical significances are indicated (asterisk P < 0.001, Student's t test)
Expression of Wt1(−KTS) and VE-cadherin is correlated in osteosarcoma cells
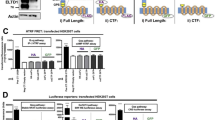
Since Wt1 exerted by far the strongest effect on VE-cadherin mRNA, we studied the interaction of these molecules in more detail. Omission of tetracycline from the culture medium of UB27 cells for 24, 48 and 72 h caused an increase of Wt1(−KTS) protein that was paralleled by a significant rise in VE-cadherin mRNA (n = 4, Fig. 2a). For comparison, a similar Wt1(+KTS) protein content in UD28 cells had no significant effect on VE-cadherin transcripts (Fig. 2a). Immunoprecipitation followed by SDS-PAGE was performed to verify the ≈130 kDa VE-cadherin protein in UB27 cells with high Wt1(−KTS) expression (Fig. 2b). Appropriate negative controls were performed with normal mouse serum instead of anti-VE-cadherin antibody (Fig. 2b). VE-cadherin could not be immunoprecipitated in uninduced UB27 cells with low Wt1(−KTS) protein (Fig. 2b). These findings indicate that VE-cadherin gene expression in osteosarcoma cells is positively correlated with Wt1(−KTS), but not with Wt1(+KTS) protein. Considering the previously recognised functions of the different Wt1 proteins [21, 27] our observations suggest a transcriptional mechanism of CDH5 gene regulation by Wt1.
VE-cadherin in osteosarcoma cells with inducible expression of different Wt1 proteins. Panel a shows the time course of VE-cadherin transcripts upon induction of the Wt1(−KTS) and Wt1(+KTS) splice variants in two clonal cell lines, UB27 and UD28, respectively. Representative immunoblots of Wt1 and β-actin proteins are depicted below. Note, that VE-cadherin transcripts were increased only in Wt1(−KTS) but not in Wt1(+KTS) expressing cells. Values are means ± S.D. of n = 4 experiments. Statistical significance vs. control (0 h) is marked by asterisks (asterisk P < 0.001, ANOVA with Bonferroni as post hoc test). The ≈130 kDa VE-cadherin protein (VE-cad) was detected in stimulated UB27 cells by immunoprecipitation (IP) and subsequent immunoblot analysis (lane 3 in panel b). Appropriate control experiments were performed with the use of mouse IgG instead of anti-VE-cadherin antibody for immunoprecipitation (lanes 2 and 4 in panel b). VE-cadherin could not be clearly verified in UB27 cells with low Wt1(−KTS) protein (lane 1 in panel b). Anti-β-actin antibody was applied to assess equal protein content in aliquots that had been removed before immunoprecipitation (panel b, lower part)
Wt1(−KTS) activates an enhancer element in the first intron of the CDH5 gene
To validate the relationship between Wt1 and VE-cadherin and to analyse the underlying regulatory mechanism, we investigated whether Wt1 can activate the promoter of the CDH5 gene directly. For this purpose, a firefly luciferase construct carrying a region between −2,790 and +99 bp relative to the transcription start site in the human CDH5 gene was transiently co-transfected into UB27 and UD28 cells along with a renilla luciferase plasmid for normalisation of transfection efficiencies. After transfection, the cells were grown for 48 h either in the presence or absence of tetracycline to adjust low and high Wt1 protein levels, respectively. Stimulation of Wt1(−KTS) in UB27 cells nearly doubled the activity of the CDH5 promoter, whereas Wt1(+KTS) in UD28 cells had no significant effect (n = 5, Fig. 3b). The relatively weak stimulatory effect of Wt1(−KTS) on reporter gene activity compared to its strong impact on VE-cadherin mRNA levels in UB27 cells (Fig. 1b) let us assume that additional, Wt1-sensitive cis-regulatory elements may exist outside of the promoter in the CDH5 gene. The CDH5 gene is organised such that the first intron, which consists of approximately 12 kb, is contained in the 5′ untranslated region (UTR), and the protein coding sequence starts in exon 2 [23] (Fig. 3a). Notably, the first intron has previously been found to be important for high-level expression of VE-cadherin [15]. To test for the presence of Wt1 inducible enhancer elements in the first intron of the CDH5 gene, reporter constructs were generated in which we combined the 2,790 bp 5′-upstream region with different parts of the first intron each comprising approximately 4,000 bp. While two of these constructs had no effect on the CDH5 promoter in Wt1(−KTS) expressing UB27 cells, a sequence extending from +4,530 to +8,811 bp significantly enhanced CDH5 promoter activity (n = 5, Fig. 3b). The same reporter plasmid did not respond to changes of Wt1(+KTS) protein in UD28 cells (Fig. 3b), but was stimulated approximately sixfold in HEK 293 cells upon co-transfection of a Wt1(−KTS) expression construct (n = 5, Fig. 3c). A reporter plasmid carrying the CDH5 promoter (from −2,790 to +99 bp) alone, e.g. without additional intron 1 sequence, was insensitive to Wt1(−KTS) in transiently transfected HEK 293 cells (Fig. 3c). We also tested whether the identified intron sequence in the CDH5 gene conferred Wt1 inducibility to a heterologous promoter. For this purpose, the respective region was ligated downsteam of the SV40 promoter in the pGL3promoter plasmid. While Wt1(−KTS) exerted no effect on the activity of the empty pGL3promoter vector in UB27 cells, a more than fourfold activation was elicited upon inclusion of the intron one sequence (data not shown). To identify the relevant cis-element in the first intron of the CDH5 gene, the ≈4.3-kb sequence was curtailed down to a 569-bp fragment (+4,530 to +5,098 bp), which was as efficient as the ≈4.3-kb piece in terms of being stimulated by Wt1(−KTS) protein (Fig. 4a). For comparison, an adjacent 522-bp sequence (+5,099 to +5,620 bp) did not significantly (P > 0.05) enhance CDH5 promoter activity in the presence of Wt1(−KTS) (Fig. 4a). Mutation of a predicted Wt1(−KTS) binding site in the 569-bp intron fragment (+4,530 to +5,098 bp) abolished the stimulatory effect of Wt1(−KTS) protein (Fig. 4a). EMSAs were performed to investigate whether the identified element could interact with Wt1 proteins (Fig. 4b). Specific retardation bands indicating the binding of recombinant Wt1(−KTS) protein were obtained with a 28-bp oligonucleotide carrying a predicted Wt1 consensus element. Interaction of Wt1(−KTS) with this sequence could be competed with unlabeled DNA including the previously identified Wt1(−KTS) binding site in the promoter of the NTRK2 gene [40]. Introducing a mutation in the oligonucleotide abolished its binding affinity for the Wt1(−KTS) protein (Fig. 4b). Notably, the Wt1(+KTS) molecule did not interact with the enhancer element in the first intron of the CDH5 gene (Fig. 4b).
Regulation of different VE-cadherin reporter constructs by Wt1. a Scheme of the 5`-flanking region of the human VE-cadherin gene. The transcription start site is marked by +1 and the start codon (ATG) in exon 2 is indicated. The position of nucleotides is given relative to the transcription start according to the reference sequence (NCBI AC012325). b Effect of Wt1(−KTS) and Wt1(+KTS) proteins on transcriptional activity of the VE-cadherin promoter alone and in combination with different parts of the first intron. Reporter constructs in pGL3-basic containing the human VE-cadherin promoter (from −2,790 to +99 bp) with and without intron 1 sequence were transiently transfected in UB27 and UD28 cells. The cells were grown either in the presence or absence of tetracycline to suppress (open bars) or stimulate (black and grey bars) Wt1 expression, respectively. c Reporter gene activity in HEK 293 cells co-transfected with empty vector (open bars) or expression constructs for Wt1(−KTS) and Wt1(+KTS) proteins (black and grey bars). A renilla luciferase vector was used for the normalisation of transfection efficiencies. All values are means ± SD of five independent experiments, each performed in triplicate. Significant differences vs. empty pGL3 reporter vector (asterisk P < 0.01, ANOVA with Bonferroni as post hoc test) and between two reporter constructs are indicated (number sign P < 0.01, Student's t test)
Identification of a Wt1(−KTS) activated cis-element in the first intron of the VE-cadherin gene. a Normalised luciferase activities of the indicated reporter constructs in UB27 (black bars) and UD28 cells (grey bars) expressing Wt1(−KTS) and Wt1(+KTS) proteins, respectively. Cells were grown in the presence of tetracycline to keep Wt1 expression suppressed (open bars). The Wt1(−KTS) sensitive region could be mapped to a 569 bp fragment extending from +4530 bp to +5098 bp relative to the transcription start site in the VE-cadherin gene. Mutation of a predicted Wt1 binding motif resulted in loss of stimulation by the Wt1(−KTS) molecule. Values represent means ± SD of five independent experiments, each performed in triplicate. Asterisks indicate significant differences vs. the effect of Wt1(−KTS) on a reporter construct containing the CDH5 promoter alone. Asterisk P < 0.01, ANOVA with Bonferroni as post hoc test. b Electrophoretic mobility shift assay demonstrating interaction of recombinant Wt1(−KTS) protein with a 28 bp oligonucleotide (oligo +5,496 to +5,524 bp) in the first intron of the human VE-cadherin gene (lane 3). The arrowhead points to the retardation band produced by interaction of Wt1(−KTS) protein with the oligonucleotide. Wt1(+KTS) protein, which fulfils a presumed role in RNA processing [21], exhibited much lower affinity for the oligonucleotide (lane 2). Interaction could be competed with unlabelled oligonucleotide containing the previously identified Wt1(−KTS) binding motif from the promoter of the human NTRK2/TrkB gene [40] (lanes 4–8). A mutation in the VE-cadherin oligoligonucleotide diminished interaction with Wt1(−KTS) protein (lanes 9–11). Note, that the same mutation prevented reporter gene activation by Wt1(−KTS) in transient transfection experiments (panel a). An oligonucleotide with the Wt1(−KTS) binding motif in the NTRK2/TrkB gene promoter was used as positive control (lanes 12–14)
ChIP was applied to explore whether Wt1 interacts with the CDH5 enhancer also in a natural chromosomal configuration. Repeated analyses were carried out with chromatin prepared from UB27 cells with high and low Wt1(−KTS) protein content, respectively. An approximately fivefold enrichment of genomic DNA from the first intron of the CDH5 gene was obtained with anti-Wt1 antibody vs. normal rabbit serum (Fig. 5a). CDH5 intron one sequence could not be enriched from UB27 cells with low Wt1 protein content (Fig. 5a). Likewise, anti-Wt1 antibody did not immunoprecipitate genomic DNA sequence from the human β-actin gene (data not shown).
Representative chromatin immunoprecipitation (ChIP) assay with anti-Wt1 antibody demonstrating an approx. Fivefold enrichment of DNA in the first intron of the CDH5 gene from UB27 cells with high Wt1 protein (panel a). VE-cadherin DNA could not be enriched from UB27 cells with low Wt1 expression. Normal rabbit serum and anti-acetylated histone 3 antibody were used as negative and positive controls, respectively. Panel b shows enhancer sequence from the first intron of the CDH5 gene. The Wt1-responsive element is marked by the box, the oligonucleotides used for PCR amplification are underlined
Wt1 and VE-cadherin are co-expressed in glomeruli of the developing kidney
Wt1 expression is particularly high in the genitourinary system of embryos [1, 30], and inactivation of Wt1 in mice caused a failure of normal formation of the kidneys and gonads [20]. We used a double immunofluorescent labelling technique to analyse the distribution of Wt1 and VE-cadherin expressing cells in mouse embryonic kidneys. After treatment with primary antibodies the frozen tissue sections were incubated with Cy3-coupled secondary antibody to visualise Wt1 protein (red fluorescence) and with a Cy2-conjugate to detect VE-cadherin-expressing cells (green fluorescence). In agreement with previous studies [1, 30] we localised Wt1 protein to the condensing mesenchyme and immature glomeruli of E12.5 foetal kidneys (Fig. 6a). No VE-cadherin expressing renal cells could be identified in this developmental stage (Fig. 6b). However, VE-cadherin was clearly visible in developing glomeruli of E19.5 mice (Fig. 6e and h). Notably, VE-cadherin positive glomeruli were distributed mainly in the deeper zones of embryonic kidneys reflecting a more advanced developmental state than superficial glomeruli (Fig. 6d–f). High power magnification revealed co-expression of Wt1 and VE-cadherin proteins in the developing podoycytes. Expectedly, Wt1 was located in the cell nuclei whereas VE-cadherin was associated with the plasma membranes (Fig. 6 g–i). In addition to the developing kidneys, an overlapping pattern of Wt1 and VE-cadherin was also seen in the epicardium of embryonic mice (data not shown). While VE-cadherin was present in the developing vasculature, we observed no general vascular expression of Wt1 protein in murine foetal tissues (data not shown).
Representative VE-cadherin expression analysis in mouse embryonic kidney. Double-immunofluorescence labelling of Wt1 (red) and VE-cadherin (green) proteins in frozen sections from murine kidneys at embryonic days E12.5 and E19.5. Cells with co-expression of nuclear Wt1 and membrane-bound VE-cadherin can be clearly identified in the high power images (g–i). Note, that Wt1 is strongly expressed in comma-shaped bodies at E12.5 (arrowheads in a), whereas VE-cadherin is absent from embryonic kidneys at this early stage (b). Co-localization of Wt1 and VE-cadherin in developing glomeruli in deeper zones of embryonic kidneys at E19.5 is clearly visible (d–i). Scale bars indicate 200 μm (a–f) and 10 μm (g–i), respectively. No specific fluorescence signal was obtained with the use of normal serum instead of primary antibody (c). Nuclei in c were stained with Dapi
VE-cadherin mRNA is reduced in the livers and hearts of Wt1-deficient mouse embryos
To examine whether Wt1 is necessary for normal expression of VE-cadherin in vivo, transcript levels were measured by real-time RT-PCR in hearts, livers and urogenital ridges [18] of wild-type (Wt1 +/+), heterozygous (Wt1 +/−) and Wt1-deficient (Wt1 −/−) mouse embryos at E12.5 (Fig. 7). Livers, in addition to urogenital ridges and hearts were chosen, because we have previously found that Wt1 is contained in CD117+ haematopoietic cells from mouse foetal liver [5, 17]. Furthermore, VE-cadherin is transiently expressed by foetal liver haematopoietic stem cells [16]. Messenger RNA measurements were not performed at later stages because most Wt1 −/− embryos fail to develop beyond 13 days post conception [20]. VE-cadherin mRNA levels were reduced by approximately 42% and 40% in the livers and hearts, respectively, of Wt1-deficient embryos compared to their wild-type (Wt1 +/+) littermates (Fig. 7). No differences in VE-cadherin transcripts were observed in the urogenital ridges of wild-type and Wt1-deficient embryos (Fig. 7). VE-cadherin transcripts were not significantly different in all tissues of wild-type and heterozygous (Wt1 +/−) embryos (Fig. 7).
VE-cadherin mRNA in different tissues of wild-type (wt, open bars), heterozygous (hz, grey bars) and homozygous Wt1-deficient (ko, black bars) embryos at E12.5. VE-cadherin transcripts were measured by real-time RT-PCR and normalised to Gapdh transcripts. Values are means ± SD, n = 5 each. Asterisks indicate significant differences vs. wild-type embryos. One asterisk, P < 0.05; two asterisks, P < 0.01, ANOVA with Bonferroni as post hoc test; n.s., not significant
Discussion
The Wilms' tumour gene, Wt1, encodes zinc finger proteins that can function as tumour suppressors and also play important roles in the development of the genitourinary system and several other tissues [32]. The purpose of this study was to identify novel putative downstream target genes of Wt1, particularly with regard to its proposed contribution to blood vessel formation [34, 37, 38, 40]. We found that Wt1 indeed stimulates the expression of vascular genes in cells derived from osteosarcoma, an aggressive bone tumour, which is capable of vasculogenic mimicry by forming a tubular network reminiscent of blood vessels [7, 9]. The observed effects were caused by the Wt1(−KTS) molecule only, but not by the Wt1(+KTS) protein suggesting that activation of a vascular gene programme was related to the transcription regulatory function of Wt1 (Figs. 1 and 2).
Furthermore, the activity of a CDH5 promoter reporter construct was markedly enhanced by the Wt1(−KTS) isoform in osteosarcoma cells (Fig. 3). VE-cadherin (CD144) is a member of the cadherin family of adhesion proteins that associates with intracellular catenins and allows contact formation among vascular endothelial cells [35]. VE-cadherin is necessary for blood vessel assembly [12, 36] and plays a pivotal role in the control of vascular permeability [10]. Previous findings indicated that the promoter of the cadherin 5 gene (CDH5, VE-cadherin) confers tissue-specific (trans)gene expression to the vasculature [29], but robust expression of VE-cadherin required interaction of the 5′-flanking region with intron one sequence [15]. In this study, we identified an enhancer element in the first intron of the CDH5 gene, which binds to the Wt1(−KTS) molecule and is required for trans-activation by Wt1(−KTS) (Figs. 3, 4, 5). Remarkably, the core binding motif (-CTCCCACA-) of the identified sequence is identical to the previously discovered Wt1-sensitive cis-regulatory element in the promoter of the NTRK2 gene, which encodes the TrkB neurotrophin receptor [40]. Activation of the TrkB receptor by its ligand, brain-derived neurotrophic factor, is a critical mechanism during blood vessel formation in the developing heart [39]. Thus, one can assume that the Wt1(−KTS) protein interacts with cis-elements that are conserved in the regulatory regions of genes involved in blood vessel configuration.
It is important to note that Wt1 is expressed in a rather restricted fashion in the developing organism including the kidneys and blood vessels of embryonic hearts [1, 40]. However, Wt1 is absent from the vasculature of most organs, and Wt1-depleted embryos exhibit no obvious vascular phenotype [20] except for a reduced blood vessel density in their hearts [40]. These observations indicate that the contribution of Wt1 to vascular organisation is confined to distinct tissues, i.e. the myocardium and possibly certain tumours [34, 37, 38, 40]. Blood vessel formation in the heart relies on a special process of EMT, which enables epicardial cells to invade the myocardium and eventually become vascular smooth muscle and endothelial cells [11]. As shown recently, Wt1 mediates EMT in epicardial cells and during embryonic stem cell differentiation, and a failure of epicardial cells to undergo this transition likely contributes to the vascular defects in Wt1-deficient hearts [25, 40]. Remarkably, the transcription factor Snail (Snai1), which is a molecular downstream target of Wt1 in EMT [25], has previously been identified as direct regulator of the CDH5 gene [22]. Furthermore, VE-cadherin transcripts were significantly reduced in Wt1-knockout compared to wild-type embryoid bodies indicating that Wt1 is necessary for normal expression of VE-cadherin during early embryogenesis [25]. The importance of Wt1 for CDH5 gene regulation in vivo is underscored by the lower VE-cadherin mRNA levels in the hearts and livers of Wt1-deficient murine embryos compared to their wild-type and heterozygous (Wt1 +/−) littermates (Fig. 7). The latter observation may reflect a reduction of VE-cadherin transcripts in haematopoietic progenitor cells [16], which normally also express Wt1 [17].
Another novel observation of this study is the robust expression of VE-cadherin in embryonic kidneys of mice (Fig. 6). Development of the mammalian kidney depends on the interaction of the ureteric bud, an outgrowth of the primary nephrogenic duct, with the surrounding metanephric mesenchyme [6]. Mesenchyme-to-epithelial conversion in vicinity of the newly formed ureteric bud tips is among the early events in nephrogenesis. Absence from embryonic mouse kidneys at E12.5 indicates that VE-cadherin is likely dispensable for the initial induction of the metanephric mesenchyme. Accordingly, no differences in VE-cadherin mRNA were found in the urogenital ridges of wild-type and Wt1-deficient embryos at E12.5 (Fig. 7). However, cellular co-expression with Wt1 suggests a possible role for VE-cadherin in the formation of the glomerular podocytes (Fig. 6). Podocytes are epithelial cells that cover the fenestrated capillaries of renal glomeruli and contribute to the kidney filtration barrier formed by the so-called “slit diaphragm” between their foot processes. Interestingly, a comparative promoter analysis identified VE-cadherin as novel slit diaphragm molecule that was co-regulated with nephrin and ZO-1, two other major components of the renal filtration barrier, in human glomerular disease [4]. Thus our findings raise the possibility that transcriptional activation of the CDH5 gene by Wt1 contributes to intercellular contact formation between glomerular podocytes.
In summary, the results of this study indicate that Wt1 is capable of activating vascular gene transcription. Furthermore, we identified the gene encoding VE-cadherin (cadherin 5, CDH5) as a novel candidate target of Wt1. Absence from vascular endothelial cells in most tissues excludes Wt1 as a general regulator of VE-cadherin in the vasculature. Instead, stimulation of VE-cadherin expression by Wt1 may rather become relevant during blood vessel assembly in the embryonic heart and possibly play a role in podocyte maturation in the developing kidney.
References
Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JB (1993) The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech Dev 40:85–97
Burwell EA, McCarty GP, Simpson LA, Thompson KA, Loeb DM (2007) Isoforms of Wilms' tumor suppressor gene (WT1) have distinct effects on mammary epithelial cells. Oncogene 26(23):3423–3430
Cai XS, Jia YW, Mei J, Tang RY (2004) Tumor blood vessels formation in osteosarcoma: vasculogenesis mimicry. Chin Med J (Engl) 117(1):94–98
Cohen CD, Klingenhoff A, Boucherot A, Nitsche A, Henger A, Brunner B, Schmid H, Merkle M, Saleem MA, Koller KP, Werner T, Gröne HJ, Nelson PJ, Kretzler M (2006) Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins. Proc Natl Acad Sci USA 103(15):5682–5687
Dame C, Kirschner KM, Bartz KV, Wallach T, Hussels CS, Scholz H (2006) The Wilms’ tumor suppressor, Wt1, is a transcriptional activator of the erythropoietin gene. Blood 107(11):4282–4290
Dressler GR (2006) The cellular basis of kidney development. Annu Rev Cell Dev Biol 22:509–529
DuBois SG, Marina N, Glade-Bender J (2010) Angiogenesis and vascular targeting in Ewing sarcoma: a review of preclinical and clinical data. Cancer 116(3):749–757
Englert C, Hou X, Maheswaran S, Bennett P, Ngwu C, Re GG, Garvin AJ, Rosner MR, Haber DA (1995) WT1 suppresses synthesis of the epidermal growth factor receptor and induces apoptosis. EMBO J 14(19):4662–4675
Folberg R, Maniotis AJ (2004) Vasculogenic mimicry. APMIS 112:508–525
Gavard J (2009) Breaking the VE-cadherin bonds. FEBS Lett 583(1):1–6
Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE (1998) Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res 82:1043–1052
Gory-Fauré S, Prandini MH, Pointú H, Roullot V, Pignot-Paintrand I, Vernet M, Huber P (1999) Role of vascular endothelial-cadherin in vascular morphogenesis. Development 126:2093–2102
Haber DA, Sohn RL, Buckler AJ, Pelletier J, Call KM, Housman DE (1991) Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc Natl Acad Sci USA 88:9618–9622
Herzer U, Crocoll A, Barton D, Howells N, Englert C (1999) The Wilms tumor suppressor gene wt1 is required for development of the spleen. Curr Biol 9(15):837–840
Hisatsune H, Matsumura K, Ogawa M, Uemura A, Kondo N, Yamashita JK, Katsuta H, Nishikawa S, Chiba T, Nishikawa S (2005) High level of endothelial cell-specific gene expression by a combination of the 5′ flanking region and the 5′ half of the first intron of the VE-cadherin gene. Blood 105(12):4657–4663
Kim I, Yilmaz OH, Morrison SJ (2005) CD144 (VE-cadherin) is transiently expressed by fetal liver hematopoietic stem cells. Blood 106(3):903–905
Kirschner KM, Hagen P, Hussels CS, Ballmaier M, Scholz H, Dame C (2008) The Wilms’ tumor suppressor Wt1 activates transcription of the erythropoietin receptor in hematopoietic progenitor cells. FASEB J 22(8):2690–2701
Klattig J, Sierig A, Kruspe D, Makki MS, Englert C (2007) WT1-mediated gene regulation in early urogenital ridge development. Sex Dev 1:238–254
Kirschner KM, Wagner N, Wagner KD, Wellmann S, Scholz H (2006) The Wilms tumor suppressor Wt1 promotes cell adhesion through transcriptional activation of the alpha4integrin gene. J Biol Chem 281(42):31930–31939
Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R (1993) WT-1 is required for early kidney development. Cell 74:679–691
Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND (1995) Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 81:391–401
Lopez D, Niu G, Huber P, Carter WB (2009) Tumor-induced upregulation of Twist, Snail, and Slug represses the activity of the human VE-cadherin promoter. Arch Biochem Biophys 482:77–82
Ludwig D, Lorenz J, Dejana E, Bohlen P, Hicklin DJ, Witte L, Pytowski B (2000) cDNA cloning, chromosomal mapping, and expression analysis of human VE-Cadherin-2. Mamm Genome 11(11):1030–1033
Martens LK, Kirschner KM, Warnecke C, Scholz H (2007) Hypoxia-inducible factor-1 (HIF-1) is a transcriptional activator of the TrkB neurotrophin receptor gene. J Biol Chem 282(19):14379–14388
Martinez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, Hall E, Reichmann J, Devenney PS, Hohenstein P, Hosen N, Hill RE, Muñoz-Chapuli R, Hastie ND (2010) Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet 42(1):89–93
Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A (1999) YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 126:1845–1857
Morrison AA, Viney RL, Ladomery MR (2008) The posttranscriptional roles of WT1, a multifunctional zinc-finger protein. Biochim Biophys Acta 1785:55–62
Park S, Bernard A, Bove KE, Sens DA, Hazen-Martin DJ, Garvin AJ, Haber DA (1993) Inactivation of WT1 in nephrogenic rests, genetic precursors to Wilms’ tumour. Nature Genet 5:363–367
Prandini MH, Dreher I, Bouillot S, Benkerri S, Moll T, Huber P (2005) The human VE-cadherin promoter is subjected to organ-specific regulation and is activated in tumour angiogenesis. Oncogene 24(18):2992–3001
Pritchard-Jones K, Fleming S, Davidson D, Bickmore W, Porteous D, Gosden C, Bard J, Buckler A, Pelletier J, Housman D, van Heyningen V, Hastie N (1990) The candidate Wilms’ tumour gene is involved in genitourinary development. Nature 346:194–197
Ratajska A, Czarnowska E, Ciszek B (2008) Embryonic development of the proepicardium and coronary vessels. Int J Dev Biol 52(2–3):229–236
Rivera MN, Haber DA (2005) Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer 5:699–712
Roberts SG (2005) Transcriptional regulation by WT1 in development. Curr Opin Genet Dev 15:542–547
Timár J, Mészáros L, Orosz Z, Albini A, Rásó E (2005) WT1 expression in angiogenic tumours of the skin. Histopathology 47:67–73
Vestweber D, Winderlich M, Cagna G, Nottebaum AF (2009) Cell adhesion dynamics at endothelial junctions: VE-cadherin as a major player. Trends Cell Biol 19(1):8–15
Vittet D, Buchou T, Schweitzer A, Dejana E, Huber P (1997) Targeted null-mutation in the vascular endothelial-cadherin gene impairs the organization of vascular-like structures in embryoid bodies. Proc Natl Acad Sci USA 94:6273–6278
Wagner N, Michiels JF, Schedl A, Wagner KD (2008) The Wilms’ tumour suppressor WT1 is involved in endothelial cell proliferation and migration: expression in tumour vessels in vivo. Oncogene 27:3662–3672
Wagner KD, Wagner N, Bondke A, Nafz B, Flemming B, Theres H, Scholz H (2002) The Wilms’ tumor suppressor Wt1 is expressed in the coronary vasculature after myocardial infarction. FASEB J 16(9):1117–1119
Wagner N, Wagner KD, Hammes A, Kirschner KM, Vidal VP, Schedl A, Scholz H (2005) A splice variant of the Wilms’ tumour suppressor Wt1 is required for normal development of the olfactory system. Development 132:1327–1336
Wagner N, Wagner KD, Theres H, Englert C, Schedl A, Scholz H (2005) Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms' tumor transcription factor Wt1. Genes Dev 19(21):2631–2642
Wagner KD, Wagner N, Vidal VP, Schley G, Wilhelm D, Schedl A, Englert C, Scholz H (2002) The Wilms’ tumor gene Wt1 is required for normal development of the retina. EMBO J 21:1398–1405
Acknowledgments
The expert technical assistance of Angelika Richter and Inge Grätsch is gratefully acknowledged. Parts of this study were supported by a grant from the Deutsche Forschungsgemeinschaft (Scho 634/6-1) and internal funds from the Charité - Universitätsmedizin Berlin.
We declare that the experiments comply with the current laws of Germany. We also indicate that we have no conflict of interest with regard to the financial support to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kirschner, K.M., Sciesielski, L.K. & Scholz, H. Wilms’ tumour protein Wt1 stimulates transcription of the gene encoding vascular endothelial cadherin. Pflugers Arch - Eur J Physiol 460, 1051–1061 (2010). https://doi.org/10.1007/s00424-010-0873-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-010-0873-6