Abstract
The cardiac action potential is primarily shaped by the orchestrated function of several different types of ion channels and transporters. One of the regional differences believed to play a major role in the progression and stability of the action potential is the transmural gradient of electrical activity across the ventricular wall. An altered balance in the ionic currents across the free wall is assumed to be a substrate for arrhythmia. A large fraction of patients with heart failure experience ventricular arrhythmia. However, the underlying substrate of these functional changes is not well-established as expression analyses of human heart failure (HF) are sparse. We have investigated steady-state RNA levels by quantitative polymerase chain reaction of ion channels, transporters, connexin 43, and miR-1 in 11 end-stage HF and seven nonfailing (NF) hearts. The quantifications were performed on endo-, mid-, and epicardium of left ventricle, enabling us to establish changes in the transmural expression gradient. Transcripts encoding Cav1.2, HCN2, Kir2.1, KCNE1, SUR1, and NCX1 were upregulated in HF compared to NF while a downregulation was observed for KChIP2, SERCA2, and miR-1. Additionally, the transmural gradient of KCNE1, KChIP2, Kir6.2, SUR1, Nav1.5, NCX1, and RyR2 found in NF was only preserved for KChiP2 and Nav1.5 in HF. The transmural gradients of NCX1, Nav1.5, and KChIP2 and the downregulation of KChIP2 were confirmed by Western blotting. In conclusion, our results reveal altered expression of several cardiac ion channels and transporters which may in part explain the increased susceptibility to arrhythmia in end-state failing hearts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ventricular arrhythmia is one of the primary causes of death in human heart failure [9]. Arrhythmia in heart failure, often being ventricular tachycardia degenerating into ventricular fibrillation [16], is likely caused either by an increased dispersion or an altered level of the different ionic currents underlying the cardiac action potential [1, 32]. Several studies have revealed that human ventricular tissue from failing hearts exhibit action potential prolongation [8, 12], which is believed to be associated with increased risk of arrhythmia (reviewed in [23]). Additionally, an increased spatial and temporal dispersion of the ventricular action potential duration has been reported in failing human hearts [7].
The cardiac action potential is shaped by the underlying ion currents and transporters, and various forms of rhythm disturbances are due to altered function of these. Several different heart failure animal models have been used in order to address the electrophysiological, translational, and transcriptional changes taking place in the ventricles. In general, these studies reveal prolongation of action potential duration, most often orchestrated by a compromised repolarizing reserve, constituted by the inward rectifier potassium current I K1 and the delayed rectifier potassium currents I Ks and I Kr, a reduced transient outward current (I to) [19, 33, 38], and a decrease in peak sodium current I Na and an increase in the late sodium current (I NaL) [33].
Changes in the heterogeneity of ion channel activities is an important substrate for arrhythmogenesis and the development of life-threatening ventricular arrhythmias is particularly associated with an increase of intrinsic transmural dispersion of repolarization. Differential expression across the ventricular wall participates in the regulation of the ventricular electrophysiological properties. This has been well established for the potassium current I to, being highly expressed in the epicardial layer, where it participates in the initial repolarization, thereby giving rise to a notch in the action potential, while it is much less expressed in the endocardial layer [3].
The molecular mechanisms underlying the electrical changes in human heart failure have only sparsely been investigated. The reduced I to observed in human heart failure [8] has been linked to downregulation of its pore-forming Kv4.3 subunit on protein level [19, 38] and of KChIP2 (KCNIP2) on mRNA level [24]. IK1 has also been reported to be functionally downregulated in human heart failure [8]. However, this functional reduction of I K1 was not found to be associated with either reduction in steady-state mRNA level or protein expression of Kir2.1 (KCNJ2), which is believed to be the primary pore forming subunit in the I K1 channel complex. Furthermore, in the failing human myocardium, a reduction in the peak I Na was shown to correspond to changed expression of Nav1.5 (SCN5A) splicing variants [29].
MicroRNAs constitute a newly discovered mechanism of transcriptional regulation, where microRNA annealing to specific three-prime untranslated region sequences of target mRNAs leads to translational suppression and/or mRNA cleavage [2]. Quantification of endogenous microRNAs, targeting specific gene products, may thereby contribute to a better understanding of the correlation between mRNA and protein expression levels.
In the present study, we have aimed at analyzing whether transcriptional changes in failing hearts can explain the increased incidence of arrhythmia. Our investigations have addressed whether down- and upregulation in the absolute expression as well as whether reduction or increase in the transmural expression profile can be established for failing (HF) vs. nonfailing (NF) hearts. mRNA quantification were performed by Taqman real-time reverse transcriptase-polymerase chain reaction (RT-PCR) on atrial and ventricular transmural samples from seven nonfailing hearts and 11 failing hearts of various ion channels and transporters as well as connexin 43 and miR-1. Transcripts encoding Cav1.2 (CACN1C), HCN2, Kir2.1 (KCNJ2), KCNE1, SUR1 (ABCC8), and NCX1 (SLC8A1) were upregulated in HF compared to NF while downregulation was observed for KChIP2 (KCNIP2), SERCA2 (AT2A2), and miR-1. The transmural expression profile of Nav1.5 (SCN5A), KCHiP2 (KCNIP2), and NCX1 (SLC8A1), and the downregulation of KChIP2 (KCNIP2) were confirmed by Western blotting.
Materials and methods
Heart failure patients and myocardial tissue
The investigation conforms to the principles outlined in the Declaration of Helsinki. The clinical characteristic of the patients whose myocardial tissue was used in the study is shown in Table 1. The patients were diagnosed with end-stage heart failure (class IV according to New York Heart Association) and underwent heart transplantation. Heart failure was secondary to either dilated cardiomyopathy (DCM; n = 6 patients) or ischemic cardiomyopathy ICM (n = 5 patients). The patients were ten males and one female aged 18–57 (mean 45). Ten patients were in sinus rhythm and two others had atrial fibrillation. Left ventricular hypertrophy was excluded in failing hearts by echocardiography in ten out of 11 patients. Heart failure was managed by beta-blocker and diuretics therapy in all the patients. Only one patient was administrated with AT1 receptor antagonist and nitrates. None of the patients was prescribed with calcium receptor antagonists.
Control (nonfailing) group consisted of seven individuals (three males and four females, mean age 42). After explantation, the hearts were stored for 4–6 h in the cardioplegic solution before being snap-frozen in the liquid nitrogen. Ventricular tissue was obtained from the anterior wall near the basis (1 cm from the mitral/tricuspidal valve ring). Transmural ventricular tissue samples were obtained by cutting 1-mm-thick slices from the epicardial and endocardial surfaces of the heart. Midmyocardial tissue was obtained from the midwall region of the ventricles.
We also quantified the mRNA in the right atria (RA) in both control and failing hearts. The results are summarized in the Supplementary materials.
RNA preparation and RT reaction
The myocardial tissue was quick-frozen in liquid nitrogen and stored at −80°C until further processing. Prior to RNA isolation, special emphasis on preserving RNA quality was taken in order to minimize RNase activity during RNA isolation. For this reason, frozen tissue specimens were thawed in RNA stabilizing solution (RNAlater®-ICE, Ambion, USA). Total RNA was purified from the homogenized tissue specimens (homogenizer Kinemtica, Buch & Holm, Switzerland) with Tri Reagent® (Sigma-Aldrich, USA) according to the manufacturer’s instructions.
Following RNA purification, the potential genomic DNA contamination was eliminated by DNase I treatment (Invitrogen, USA). RNA was quantified by photometry (A = 260 nm) and purity verified by A260/280 ratio by NanoDrop (ND-1000, USA). The integrity of total RNA was assessed by ethidium bromide staining, where the presence of 28S and 18S was confirmed for all samples.
Total cDNA RT
DNA was synthesized from 2 µg total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem, USA) with random hexamer primers following the manufacturer’s instructions.
MicroRNA-specific RT
Total RNA was not enriched for low molecular weight RNA before microRNA-specific reverse transcription. TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystem, USA) was used for multiplex reverse transcription reactions carried out with the primer sets for miR-1 and RNU44 (used as en endogenous control). Each miR-specific RT reaction mixture contained 0.15 µg of total RNA, 0.15 µl 100 nM dNTPs, 1.5 µl 10× RT buffer, 0.18 µl 20 U/µl RNase inhibitor, 1 µl 50 U/µl MultiScribeReverse Transcriptase, and 1 µl of each three specific 5× stem-loop primers (supplied with TaqMan MicroRNA assays) in total volume of 15 µl.
Real-time PCR
Real-time RT-PCR was performed on the 7300 RT-PCR System (Applied Biosystem, USA), and data were collected by SDS1.2 software. The genes selected for quantification were investigated using predesigned TaqMan assays (TaqMan® MGB probe and primers; see Table in supplementary data). Only the primers and the probe targeting the GJAJ (Cx43) gene were designed manually using Primer Express 3.0 software and synthesized by Applied Biosystem.
Thermal profile for all the real-time PCR reactions was as followed: initial steps 50°C (2 min), 95°C (10 min), and then 40 cycles with 95°C (15 s) and 60°C (1 min).
Prior to the experiments, PCR efficiency for each assay was calculated using a standard curve constructed by plotting range of log cDNA input against C t (threshold cycle) value. The slope of the plot was used to calculate the percentage amplification efficiency (PE). PE values were all ranged between 90% and 110%, and an amplification efficiency of 2 (100%) per cycle has therefore been used in all the calculations. For each tissue specimen, each gene was quantified in triplicates.
Three reference genes were tested: 18sRNA, HPRT, and PPIA (Supplementary data), but only the latter one (encoding cyclophilin A) was chosen for normalization due to its highly uniform expression in our tissue. Moreover, RNU44 (C/D box 44, small nucleolar RNA) was used as a reference control transcript for miR-1 quantification.
The comparative threshold cycle (C t) relative quantification method [20] was used as computation method. First, delta C t (dC t) values were calculated by subtracting mean C t value of a target gene and cyclophilin A (PPIA) for each sample. Then, the relative expression of each gene vs. cyclophilin A was calculated by \( 2^{{ - dC_{\text{t}} \; \times \;100}} \) (± standard error of the mean).
Western blotting
Total membrane proteins were purified using a method modified after Han et al. [14]. Briefly, the snap-frozen tissue was pulverized in liquid nitrogen and suspended in 600 µl of cold TE buffer (containing Tris 20 mM, EDTA 1 mM supplemented with a cocktail of protease inhibitors, 10 µM 4-(2-aminoethyl)benzenesulfonyl fluoride, 0.2 µM leupeptin, 0.4 µM bestatin, 0.15 µM pepstatin A, 0.14 µM M E-64, and 8 pM aprotinin, all from Sigma). The tissue suspension was then quickly homogenized (5 s) and 2% of Triton X-100 was added. The suspension was left on ice for 2 h to allow effective membrane solubilization. Spinning down the suspension (15,000×g, 15 min, 4°C) resulted in a soluble supernatant of tissue homogenate. Protein concentration was assessed by the Bradford method (DC Protein Assay, SIGMA Aldrich, USA) which is compatible with 2% Triton-X 100.
Tissue homogenate (50 μg/lane) was separated on 4–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (for blotting against KChiP2 and SCN5A) and 7.5% gels (for blotting against NCX1). Separated proteins were transferred onto the hybond-P polyvinylidene fluoride transfer membranes (Amersham Biosciences, 0.45 µm). Membranes were blocked with 5% nonfat milk in TTBS (0.1% Tween 20 plus Tris-buffered saline solution) for 1 h at RT and then incubated with the primary antibodies against KChiP2 (1:200, clone K60/73, Neuromab, USA), Nav1.5 (1:500, ASC005, Alomone Labs, Israel), and NCX1 (1:1500, R3F1 Swant, Switzerland) in 4°C overnight. The monoclonal antibody against KChiP2 (clone K60/73) was obtained from NeuroMab Facility, supported by NIH grant U24NS050606 and maintained by the Department of Pharmacology, School of Medicine, University of California, Davis, CA 95616, USA.
The proteins were detected by horseradish peroxidase (HRP)-conjugated donkey antimouse antibody (1/10,000, Jackson Immunosearch Laboratories, UK) and HRP-conjugated donkey antirabbit (1:10,000, SIGMA Aldrich, USA). Enhanced chemiluminescence (ECL) staining was performed using the standard procedure (Supersignal West Pico Chemiluminescent detection system, Pierce). Immunoblots were exposed on hyperfilm ECL (Amersham Biosciences). Note that endogenous controls are not included. As primarily surface membrane associated proteins are harvested by this extraction procedure, we have not found neither cytochrome C nor β-tubulin, nor actin suitable as loading controls. However, great care has been taken in loading exactly the same amount of proteins on the gels, and the Western blotting experiments have been repeated several times. Band density was quantified by Quantity-One software as Gaussian trace quantity on the films exposed and processed equally.
Statistical analysis
Statistical analysis was performed with unpaired, two-tailed Student’s t test, Mann–Whitney test, and repeated measures analysis of variance (ANOVA) test (followed by posttest: Bonferroni’s multiple comparison test). Unpaired two-tailed t test was applied to compare the RNA expression both between each ventricular wall layer (Figs. 2, 3, 4, and 5) and between the averaged ventricular values (Fig. 1) of the NF and the HF hearts. The transmural gradient of expression in the NF and HF left ventricles was assessed by the repeated measures ANOVA test (followed by posttest: Bonferroni’s multiple comparison test; Figs. 2, 3, 4, and 5). Mann–Whitney test was used to compare protein level of KChiP2 between NF and HF hearts (Fig. 6b).
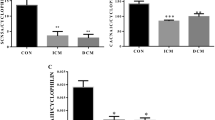
Transcriptional expression in nonfailing vs. failing human hearts. In order to compare the expression of the genes between left ventricles (LV) of NF and HF the data from epi-, mid-, and endocardium of LV were pooled. Note that since the level of miR-1 was not normalized to PPIA (but to RNU44), the data for miR-1 are not included in this overview. HCN1 transcript was not detected in the LV, but only in the RA. Statistically significant differences in gene expression between HF and HF are marked with asterisks (*p < 0.05; **p < 0.01; ***p < 0.001)
KChiP2 protein expression in tissue samples from nonfailing and failing hearts. a Transmural gradient of KChIP2 protein expression was preserved, albeit at a lower level, in HF. b Downregulation of the KChIP2 protein level in HF was investigated by Western blotting of membrane extracts. In epicardium, a trend toward downregulation is observed, and for midmyocardium, significance is reached (p < 0.05)
Results
The progression of the human heart into a failing heart condition is known to involve electrical remodeling. In order to investigate the transcriptional changes, potentially explaining this electrical remodeling, tissue from the explanted NF and failing (HF) hearts were dissected from right atria as well as endo-, mid-, and epicardium of the left ventricle, RNA was extracted, and expression was quantified by real-time RT-PCR (“Materials and methods” section and Supplementary data). The diseased tissues originate from six DCM and five ischemic cardiomyopathy (ICM) end-stage failing hearts. Heart failure (class IV according to New York Heart Association) was established from changes in hemodynamic parameters (Table 1). The average age was 45 years. The control myocardial tissue (NF) was obtained from seven donors with an average age of 42 years.
The expression level of the mRNAs, encoding proteins of importance in generating the cardiac action potential, was investigated in both failing and nonfailing hearts, making it possible to compare the relative expression level as well as to examine transmural gradients and potential changes in this gradient.
Although heart failure is considered to be a final common phenotype independent of the etiology, the differential changes in some ionic currents (I K1 and I f) in cardiomyocytes obtained from either DCM or ICM hearts were reported [11, 17]. However, these studies have not addressed whether transcriptional changes of ion channel subunits underlie this phenomenon. In our study, no significant expression differences of the studied genes were found between the DCM and ICM hearts, and we have therefore pooled the data from these two kinds of failing hearts and compared them to nonfailing hearts.
Relative expression in nonfailing hearts
Quantification of the transcriptional level in atria (Supplementary data) and ventricle of nondiseased hearts (overview in Fig. 1 and exact values in Supplementary data) reveal an expression profile similar to what was previously reported by Gaborit and colleagues [13]. In general, the ion channel transcripts, apart from the calcium channel transcripts encoding RyR2 and Cav1.2 (CACN1C), were expressed at lower levels than the ion pumps, SERCA2 (ATP2A2), Na/K ATPase (ATP1A1), and the sodium/calcium exchanger NCX1 (SLC8A1). Within the group of potassium channels, notable differences are observed for the components underlying the slow delayed rectifier current I Ks, where Kv7.1 (KCNQ1), being the pore-forming subunit, is expressed several times more abundantly than KCNE1, which is believed to be its primary auxiliary subunits. Furthermore, in ventricle, the expression level of the KATP regulatory subunit SUR2A (ABBC8) is much greater than SUR1 (ABBC8). Among the hyperpolarization-activated cyclic nucleotide-gated (HCN) family of ion channel subunits (HCN1, HCN2, HCN4), HCN4 was found to be the predominant subunit in the right atria (expressed at a higher level than HCN1 and HCN2). Ventricular expression of HCN2 and HCN4 was comparable, whereas HCN1 transcript was not detectable in the left ventricle.
Steady-state mRNA differences between NF and HF
Figure 1 provides a transcriptional overview, illustrating the average values obtained from the three ventricular layers, of both NF and HF. As can be observed, then it is only the level of four transcripts which was found to be different when comparing the summarized values of the left ventricle. However, as it is well-established that electrical remodeling takes place in failing hearts, it can be speculated that the mRNA level of many genes involved in ion conductance can be altered. Furthermore, as transmural heterogeneity has been suggested to be involved in the increased susceptibility for arrhythmia in HF, a more detailed analysis was accomplished to establishing the transcriptional level through the left ventricular free wall. For this investigation, tissue was divided into endo-, mid-, and epicardial samples.
To confirm diseased status of the failing hearts, we used the Brain Natriuretic Peptide Precursor BNP (NPPB) transcript, which is known to be elevated in failing hearts, as a positive marker (Fig. 2). The mRNA quantity of BNP (NPPB) was significantly higher in the failing left ventricles as compared to the nondiseased left ventricles (4-fold, p < 0.01), and this difference was even more pronounced in the atria (10-fold, p < 0.001; Supplementary data).
The microRNA miR-1 has been reported to downregulate Cx43 (GJA1) and Kir2.1 (KCNJ2) [37], as well as HCN2/4 [21] and KCNE1 [22]. We found miR-1 level to be drastically reduced in HF (Fig. 2). However, transcripts encoding connexin 43 (GJA1) was not found altered in HF, but an increased amount of Kir2.1 (KCNJ2), HCN2, and KCNE1 transcripts was established (see Fig. 5).
One of the hallmarks in heart failure is a functional downregulation of the ion pumps SERCA2 (ATP2A2) and Na/K ATPase (ATP1A1) [28, 35]. We find a reduced level of Na/K ATPase α1 (ATP1A1) level in all three ventricular layers and of SERCA2 (ATP2A2) in endocardium (Fig. 3). For the sodium calcium exchanger NCX1, a significant upregulation in failing hearts was established in epi- and endocardium, while for the ryanodine receptor RyR2 no difference in expression between NF and HF could be observed (Fig. 3).
The HCN channels, which are pivotal in the process of generating new action potentials in the sinus node, are also present in the ventricles where they contribute to the intrinsic automaticity observed at low pacing rates. HCN4 expression is unaltered in HF, but for HCN2, a significant increased mRNA level is revealed in endocardium and a trend toward an increased level is found in midmyocardium (Fig. 4). The expression level of the major cardiac sodium channel Nav1.5 (SCN5A) was unaltered, while the transcript encoding the major cardiac L-type calcium channel Cav1.2 (CACN1C) was approximately 3-fold upregulated in epi- and endocardium (Fig. 4).
For the potassium channels, a similar expression level was found for Kv4.3 (KCND3), hERG1 (KCNH2), Kv7.1 (KCNQ1), and Kir6.2 (KCNJ11) in NF and HF (Fig. 5). Kir2.1 (KCNJ2), KCNE1, and SUR1 (ABCC8) were upregulated in epi- and endocardium in HF. The regulatory subunit of the Kv4.3 channel, KChIP2 (KCNIP2), was found reduced in midmyocardium.
Transmural expression
For a number of the transcripts, we observe a difference in the expression level between the different myocardial layers in the NF hearts. This regional variation in expression contribute to the differential action potential morphology recognized across the ventricular wall, and subtle changes in these expression levels may lay the ground for an increased susceptibility to arrhythmia. A transmural transcriptional gradient in NF hearts was found for KCNE1, KChIP2 (KCNIP2), Kir6.2 (KCNJ11), SUR1 (ABCC8), Nav1.5 (SCN5A), NCX1 (SLC8A1), and RyR2 (Figs. 2, 3, and 5). Remarkably, this gradient was not found for KCNE1, Kir6.2 (KCNJ11), SUR1 (ABCC8), NCX1 (SLC8A1), and RyR2 in the samples from HF patients, and in addition, then a gradient for SERCA2, which is not present in NF, appeared in the HF samples.
Reduced expression of KChIP2 protein
A pivotal component in constituting the transient outward potassium current (I to) channel complex in humans is KChIP2, regulating the pore-forming Kv4.3 subunit. On mRNA level, we observed, like several others, a transmural gradient of KChIP2 (KCNIP2). Whether this mRNA gradient is translated on to the protein level was investigated by comparing endo- and epicardium from two NF and two HF hearts (Fig. 6a). Here, we find, for all four analyzed hearts, a higher expression in epicardium than in endocardium, confirming previous published results as well as the herein performed mRNA quantifications.
I to has been reported to be functionally downregulated in human heart failure [19], and the herein quantitative transcriptional analysis has found a reduction of KChIP2 (KCNIP2) mRNA. In order to investigate this downregulation further, Western blotting of epi- and midmyocardium samples was performed (Fig. 6b). Similar to the mRNA results, we found a significant downregulation in midmyocardium, while there is a trend towards downregulation in epicardium.
Transmural gradient of NCX1 and Nav1.5
The transcriptional profiling of the sodium calcium exchanger NCX1 reveals in NF hearts a bell-shaped expression with the highest expression in midmyocardium. Western blotting of samples from two NF hearts confirmed this expression pattern (Fig. 7a). However, in HF, a similar pattern could not be observed which is in concordance with the obtained mRNA data. Another interesting observation, which is exposed on the Western blot, is the fact that the bands appearing between 80 and 100 kDa, previously suggested to be either splice variants or proteolytic fragments [34], cannot be detected in the HF samples.
Transmural protein expression of NCX1 and Nav1.5. a Western blotting of transmural samples from two NF and one HF heart. For the two NF hearts, a bell-shaped expression profile of NCX1, alike the one found on mRNA level, can be seen. This gradient is not present in the HF samples. b Western blotting of Nav1.5 in two pairs of endo- and epicardium LV samples and one pair of endo- and epicardium samples from both RV and LV. All samples reveal an increase in expression from epi- to endocardium
In both NF and HF, a transmural gradient of Nav1.5 (SCN5A), with the lowest expression in epicardium, was found on mRNA level. Western blotting of endo- and epicardial samples confirms that this difference is also present on protein level (Fig. 7b). As we furthermore had access to transmural tissue from right ventricle from HF hearts, we have also performed quantifications of Nav1.5 (SCN5A) on these samples in order to investigate whether such a gradient is present in the free wall of both ventricles. We find a similar Nav1.5 (SCN5A) mRNA gradient (data not shown), and Western blotting on protein extracted from right ventricular samples also indicates higher expression of Nav1.5 in endo- vs. epicardium (Fig. 7b, right lanes).
Discussion
Tight management of the cardiac ionic currents is necessary in order to control the activation, progression, and termination of the cardiac action potential. This tight electrical control is obtained through differential expression of a number of membrane proteins conducting and transporting ions, their auxiliary proteins, as well as through posttranslational modifications of these. Changes in the functionality of the ionic currents can be a substrate for arrhythmia. Heart failure is a detrimental disease associated with a high incidence of fatal ventricular arrhythmias. Arrhythmia in heart failure is known to be associated with functional changes in the electrical currents underlying the cardiac action potential. A number of studies have analyzed the changes in the ionic currents taking place in human failing cardiomyocytes, while thorough quantification of the transcripts underlying the cardiac currents is lacking. In the current study, we address whether transcriptional changes, both in absolute value and in the transmural profile, can explain the high arrhythmogenic risk observed in failing hearts. We have therefore performed mRNA quantifications, using real-time PCR, of the most relevant transcripts encoding proteins involved in generating, progressing, and shaping the cardiac action potential.
Nonfailing hearts
Gaborit and colleagues [13] recently published a very comprehensive study regarding the gene expression profile of several of the genes involved in generating the electrical impulse in nondiseased human hearts. Our data from the nonfailing hearts largely coincide, both concerning relative expression level and differential expression between atria and ventricle, with this report. Gaborit et al. [13] did also quantify potential transmural differences in expression between endo- and epicardium in left ventricle, and they do, as we, observe a higher expression of Nav1.5 (SCN5A) in endo- vs. epicardium and a lower expression of KCNIP2 in endo- vs. epicardium (where we see a trend). However, we do not detect the small, but significant, differences for Cav1.2 (CACN1C), Kv4.3 (KCND3), Kv7.1 (KCNQ1), SERCA2 (ATP2A2), and NCX1 (SLC8A1) as they do [13].
Potassium subunit regulation in HF
Heart failure has often been found to be associated with prolonged action potential duration (APD). This prolongation has, at least partly, been linked to a decrease in some of the late potassium currents. Our results do not reveal a downregulation of the transcripts underlying the pore-forming subunit of the major cardiac K+ currents, but do in contrast show an upregulation of Kir2.1 (KCNJ2). This upregulation could be explained by the drastic downregulation of miR-1, which has been reported to negatively regulate Kir2.1 (KCNJ2) transcripts [37]. Additionally, upregulation of KCNE1, which is an essential subunit of the I Ks channel complex, was also found in epi- and endocardium. Watanabe et al. [36] recently reported a similar upregulation of KCNE1 as well as no change in expression of hERG1 (KCNH2) and Kv7.1 (KCNQ1) in mild to moderate chronic heart failure, and Radicke et al. [24] reported an upregulation of KCNE1 in end-stage HF.
Thereby, in respect to potassium channel regulation, our results cannot explain the APD prolongation observed in the ventricles in failing hearts, and it may therefore be speculated that other regulation mechanisms, such as miRNA silencing and posttranslational modifications, could play a role in the electrical remodeling. I to has previously been found to be downregulated in HF [19]. However, the molecular mechanisms underlying this downregulation have been reported to be caused both by changes in the expression of Kv4.3 (KCND3) and KChIP2 (KCNIP2) [19, 24]. Our experiments do not reveal any change in Kv4.3 (KCND3) expression, but do show a downregulation of KChIP2 (KCNIP2), which is required for proper trafficking of Kv4.3 channels to the cell membrane [4, 10, 18]. This observed downregulation of KChIP2 (KCNIP2) was confirmed on protein level where a significant reduced expression of KChIP2 was found.
Pacemaker and sodium currents in HF
We report an increase in the expression of the pacemaker current (I f) subunit HCN2 in endocardium of the failing hearts. Other reports have found an increased I f in ventricular myocytes from ischemic cardiomyopathy [11] coinciding with an increased transcriptional and translational level of HCN2 and HCN4 in ventricle [31]. An increase in the expression of pacemaker channels could contribute to an increased electrical instability of the myocardium by an augmentation of triggered activity and thereby potentially be a pro-arrhythmic parameter.
The peak sodium current is both responsible for the depolarization of the cardiac myocytes as well as the major determinant of the conduction velocity, while the late (sustained) sodium current, also generated by Nav1.5 (SCN5A), is implicated in action potential duration [33]. Both decrease and increase in peak I Na has been reported in the human failing cardiomyocytes [29, 33]. An altered expression of Nav1.5 (SCN5A) splice variants, resulting in a decrease of the native SCN5A mRNA, was recently described in the ventricles of the failing hearts [29]. We do not observe a difference between NF and HF in left ventricle.
Sodium, potassium, and calcium handling in HF
Adverse molecular remodeling of the Na/K ATPase and of SERCA2 in heart failure has previous been shown [28, 35]. We find a significant reduction in the expression level of both Na/K ATPase α1 (ATPA1) and SERCA2A (ATP2A2) in left ventricle. Our data thereby confirm that one of the hallmarks of heart failure seems to be a reduction in the activity of the sodium/potassium ATPase and of sarcoplasmic reticulum calcium ATPase2α. Reduction of SERCA expression is believed to lead to an increase in intracellular Ca2+ and thereby also an increase in contractile force. The same, but indirect, effect would be the consequence of reduced sodium/potassium ATPase expression, with increased intracellular Na+ forcing the Na+/Ca2+ exchanger in reverse mode and thereby increasing intracellular Ca2+. In addition to this, NCX1 (SLC8A1) expression is increased, thereby further supporting the theory of an augmented inotropic response in order to compensate for the lost contractility of the failing ventricle [15, 30]. The L-type calcium current (I Ca-L) in HF has been reported both to be up- and downregulated in end-stage heart failure [26, 27]. We observe a significant increased level of Cav1.2 (CACN1C) transcripts, which is the primary pore-forming subunit underlying ICa-L, in epi- and endocardium. This augmented mRNA level will further enhance the intracellular Ca2+ concentration. A concomitant effect of the change in Ca2+ homeostasis is an increased risk for triggered activity that can serve as the starting point for arrhythmias. The increased proclivity for arrhythmias in HF patients might therefore be related to the observed transcriptional changes for Ca2+ handling proteins.
Transmural expression
As altered transmural dispersion has been associated with an increased susceptibility to arrhythmia [5], it has been one of the major tasks of this study to establish whether the relative transmural expression levels are changed. Up to now, the gene expression of only few ion channel subunits (Kv4.3, KChIP2, and KCNE1-5) has been studied in epi- and endocardial layer of failing ventricles [24, 38]. In the samples from NF hearts, a difference in expression level between the three layers were found for KCNE1, Kir6.2 (KCNJ11), SUR1 (ABCC8), Nav1.5 (SCN5A), NCX1 (SLC8A1), and RyR2. This pattern of expression was lost for KCNE1, Kir6.2 (KCNJ11), SUR1 (ABCC8), NCX1 (SLC8A1), and RyR2 and thereby only preserved for Nav1.5 (SCN5A). Dispersion in expression of SERCA2 (ATP2A2) was in addition found in HF. Further, in HF, a KChIP2 gradient appears, which was only revealed as a trend (p = 0.06) in NF. However, as a gradient of KChIP2 (KCNIP2) in NF hearts is well-established, it can be presumed that this transmural gradient is preserved between NF and HF [5, 13, 24, 38]. Expression of the sodium channel transcript Nav1.5 (SCN5A) was markedly higher in endo- and midmyocardium as compared to epicardium in both NF and HF. This has already been established on mRNA level in nondiseased hearts [13] and corresponds well with an augmented sodium current observed in endo- vs. epicardium of rat and mice [6, 25].
Study limitations
Myocardial tissue form failing hearts was obtained from patients with heart failure due to two different etiologies (dilated and ischemic cardiomyopathy) in order to obtain an adequate number of samples. However, analyses of the transcription level of the investigated genes have not been found to be differently expressed between ICM and DCM (data not shown). Furthermore, when doing investigations of tissue from human hearts, it should be bear in mind that patients, in contrast to animals, are neither environmentally nor genetically controlled, and HF patients have additionally been subjected to pharmacological therapy. Hence, multiple factors can potentially influence the transcriptional regulation.
Conclusion
The high frequency of arrhythmia observed in HF involves ionic remodeling. The performed quantifications establish major changes in the transcripts encoding the proteins involved in calcium handling and thereby confirm one of the hallmarks of HF. Further, by including analyses of transmural samples, we have detected an altered transmural expression pattern for several of the gene transcripts. These changed gradients may be one of the important parameters, together with global changes in the expression of specific genes, in transforming the myocardium into a state where it is more prone to arrhythmia.
The prolonged APD observed in HF have in many animal HF models been found to correlate with a reduced amount of the transcripts encoding the repolarizing currents. We do not see a similar downregulation in these end-stage human failing hearts. We do find an increase in the Cav1.2 and NCX encoding mRNA and a decrease in KChIP2 (KCNIP2) mRNA and protein, which at least partly could explain an increased APD. It is also possible that auxillary proteins and posttranslational modifications, e.g., inducing an increase sustained sodium current, plays a larger role in humans where HF has developed over several years.
Abbreviations
- HF:
-
Heart failure
- NF:
-
Nonfailing
- DCM:
-
Dilated cardiomyopathy
- ICM:
-
Ischemic cardiomyopathy
- BMI:
-
Body mass index
- SR:
-
Sinus rhythm
- AF:
-
Atrial fibrillation
- LVEF:
-
Left ventricular ejection fraction
- LVEDP:
-
Left ventricular end-diastolic pressure
- PH:
-
Pulmonary hypertension
- LA d:
-
Left atrium dimension
- LVED d:
-
Left ventricular end-diastolic dimension
- IVS d:
-
Intraventricular septum diameter
- LVPW d:
-
Left ventricular posterior wall diameter
- ACE:
-
Angiotensin-converting enzyme
- AT1Bl:
-
Angiotensin receptor blockers
References
Akar FG, Rosenbaum DS (2003) Transmural electrophysiological heterogeneities underlying arrhythmogenesis in heart failure. Circ Res 93:638–645
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Amos GJ, Wettwer E, Metzger F, Li Q, Himmel HM, Ravens U (1996) Differences between outward currents of human atrial and subepicardial ventricular myocytes. J Physiol 491(1):31–50
An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ (2000) Modulation of A-type potassium channels by a family of calcium sensors. Nature 403(6769):553–556
Antzelevitch C (2007) Heterogeneity and cardiac arrhythmias: an overview. Heart Rhythm 4:964–972
Ashamalla SM, Navarro D, Ward CA (2001) Gradient of sodium current across the left ventricular wall of adult rat hearts. J Physiol 536:439–443
Barr CS, Naas A, Freeman M, Lang CC, Struthers AD (1994) QT dispersion and sudden unexpected death in chronic heart failure. Lancet 343:327–329
Beuckelmann DJ, Näbauer M, Erdmann E (1993) Alterations of K + currents in isolated human ventricular myocytes from patients with terminal heart failure. Circ Res 73:379–385
Brown DW, Giles WH, Croft JB (2000) Left ventricular hypertrophy as a predictor of coronary heart disease mortality and the effect of hypertension. Am Heart J 140:848–856
Bähring R, Leicher T, Pongs O, Isbrandt D, Dannenberg J, Peters HC (2001) Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2.2 on channel expression and gating. J Biol Chem 276(26):23888–23894
Cerbai E, Sartiani L, DePaoli P, Pino R, Maccherini M, Bizzarri F (2001) The properties of the pacemaker current I(F)in human ventricular myocytes are modulated by cardiac disease. J Mol Cell Cardiol 33:441–448
Coltart DJ, Meldrum SJ (1972) Intracellular action potential in hypertrophic obstructive cardiomyopathy. Br Heart J 34:204
Gaborit N, Le Bouter S, Szuts V, Varro A, Escande D, Nattel S (2007) Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol 582:675–693
Han W, Bao W, Wang Z, Nattel S (2002) Comparison of ion-channel subunit expression in canine cardiac Purkinje fibers and ventricular muscle. Circ Res 91(9):790–797
Houser SR, Piacentino V 3rd, Mattiello J, Weisser J, Gaughan JP (2000) Functional properties of failing human ventricular myocytes. Trends Cardiovasc Med 10(3):101–107
Kjekshus J (1990) Arrhythmias and mortality in congestive heart failure. Am J Cardiol 65:421–481
Koumi S, Backer CL, Arentzen CE (1995) Characterization of inwardly rectifying K+ channel in human cardiac myocytes. Alterations in channel behavior in myocytes isolated from patients with idiopathic dilated cardiomyopathy. Circulation 92:164–174
Kuo HC, Cheng CF, Clark RB, Lin JJ, Lin JL, Hoshijima M, Nguyêñ-Trân VT, Gu Y, Ikeda Y, Chu PH, Ross J, Giles WR, Chien KR (2001) A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of I(to) and confers susceptibility to ventricular tachycardia. Cell 107(6):801–813
Kääb S, Dixon J, Duc J, Ashen D, Näbauer M, Beuckelmann DJ (1998) Molecular basis of transient outward potassium current downregulation in human heart failure: a decrease in Kv4.3 mRNA correlates with a reduction in current density. Circulation 98:1383–1393
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z (2008) Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem 283(29):20045–20052
Luo X, Xiao J, Lin H, Li B, Lu Y, Yang B, Wang Z (2007) Transcriptional activation by stimulating protein 1 and post-transcriptional repression by muscle-specific microRNAs of IKs-encoding genes and potential implications in regional heterogeneity of their expressions. J Cell Physiol 212(2):358–367
Nattel S, Maguy A, Le Bouter S, Yeh YH (2007) Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation. Physiol Rev 97:425–456
Radicke S, Cotella D, Graf EM, Banse U, Jost N, Varró A (2006) Functional modulation of the transient outward current Ito by KCNE beta-subunits and regional distribution in human non-failing and failing hearts. Cardiovasc Res 71:695–703
Remme CA, Verkerk AO, Hoogaars WM, Aanhaanen WT, Scicluna BP, Annink C, van den Hoff MJ, Wilde AA, van Veen TA, Veldkamp MW, de Bakker JM, Christoffels VM, Bezzina CR (2009) The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res Cardiol 104(5):511–522
Richard S, Leclercq F, Lemaire S, Piot C, Nargeot J (1998) Ca2+ currents in compensated hypertrophy and heart failure. Cardiovasc Res 37(2):300–311
Schröder F, Handrock R, Beuckelmann DJ, Hirt S, Hullin R, Priebe L, Schwinger RH, Weil J, Herzig S (1998) Increased availability and open probability of single L-type calcium channels from failing compared with nonfailing human ventricle. Circulation 98(10):969–976
Schwinger RH, Bundgaard H, Müller-Ehmsen J, Kjeldsen K (2003) The Na, K-ATPase in the failing human heart. Cardiovasc Res 57:913–920
Shang LL, Pfahnl AE, Sanyal S (2007) Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel. Circ Res 101:1146–1154
Sipido KR, Volders PG, Vos MA, Verdonck F (2002) Altered Na/Ca exchange activity in cardiac hypertrophy and heart failure: a new target for therapy? Cardiovasc Res 53:4–782
Stillitano F, Lonardo G, Zicha S, Varro A, Cerbai E, Mugelli A, Nattel S (2008) Molecular basis of funny current (If) in normal and failing human heart. J Mol Cell Cardiol 45(2):289–299
Tomaselli GF, Marbán E (1999) Electrophysiological remodeling in hypertrophy and heart failure. Cardiovasc Res 42:270–283
Valdivia CR, Chu WW, Pu J, Foell JD, Haworth RA, Wolff MR (2005) Increased late sodium current in myocytes from a canine heart failure model and from failing human heart. J Mol Cell Cardiol 38:475–483
Van Eylen F, Kamagate A, Herchuelz A (2002) Characterization and functional activity of a truncated Na/Ca exchange isoform resulting from a new splicing pattern of NCX1. Ann N Y Acad Sci 976(81):84
Vanderheyden M, Mullens W, Delrue L, Goethals M, de Bruyne B, Wijns W (2008) Myocardial gene expression in heart failure patients treated with cardiac resynchronization therapy responders versus nonresponders. J Am Coll Cardiol 51:129–136
Watanabe E, Yasui K, Kamiya K, Yamaguchi T, Sakuma I, Honjo H, Ozaki Y, Morimoto S, Hishida H, Kodama I (2007) Upregulation of KCNE1 induces QT interval prolongation in patients with chronic heart failure. Circ J 71(4):471–478
Yang B, Lin H, Xiao J, Lu Y, Luo X, Zhang Y (2007) The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med 13:486–491
Zicha S, Xiao L, Stafford S, Cha TJ, Han W, Nattel S (2004) Transmural expression of transient outward potassium current subunits in normal and failing canine and human hearts. J Physiol 561:735–748
Acknowledgments
This work was financially supported by The Danish National Research Foundation, The Danish Heart Foundation (ES), The Lundbeck Foundation (TJ), The Novo Nordisk Foundation (TJ and MG), The Aase and Ejnar Danielsen Foundation (MG), and The National Danish Research Council (MG). The authors wish to thank Ursula Ravens for manuscript revision and Victoria Szuts for supplying samples from nonfailing hearts.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary data
(DOC 30 kb)
Supplementary Table 1
(DOC 79 kb)
Supplementary Table 2
(DOC 74.5 kb)
Rights and permissions
About this article
Cite this article
Soltysinska, E., Olesen, SP., Christ, T. et al. Transmural expression of ion channels and transporters in human nondiseased and end-stage failing hearts. Pflugers Arch - Eur J Physiol 459, 11–23 (2009). https://doi.org/10.1007/s00424-009-0718-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-009-0718-3