Abstract
CA IX is an active transmembrane carbonic anhydrase isoform functionally implicated in cell adhesion and pH control. Human CA IX is strongly induced by hypoxia and frequently associated with various tumors. In this study, we investigated the expression of the rat CA IX in response to chronic hypoxia and to treatment with chemical compounds that disrupt oxygen sensing, including dimethyloxalylglycine, dimethylester succinate, diazoxide, and tempol. We brought the evidence that expression of CA IX is regulated by hypoxia and hypoxia-mimicking compounds in immortalized Rat2 fibroblasts and BP6 rat fibrosarcoma cells in a cell-type-specific manner. We also demonstrated, for the first time, that CA IX is expressed in hypoxic primary rat cardiomyocytes and in immortalized H9c2 cardiomyocytes exposed to physiological or chemical hypoxia and that CA IX expression is increased in hypoxic rat tissues in vivo. Our findings suggest that CA IX expression is not limited to cancer but may be also induced in other pathological situations associated with ischemia or metabolic disturbances leading to activation of the HIF pathway. These data support the view that rats can represent useful model for studies of CA IX as a component of endogenous protection mechanisms associated with hypoxia or perturbed oxygen sensing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carbonic anhydrase IX (CA IX) belongs to a large family of α-carbonic anhydrases, widely distributed zinc metalloenzymes that participate in ion transport and pH control in mammals. Sixteen α-CA isoforms display a considerable variability in their subcellular localization, distribution within the body, enzyme activity, affinity for inhibitors, and functional attributes. Twelve active isoenzymes catalyze reversible conversion of CO2 to bicarbonate ion and proton. This reaction takes place in their conserved catalytic domain that contains three histidine residues for the coordination of a zinc ion and a fourth histidine that functions as a proton shuttle [36]. Membrane-associated and cytoplasmic carbonic anhydrases cooperate with ion exchangers to facilitate efficient ion transport across the plasma membrane by forming protein complexes called metabolons [5].
CA IX is a transmembrane isoform mainly associated with tumors, in contrast to other CAs that are mostly present in differentiated cells of normal tissues. Under physiological conditions, CA IX expression is predominantly confined to the gastrointestinal epithelia [25]. On the other hand, CA IX is expressed in a broad range of tumors including carcinomas of the cervix uteri, breast, lung, colon, brain, kidney, and other types of neoplasia that are generally derived from tissues, which contain no or low level of CA IX [46, 27]. Moreover, the presence of CA IX in most tumor types is linked to poor prognosis [30], and the protein appears to be functionally implicated in tumorigenesis as a part of regulatory pathways that control pH and cell adhesion [38–40].
Expression pattern of CA IX is principally determined by strong activation of the CA9 gene transcription via hypoxia-inducible factor 1 (HIF-1) that binds to the hypoxia responsive element (HRE) localized in the CA9 promoter proximal to the transcription start site at the −10/−3 position [44].
HIF-1 is the main regulator of molecular responses to hypoxia [11]. It is a heterodimer that consists of an oxygen-sensitive α subunit and a constitutively expressed β subunit. Under normoxic conditions, HIF-1α is hydroxylated by prolyl hydroxylases (PHDs) on specific proline residues localized within an oxygen-dependent degradation domain. PHDs are able to transmit “pO2 information” directly to HIF-1α in the form of covalently attached OH− groups, which regulate the stability of HIF-1α [14, 15]. Von Hippel Lindau protein (pVHL) recognizes the prolyl hydroxylated form of HIF-1α and recruits ubiquitin ligase for its ubiquitination. HIF-1α is then destroyed in the proteasome. In hypoxia, HIF-1α subunit cannot be efficiently hydroxylated by PHDs, and thus, it is able to escape pVHL-mediated degradation in the proteasome. HIF-1α is then accumulated in the cytoplasm and translocated to the nucleus where it dimerizes with the constitutive β subunit to form the HIF transcription factor that binds to HRE elements in the promoters of hypoxia-inducible genes and induces their transcription [35]. The HIF pathway significantly changes the expression profile of weakly oxygenated tumor cells by activating genes that either support their survival and adaptation to hypoxia (like CA IX) or lead to their death. As a result, hypoxia selects more aggressive tumor cells that have increased capability to invade and metastasize and is therefore inherently associated with bad prognosis and poor response to anticancer therapy [11].
Increased level and transactivation capacity of HIF can also result from a genetic event, such as activation of oncogenic pathways (MAPK and/or PI3K) or inactivation of tumor suppressors (particularly the VHL gene) [35]. Furthermore, hypoxia-like response can be induced in normoxia by hypoxia mimetics (e.g., dimethyloxalylglycine) as well as by tricarboxylic acid cycle intermediates (e.g., succinate) and inhibitors (e.g., diazoxide) that interfere with PHD-mediated oxygen sensing [42]. Inactivation of PHDs with these chemical compounds leads to stabilization of HIF and to the transcriptional activation of its target genes.
CA IX belongs to the most hypoxia-responsive genes and was therefore proposed to serve as an intrinsic marker of hypoxia [30]. Studies of CA IX regulation revealed that its intratumoral expression pattern reflects the transcriptional activation of the CA9 gene by hypoxia, as well as the high posttranslational stability of the CA IX protein in reoxygenated cells, and the shedding of its ectodomain [31, 45]. It has been clearly shown that the HIF-mediated induction of CA IX involves transcriptional responses to chronic hypoxia and to activation of PI3K/MAPK pathways [44, 17, 21].
So far, hypoxic regulation of CA IX has been analyzed mostly in human cells and tissues in connection with cancer. Few papers describe CA IX expression in rat tissues and cultured cells, but its relationship to hypoxia has remained unexplored [46, 25]. In this paper, we show that rat fibroblast cell lines and cardiomyocytes express CA IX in response to hypoxia similarly to human cells. In addition, we proved that CA IX can be induced by chemical compounds that disrupt oxygen sensing by PHDs and that these compounds operate in a cell-type-specific manner. Finally, we also demonstrated that hypoxia induces expression of CA IX in rat tissues in vivo.
Experimental procedures
Cell culture and treatment
Rat2 thymidine kinase-deficient immortalized fibroblasts, BP6 fibrosarcoma cells (kindly provided by Dr. J. Zavada, Institute of Molecular Genetics, Prague, Czech Republic), and H9c2 cardiomyocytes (kindly provided by Prof. I. Morano, Charité University Medicine, Berlin, Germany) were cultivated in high glucose (4.5 g l−1) Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS; BioWhittaker, Verviers, Belgium) and 40 μg ml−1 gentamicin (Lek, Slovenia) in a humidified 5% CO2 incubator at 37°C and routinely split when 80–90% confluent. The cells were plated at 250,000 per 35-mm dish and allowed to adhere for 24 h. For hypoxic treatment, cells were incubated within a hypoxic workstation (Ruskin Technologies, Bridgend, UK) in a mixture of gases containing 2% O2, 5% CO2, 10% H2, and 83% N2 at 37°C for 48 h. Dimethyloxalylglycine (DMOG), an inhibitor of PHDs, was purchased from Frontier Scientific (Logan, UT) and used at 1 mmol l−1 concentration. All other chemical compounds were from Sigma (St. Louis, MO, USA) and were used at the following concentrations: 20 mmol l−1 dimethylester succinate (DMS), 0.5 mmol l−1 7-chloro-3-methyl-2H-1,2,4-benzothiadiazine 1,1-dioxide (diazoxide, DZO), and 2.5–10 mmol l−1 4-hydroxy-2,2,6,6-tetramethyl piperidinoxyl (tempol, TL). The compounds were added to cells 24 h after plating, and the incubation time was 48 h.
Preparation and cultivation of primary rat cardiac myocytes
Preparation of isolated rat cardiomyocytes has been described previously [24]. Briefly, the heart was removed from adult Wistar rat anesthetized with diethyl ether, immediately attached to an aortic cannula, and perfused at constant pressure (80 mmHg) with Krebs–Henseleit physiological salt solution (118 mmol l−1 NaCl, 4.5 mmol l−1 KCl, 24 mmol l−1 NaHCO3, 5 mmol l−1 glucose, 1.2 mmol l−1 KH2PO4, 1.2 mmol l−1 MgCl2, 20 mmol l−1 taurine, and 1.2 mmol l−1 CaCl2) that was equilibrated in an atmosphere of 95% O2 and 5% CO2 and maintained at 37°C throughout the experiment. After 5 min, the solution was replaced by Krebs–Henseleit solution containing 5 mmol l−1 CaCl2 for 5 min, and subsequently to a solution containing 5 mmol l−1 CaCl2 and 0.9% liberase (Roche Applied Science, Mannhein, Germany) for 15 min. The ventricle was then removed, cut into small pieces, and shaken in Krebs–Henseleit physiological salt solution containing 0.8% albumin and 50 mmol l−1 CaCl2. The released cells were collected by centrifugation in Krebs–Henseleit salt solution containing 0.8% albumin and 1.2 mmol l−1 CaCl2.
The isolated cells were placed into flasks that had been pre-coated with 0.02% gelatin/fibronectin (Sigma) in an incubator for 2 days and kept in EX-CELL 320 medium with HEPES and l-glutamine designed for cardiomyocytes (JRH Biosciences, Lenexa, KS, USA). Insulin (15 μg ml−1), Penstrephten (100 U ml−1), non-essential amino acids (0.1 mmol l−1), retinoic acid (1 mmol l−1), and 10% FCS were added (all from Invitrogen, Carlsbad, CA, USA). The cells were allowed to settle for 2 days, and then the medium was changed every second day. The cells were able to grow continually for several weeks. Experiments were performed using cultures grown for at least 3 weeks in strictly controlled conditions. At that moment, only cardiomyocytes were present in the culture.
Animals
Adult male Wistar rats were kept at room temperature for 6 h either outside or inside a hypoxic workstation (Ruskin Technologies, Bridgend, UK) in a mixture of gases containing 8% O2, 1% CO2, 10% H2, and 81% N2. The animals were killed, perfused transcardially with 4% paraformaldehyde in 0.1 mol l−1 phosphate buffered saline pH 7.2, and either frozen in liquid nitrogen for RNA isolation and reverse transcription–polymerase chain reaction (RT-PCR) analysis, or fixed in paraformaldehyde, embedded in paraffin, and cut on Leica RM2125 microtome for immunohistochemistry.
Antibodies and plasmids
M75 mouse monoclonal antibody specific for the N-terminal proteoglycan-like region of the human and rat CA IX protein was described previously [25, 47]. Anti β-actin mouse monoclonal antibody was purchased from Sigma. Secondary anti-mouse antibody conjugated with horseradish peroxidase was purchased from Sevapharma (Prague, Czech Republic). Anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody was from Vector Laboratories (Burlingame, CA, USA). Rabbit anti HIF-1α antibody was purchased from Novus Biologicals (Littleton, CO, USA), and secondary anti-rabbit antibody conjugated with horseradish peroxidase was from Sevapharma.
The PR1-HRE-luc plasmid containing the human Ca9 core promoter was generated by an insertion of a −50/+37 CA9 genomic region amplified by PCR upstream of the firefly luciferase gene in pGL3-Basic luciferase reporter vector (Promega, Madison, WI, USA) [21]. Renilla pRL-TK vector (Promega) served as a control for transfection efficiency. Human HIF-1α complementary DNA (cDNA) and mouse HIF-2α cDNA, respectively, cloned in pcDNA1/Neo expression plasmid were kindly provided by Prof. Patrick Maxwell, Imperial College of Science, Technology and Medicine, London, UK. Empty pcDNA1/Neo plasmid was used as a negative control for mock transfection.
Expression plasmid pcDNA1-mutHIF1α coding for deletion mutant of HIF1α was created by PCR from the original expression plasmid pcDNA1-HIF1α. Part of the oxygen-dependent degradation domain (aa 402–564) was deleted using the following primers: F 5′-TATATCCCAATGGATGATGACTTCC-3′ and R 5′-GGCCAGCAAAGTTAAAGCATC-3′. Resulting PCR product was gel-purified, phosphorylated, and ligated. Before transfection experiments, the plasmid was sequenced to ensure that the open reading frame of the deletion mutant of HIF1α was unaltered.
Transient transfection and reporter gene assay
Cells were plated into 35-mm Petri dishes to reach approximately 70% monolayer density on the following day. Transfection was performed with 1 μg of PR1-HRE-luc and 50 ng of pRL-TK plasmid DNAs using GenePorterII reagent (Genlantis, San Diego, CA, USA) according to the manufacturer’s recommendations. To analyze the effect of the wild-type HIF1α and its deletion mutant, the plasmids containing corresponding cDNAs were co-transfected with PR1-HRE-luc and pRL-TK. On the following day after transfection, transfected cells were trypsinized and plated in quadruplicates into 24-well plates. Cells were allowed to attach for 20 h, and then they were transferred to hypoxia for additional 24 h. Reporter gene expression was assessed using Dual-Luciferase Reporter Assay System (Promega), and luciferase activity was normalized against renilla expression.
RNA isolation and reverse transcription PCR
Total RNA was isolated from cultured cells or frozen tissues using InstaPure (Eurogentec, Seraing, Belgium) according to the manufacturer’s instructions. Reverse transcription was performed with M-MuLV reverse transcriptase (Finnzymes, Espoo, Finland) and random heptameric primers (400 ng μl−1). The mixture of 3 μg total RNA and random primers (400 ng μl−1) was heated for 10 min at 70°C, cooled on ice, and supplemented with dNTPs (each at 0.5 mmol l−1 concentration, Finnzymes), M-MuLV reverse transcriptase buffer (Finnzymes), and with 200 U of reverse transcriptase M-MuLV, incubated for 1 h at 42°C, heated to 70°C for 15 min, and stored at −80°C until further use.
PCRs were performed with Taq polymerase (Promega) in an automatic DNA thermal cycler (Eppendorf AG) using gene-specific primers and primers for β-actin that served as internal standards (Table 1). The amplified products were obtained in the exponential phase (30–35 cycles) of PCR. The PCR protocol consisted of 94°C for 3 min followed by 30–35 cycles of: denaturation at 94°C for 30 s, annealing for 40 s (T m in Table 1) and extension at 72°C for 40 s, followed by final extension at 72°C for 5 min. Resulting PCR fragments were run on 1.2% agarose gels, and their identity was verified by sequencing. The intensity of bands corresponding to individual PCR products was evaluated with ImageJ 1.34s software (Rasband, W.S., ImageJ, US National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997–2007). ImageJ can create density histograms and calculate area and pixel value statistics of user-defined selections. Amount of gene-specific PCR products was semiquantitatively expressed as the ratio of the intensity of each band to the intensity of the related β-actin internal standard.
Immunofluorescence
Cells grown on glass coverslips were fixed in ice-cold methanol at −20°C for 5 min. Non-specific binding was blocked by incubation with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA; PBS–BSA) for 30 min at 37°C. Subsequently, the cells were incubated with M75 antibody (undiluted hybridoma medium) for 1 h at 37°C, washed four times in PBS with 0.2% Tween 20 (Sigma) for 10 min, incubated with anti-mouse FITC-conjugated horse antibody diluted 1:300 in PBS–BSA for 1 h at 37°C, and washed as before. Finally, the cells were mounted onto slides in the Fluorescent Mounting Media (Calbiochem, Darmstadt, Germany), analyzed with Leica DM4500B microscope and photographed with Leica DFC480 camera.
Immunoblotting
For analysis of CA IX expression, proteins were extracted from cell monolayers with RIPA buffer (1% Triton X-100, 0.1% sodium deoxycholate, 1 mmol l−1 phenylmethylsulfonyl fluoride in PBS) containing inhibitors of proteases Complete mini (Roche Applied Science) for 30 min on ice. The extracts were then centrifuged for 15 min at 13,000 rpm in Eppendorf microcentrifuge, and total protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce, Rockford, IL, USA) according to the manufacturer’s instructions. The extracts (equal aliquots containing 40–80 μg of total proteins depending on cell type) were separated in 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). After separation, the proteins were blotted onto polyvinylidene fluoride (PVDF) membrane (Immobilon™-P, Millipore, Billerica, MA, USA). The membrane was treated with blocking buffer containing 5% non-fat milk in PBS with 0.2% Nonidet P-40 (Sigma) for 1 h and then incubated for 1 h with the primary antibody diluted in blocking buffer (M75 monoclonal antibody in hybridoma medium diluted 1:2). After treatment, the membrane was washed in PBS with 0.2% Nonidet P-40 three times for 10 min, incubated for 1 h with swine anti-mouse antibody conjugated with horseradish peroxidase (Sevapharma) diluted 1:7,500 in blocking buffer. After repeated washing, the membrane was developed with ECL detection system.
For HIF-1α analysis, proteins were extracted from cell monolayers with urea buffer (10% glycerol, 1% SDS, 5 mmol l−1 DTT, 1 mmol l−1 Tris pH 6,8, 1 mmol l−1 phenylmethylsulfonyl fluoride) containing inhibitors of proteases Complete mini (Roche Applied Science) for 5 min on ice. The extracts were homogenized with IKA T10 basic homogenizer, centrifuged for 15 min at 13,000 rpm, and total protein concentration was determined by BCA assay (Pierce). The extracts (equal aliquots containing 100 μg of total proteins) were separated in 8% SDS-PAGE, and proteins were blotted onto PVDF membrane. The membrane was treated as above, except using 0.2% Tween 20 (Sigma) instead of Nonidet P-40. Primary anti-HIF-1α antibody was diluted 1:500 in 3% BSA in PBS with 0.2% Tween 20, and goat anti-rabbit antibody conjugated with horseradish peroxidase (Sevapharma) was diluted 1:5,000. After treatment, the membrane was washed and developed with ECL detection system. The intensity of protein bands was evaluated with ImageJ 1.34s software as described above.
Enzyme-linked immunosorbent assay
Microplate wells were coated overnight at 37°C with RIPA extracts from control and treated cells and diluted in PBS so that the individual samples contained equal amount of total proteins. Due to a high basal expression of CA IX in Rat2 cells, Rat2 samples contained half the amount of total proteins of BP6 and H9c2 samples. After blocking with 10% FCS in PBS for 30 min, the coated wells were incubated with peroxidase-labeled M75 monoclonal antibody for 1 h at room temperature and then washed with PBS containing 0.05% Tween 20. The amount of bound antibody was determined with a peroxidase substrate orthophenylene diamine (Sigma).
Immunohistochemistry
Deparaffinized 4-μm tissue sections were stained using the M75 antibody (dilution 1:50), and immunoreaction was visualized with UltraTech HRP streptavidin–biotin universal detection system (Immunotech, France). The specimens were counterstained with hematoxylin, examined with Leica DM4500B microscope and photographed with Leica DFC480 camera.
Results
Hypoxia-induced expression of CA IX in rat fibroblasts
Before the analysis of hypoxia-inducible expression of rat CA IX, we examined the capability of two rat fibroblast cell lines, namely Rat2 immortalized fibroblasts and BP6 fibrosarcoma cells, to sense hypoxia. For this purpose, we used the PR1-HRE-luc luciferase construct containing core promoter of the human CA9 gene that has been thoroughly studied before and was proven to bind HIF and display strong transcriptional activation by hypoxia [44, 21]. Moreover, human and rat promoters are almost identical in respect to sequence and position of the binding sites for the critical transcription factors (data not shown). The cells were transiently transfected with PR1-HRE-luc plasmid and incubated in parallel under normoxic and hypoxic conditions for 24 h. Hypoxia significantly induced luciferase activity in both cell lines, suggesting that they do sense low oxygen and are competent to trigger a transcriptional response (Fig. 1a). BP6 fibrosarcoma cells showed strong responsiveness (more than fivefold), whereas the induction of luciferase activity driven by the CA9 promoter in Rat2 cells was weaker (less than twofold). On the other hand, Rat2 cells exhibited approximately six times higher normoxic CA9 promoter activity (calculated as luciferase to renilla ratio) when compared to BP6 cells. Despite their lower hypoxic induction, the final transcriptional activity of the CA9 promoter was four times higher in Rat2 cells (data not shown).
Hypoxia-induced CA IX expression in rat fibroblast cell lines. a Rat2 and BP6 cells were transiently co-transfected with a luciferase construct containing hypoxia-responsive core promoter of the human CA9 gene and renilla plasmid. The cells were incubated in normoxia and hypoxia, respectively, for 24 h. The luciferase activity was normalized by renilla values and then expressed as fold of induction by hypoxia. b Transcription of Car9 gene was analyzed in normoxic (N) vs hypoxic (H) Rat2 and BP6 cells by reverse transcription PCR analysis in comparison with β-actin gene that was used as an internal standard and with HIF-1α and HIF-2α genes. c Immunoblotting and d immunofluorescence analysis of CA IX protein expression in Rat2 and BP6 cells incubated in normoxia (N) and hypoxia (H) for 48 h. Scale bars correspond to 50 μm. Relative levels of Car9 transcript and CA IX protein are given below the relevant figures
Results from RT-PCR analysis of the rat Car9 gene transcription in normoxia and hypoxia were in line with the above data. Rat2 cells expressed Car9 gene also under normoxia, and its hypoxic induction was less pronounced than the induction observed in the BP6 cell line that showed no or very low normoxic level of the Car9 transcript (Fig. 1b). Interestingly, Rat2 cells expressed lower level of HIF-1α transcript than BP6 cells. HIF-2α transcription could not be detected in any of these two fibroblast cell lines.
CA IX protein expression was examined by immunoblotting using the M75 monoclonal antibody. In accord with earlier data, rat CA IX was detected as a single 53.5-kDa band [46]. Immunoblotting results corresponded well with the levels of RT PCR Car9 products in both cell lines (Fig. 1c). Expectedly, Rat2 cells also displayed normoxic expression of CA IX and its increase under hypoxia was relatively weak, whereas BP6 cells were CA IX-negative in normoxia and hypoxia strongly induced their CA IX expression.
Immunofluorescence analysis demonstrated that the protein is localized on the cell membrane of both cell lines (Fig. 1d), although cytoplasmic staining was also visible similarly as in various hypoxia-treated human tumor cells.
Altogether, these results show that hypoxia can induce the expression of rat CA IX in the fibroblast cell lines.
Expression of CA IX in rat fibroblast cell lines treated with compounds that disrupt oxygen sensing
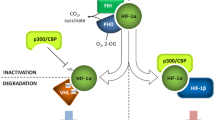
Stability of HIF-1α is principally controlled by PHDs that utilize molecular oxygen as a co-substrate for hydroxylation of critical prolines in its oxygen-dependent degradation domain. Low oxygen supply under hypoxia reduces the catalytic activity of PHDs leading to accumulation of HIF-1α and activation of its downstream target genes. However, activation of the HIF-1 pathway can be also achieved in the presence of oxygen by chemical inhibition of PHDs. Here, we wanted to compare the effects of selected PHD inhibitors on expression of rat CA IX. Based on different mode of their action, we chose the compounds depicted in Fig. 2a. These included DMOG, an analogue of the PHD cofactor 2-oxoglutarate (2-OG), [22], DMS, a membrane-permeable derivative of succinate that blocks decarboxylation of 2-OG [34], DZO that inhibits conversion of succinate to fumarate by succinate dehydrogenase leading to increased levels of succinate [4], and TL, a superoxide dismutase mimetic proposed to act towards reactive oxygen species (ROS) that were reported to inhibit PHD activity by oxidizing the PHD cofactors ferrous iron and ascorbate [8].
Mechanisms of action of the selected chemical compounds that disrupt oxygen sensing and their effect on transcription of Car9 gene in Rat2 and BP6 cell lines. a Schematic illustration of the HIF pathway and its modulation by chemical compounds. Under low oxygen supply (small O2), prolyl hydroxylases (PHD) cannot hydroxylate HIF-α subunit, which accumulates in cytoplasm, translocates to nucleus, dimerizes with HIF-β, binds to Car9 promoter, and induces its expression. Normal oxygen supply (large O2) causes hydroxylation of HIF-α and its degradation in proteasome. Treatment of normoxic cells with diazoxide (DZO) inhibits succinate dehydrogenase, leading to high level of succinate that inhibits PHDs. Inhibition of PHDs can be achieved also by dimethylester succinate (DMS), a membrane-permeable derivative of succinate that blocks decarboxylation of a PHD cofactor 2-oxoglutarate (2-OG), as well as by dimethyloxalglycine (DMOG), an analogue of 2-OG. Finally, tempol (TL) was proposed to modulate PHDs as a scavenger of oxygen and hydroxyl radicals. b RT-PCR analysis of Car9 transcription in Rat2 and BP6 cells treated for 48 h by hypoxia-mimicking chemical compounds. β-actin was used as an internal control. Numbers given below the figures represent relative levels of PCR products
An insight into transcription of Car9 in rat cell lines treated with these inhibitors revealed remarkable cell-type-related differences (Fig. 2b). Immortalized Rat2 fibroblasts responded with an increased Car9 transcription to all examined inhibitors. The increase in Car9 messenger RNA (mRNA) levels was similar in all cases and roughly corresponded to its induction by physiological hypoxia. A particularly strong response was obtained with TL and a combination of TL with DZO. In contrast, BP6 fibrosarcoma cells responded well only to DMOG and TL, whereas inhibitors of mitochondrial tricarboxylic acid (TCA) cycle did not affect the expression of Car9. These data suggest that the cell lines differ by their sensitivity to compounds that disrupt sensing of oxygen.
CA IX protein levels visualized by immunoblotting and quantitatively assessed by enzyme-linked immunosorbent assay (ELISA) principally reflected the amounts of RT-PCR products (Fig. 3a, b). Analysis of a dose response in Rat2 cells showed that increased concentrations of the inhibitors generally induced higher levels of CA IX with an exception of 0.5 mmol l−1 DZO combined with 10 mmol l−1 TL that induced CA IX to a lesser extent than less concentrated combinations of these compounds, apparently due to cell toxicity (Fig. 3c). Figure 3d, e shows that the changes in the levels of HIF-1α protein detected by immunoblotting roughly corresponded with the induction of CA IX protein, although few discrepancies could be observed. Noteworthy, CA IX protein was expressed in the normoxic Rat2 cells despite no detection of HIF-1α protein (in accord with the low level of HIF-1α transcript in these cells), supporting the view that hypoxia and inhibitors do influence CA IX via HIF pathway, but other mechanisms also seem to play a role.
CA IX protein and HIF-1α subunit expression in rat fibroblasts. Expression of CA IX protein in Rat2 (a) and BP6 (b) cells treated for 48 h by chemical compounds was assessed by ELISA and simultaneously evaluated by immunoblotting. The amount of CA IX protein in the individual samples of ELISA was expressed as folds of induction of CA IX level compared to normoxia. Relative levels of CA IX protein in immunoblotting (calculated toward β-actin internal control) are given below the relevant figures). c Dose-response analysis of CA IX expression in Rat2 cells. Numbers given after abbreviations indicate the concentrations (mmol l−1) in which the compounds were applied. Immunoblotting analysis of HIF-1α protein levels in Rat2 (d) and BP6 cells (e) treated for 48 h with the chemical compounds. Relative levels of HIF-1α protein calculated to β-actin are shown below the blots. The experiments were repeated three to five times
Expression and induction of other HIF target genes at the level of transcription
We also wanted to learn about the response of additional HIF targets to treatment with chemical compounds that disrupt oxygen sensing. Thus, the RT-PCR analysis was extended to some other genes known to be regulated by hypoxia via HIF. This experiment disclosed further differences between the studied cell lines. Immortalized Rat2 cells exhibited relatively high normoxic expression and moderate hypoxic induction of vascular endothelial growth factor (VEGF) and glucose transporter (GLUT-1). VEGF was slightly induced also by DMOG, but was well induced by TL and DZO combined with TL. DMS and DZO alone did not show any effect on VEGF transcription. GLUT-1 responded only to physiological hypoxia, TL, and DZO + TL. No induction with any inhibitor was observed in the case of GLUT-4. Normoxic level of the inducible nitric oxide synthase (iNOS) transcript was very low, but its expression was slightly induced by physiological hypoxia, DMOG, TL, and DZO + TL. Finally, monocaboxylate transporter (MCT4) was undetectable in Rat2 cells and could not be induced by any of the compounds (Fig. 4a).
Expression of hypoxia-inducible genes in response to treatment with hypoxia-mimicking chemical compounds. The cDNAs isolated from Rat2 (a) and BP6 (b) cells, analyzed above for expression of Car9 as shown in Fig. 2b, were also used for amplification of additional hypoxia-regulated genes. The data show cell- and gene-specific differences in the normoxic levels as well as in responses to various hypoxia mimetics. Numbers given below the figures represent relative levels of PCR amplicons
BP6 cells exhibited a different expression pattern. They contained relatively high levels of normoxic VEGF as well as iNOS (but not GLUT-1, GLUT-4, and MCT4), and their hypoxic induction was weak. In contrast to CA IX, VEGF did not respond to any of the treatments. GLUT-4 was weakly induced by DZO + TL. MCT4 was detectable under normoxia, and its levels were increased in cells treated by hypoxia, DMOG, and TL. Furthermore, transcription of iNOS appeared reduced in response to DMS, DZO + TL, and TL (Fig. 4b).
These results indicate that the cell lines display a differential expression pattern with respect to basal and induced levels of hypoxia-regulated genes and that they respond inconsistently to various hypoxia mimicking compounds.
Hypoxia-regulated expression of CA IX in rat cardiomyocytes
As an aerobic organ, the heart is very susceptible to lack of oxygen. Therefore, we decided to examine CA IX expression in primary cardiomyocytes isolated from the adult rat heart and maintained in culture for 3 weeks before the experiment. The myocytes were incubated in parallel in normoxia and hypoxia (2% O2) for 48 h, and CA IX expression was analyzed by RT-PCR, immunoblotting, and immunofluorescence (Fig. 5). As demonstrated by the first two methods, CA IX was present already in the normoxic cardiomyocytes, while the exposure to 48-h hypoxia resulted in its roughly twofold induction (Fig. 5a, c). Immunofluorescence analysis of the hypoxic culture revealed a sporadic but clearly visible plasma membrane staining, whereas no significant staining signal was detectable in the normoxic control (Fig. 5d). Hypoxic induction of Car9 transcription was accompanied by a considerably increased expression of GLUT-1, iNOS, and MCT4, whereas VEGF was relatively well expressed also under normoxia and only slightly elevated in hypoxia (Fig. 5b). In contrast to Rat2 and BP6 fibroblasts, both HIF-1α and HIF-2α were present in isolated cardiomyocytes (Fig. 5a). Interestingly, HIF-1α transcription was also increased in hypoxic conditions.
Expression of CA IX in the primary cardiomyocytes isolated from adult rat heart, cultured for 3 weeks and subjected to 48-h hypoxia. a RT-PCR analysis of Car9 transcription in response to hypoxia and in relationship to mRNAs of HIF-α subunits. β-actin was used as an internal control. b RT-PCR analysis of expression of additional hypoxia-regulated genes using the same cDNAs as in a. c Immunoblotting analysis of CA IX protein expression in normoxic (N) and hypoxic (H) primary cardiomyocytes. Numbers given below the figures represent relative levels of CA IX protein calculated to β-actin. d Immunofluorescence analysis of subcellular localization of CA IX protein. The cells were stained with M75 monoclonal antibody followed by FITC-conjugated secondary antibody (green signal). Cell nuclei were stained with propidium iodide (red staining). The CA IX protein was clearly localized at the cell membrane of hypoxic cardiomyocytes (see the magnified area). Scale bars correspond to 50 μm
We also analyzed CA IX expression in H9c2 cell line, which is an established model for physiological and molecular studies of cardiomyocytes. These cells exhibited strong hypoxic induction of CA IX that was clearly visible at the level of promoter activity (Fig. 6a), transcription (Fig. 6b, c), and protein (Fig. 6d). The induction appeared to be mediated by HIF-1α, as the intrinsic level of HIF-2α transcript was almost undetectable. Car9 transcription and CA IX protein expression in H9c2 cells were induced also by treatment with hypoxia mimetics such as DMOG and, to a lesser extent, with TL (Fig. 6c, d). Similarly to the other cell models, hypoxic H9c2 cells exhibited cell surface localization of the rat CA IX protein (Fig. 6e). Finally, RT-PCR analysis of the other selected hypoxia-regulated genes revealed that H9c2 cells display relatively high level and no or weak induction of VEGF and GLUT-4, very low normoxic level and clear induction of GLUT-1, no normoxic expression and strong induction of MCT4 (but only weak induction of MCT4 by DMOG and TL), and undetectable iNOS (Fig. 6f). These data indicate that in cardiomyocytes, CA IX represents an integral part of molecular responses to physiological hypoxia and to chemical disruption of oxygen sensing.
Expression of CA IX in H9c2 immortalized cardiomyocytes treated by hypoxia and hypoxia mimetics. a H9c2 cells were transiently co-transfected with a luciferase construct containing hypoxia-responsive core promoter of the human CA9 gene and renilla plasmid. The cells were incubated in normoxia and hypoxia, respectively, for 24 h. The luciferase activity was normalized by renilla values and then expressed as fold of induction by hypoxia. b Transcription of Car9 and HIF-α subunits was analyzed in normoxic (N) vs hypoxic (H) H9c2 cells by reverse transcription PCR analysis in comparison with β-actin gene that was used as an internal standard. Car9 gene expression was analyzed by RT-PCR (c), ELISA and immunoblotting (d) where CA IX levels are expressed in relative terms as fold of induction related to normoxia (ELISA) or in numbers relative to β-actin. e Immunofluorescence in H9c2 cells incubated in normoxia (N) and hypoxia (H) for 48 h (scale bars correspond to 50 μm). f RT-PCR analysis of hypoxia-inducible genes in response to treatment with hypoxia-mimicking chemical compounds. The experiments were repeated three times. Numbers given below the figures represent relative levels of PCR products or CA IX protein calculated to β-actin
HIF-1α-dependent hypoxic induction of Car9 gene in rat cells
As shown on Fig. 1b, inspection of intrinsic HIF-1α expression disclosed lower level of HIF-1α transcript in Rat2 cells compared to BP6 and H9c2 cells, but HIF-2α transcription could not be detected in any of these cell lines. This fact suggested that HIF-1α component of the HIF transcription factor is responsible for the hypoxic induction of Car9 gene in these cells. This assumption was verified by transfection of Rat2 cells with HIF-1α and HIF-2α cDNA, respectively. In this experiment, Car9 transcription was induced only after transfection of HIF-1α cDNA, but not of HIF-2α cDNA (Fig. 7a).
Effect of HIF-1α on Car9 transcription and promoter activity in rat cell lines. a Role of HIF-α subunits in hypoxic induction of Car9 gene examined by RT PCR in Rat2 cells transiently transfected with HIF-1α and HIF-2α cDNA, respectively. Numbers given below the figures represent relative levels of PCR products or CA IX protein calculated to β-actin. b Schematic drawing of the wild-type HIF-1α (wt HIF-1α) domain composition with indicated region deleted in the mutant (mut HIF-1α). c CA9 promoter activity analyzed in Rat2, BP6, and H9c2 cells transiently co-transfected with a luciferase construct containing hypoxia-responsive core promoter of the human CA9 gene, renilla plasmid, and with wt HIF-1α and mut HIF-1α cDNA, respectively. The cells were incubated in hypoxia for 24 h. The luciferase activity was normalized by renilla values and then expressed as fold of induction mediated by wt HIF-1α or the transactivation-defective mutant (mut HIF-1α)
To obtain further evidence supporting the HIF-1α role in regulation of rat Car9 gene, we have generated cDNA encoding the mutated form of HIF-1α with deletion of a large part of oxygen-dependent degradation domain including the N-TAD domain (Fig. 7b). As this HIF-1α mutant can still dimerize with HIF-1β and bind to DNA but cannot properly transactivate the HIF-1α target genes, it should abolish or at least reduce the induction of CA9 promoter. Indeed, in all three rat cell lines incubated under hypoxia, transfection of the wild-type HIF-1α cDNA resulted in increased promoter activity, whereas the mutated HIF-1α returned the promoter activity even below the control mock-transfected values (Fig. 7c).
Hypoxia-induced expression of CA IX in rat tissues
All the above experiments showing hypoxic induction of CA IX were performed using cultured cells. To prove that CA IX can be induced in normal rat tissues in vivo, we performed RT-PCR analysis of the heart, stomach, kidney, and brain obtained from the rat kept for 6 h in a hypoxic workstation containing 8% of oxygen in comparison with the tissues from the control normoxic rat. All examined hypoxic tissues revealed higher levels of Car9-specific PCR product when compared to the normoxic counterparts (Fig. 8a). In addition, immunohistochemical staining of the tissue sections from the heart and stomach clearly showed an increased CA IX-specific staining signal in response to hypoxia. In agreement with previous immunohistochemical data obtained in normoxic rat tissues [25], CA IX was well expressed in the membranes of the stomach epithelial cells already under normoxia, but the intensity of the staining signal was much stronger in the hypoxic stomach (Fig. 8b). On the other hand, CA IX was undetectable in the normoxic heart, but the faint CA IX-specific signal was obtained in the hypoxic heart and appeared predominantly associated with the membranes of the cardiac myocytes.
Expression of CA IX in normal tissues from rats exposed to hypoxia vs normoxia. a RT-PCR analysis of Car9 transcription in tissues from rat exposed for 6 h to systemic hypoxia (8% O2) in comparison to normoxic rat tissues. Numbers given below the figures represent relative levels of Car9-specific PCR products calculated to β-actin that was used as an internal control. b Immunohistochemical staining of CA IX protein in the stomach and heart. Tissue sections were stained with M75 monoclonal antibody. CA IX protein was clearly localized at the cell membrane of gastric epithelial cells (arrows) in both stomach specimens, but the intensity of signal is higher in the hypoxic tissue. On the other hand, no signal is visible in the normoxic heart, but the hypoxic cardiac myocytes show a weak membrane staining (see the magnified area)
Discussion
During the past period, carbonic anhydrase IX has been intensely studied as a marker of tumor hypoxia with a potential diagnostic and prognostic value [30, 26, 27]. Numerous papers describe CA IX distribution in various human tumors and its relationship to clinical parameters. CA IX expression pattern has been confronted with that of other hypoxia-regulated proteins, such as HIF-1α, VEGF, GLUT-1, etc. and with hypoxia-related phenomena, such as tumor necrosis and microvascular density. In addition, a considerable attention has been paid to molecular aspects of CA IX, particularly to its regulation at the transcriptional and posttranslational levels, and to its functional attributes in cell adhesion and in pH regulation [38–40]. All these studies contributed to the recent view of CA IX as a cancer-associated protein that is strongly induced by severe-to-moderate chronic hypoxia and exhibits high posttranslational stability during reoxygenation. They also shed light on the biological relevance of the catalytic activity of CA IX that participates in the bicarbonate transport metabolon and helps to acidify extracellular pH and to maintain neutral intracellular pH in hypoxic conditions [39, 40, 23].
Although to a much lesser extent, CA IX has been also investigated in rodents, mostly with respect to its distribution in the normal rat and mouse tissues, to its expression in rodent cell lines, and to biological consequences of targeted disruption of the Car9 gene in mouse [25, 10, 12, 13, 16]. The data obtained in this area revealed some differences in expression patterns in the normal human vs rodent tissues and demonstrated a physiological significance of CA IX in correct development of the mouse stomach, especially in the control of differentiation and proliferation of the gastric cell lineages [10]. However, the information on hypoxic regulation of CA IX in cells of rat origin and on its possible relationship to cancer-independent pathologies is completely missing. This fact motivated us to perform the present study in which we wanted to investigate: (1) whether CA IX responds to hypoxia in rat cell lines, (2) whether CA IX is expressed and hypoxia-regulated directly in cardiomyocytes representing a non-cancerous cell type frequently affected by ischemia-associated low oxygen supply, (3) whether CA IX expression can be induced in normoxia by chemical compounds that disrupt oxygen sensing, (4) whether physiological and chemical induction of CA IX is accompanied by induction of other hypoxia-regulated genes, and (5) whether hypoxia induces CA IX expression in non-cancerous rat tissues in vivo.
CA IX responsiveness to chronic hypoxia was proven in three rat cell lines at the level of promoter activity, transcription, and protein expression. However, the cell lines differed in the normoxic expression of CA IX and in magnitude of its hypoxic induction. In the latter respect, H9c2 cardiomyocytes were more similar to BP6 fibrosarcoma cells than to Rat2 immortalized fibroblasts possibly due to higher intrinsic levels of HIF-1α transcripts. Transfection experiments using transactivation-defective HIF-1α mutant with deletion in ODDD and N-TAD regions further supported role of HIF-1α in hypoxic induction of CA IX in rat cell lines. This is in accord with the findings that human CA IX is an exclusive HIF-1 target [9, 32] and suggests that this relationship is evolutionarily conserved. However, disagreement between absence of HIF-1α and presence of CA IX in the normoxic Rat2 cells suggests involvement of additional pathway(s) in CA IX regulation.
It is well known that degradation of HIF-1α under normoxia is mediated by PHDs that function as oxygen sensors, and that their inhibition due to lack of oxygen leads to HIF-1α accumulation and transactivation of its target genes [14, 15]. However, inhibition of PHDs and activation of the HIF pathway can occur not only due to hypoxia but also due to genetic and metabolic diseases that perturb mitochondrial TCA cycle. The most obvious examples represent paragangliomas, hereditary leiomyomatosis, and renal cancer syndrome that result from inactivation of two tumor suppressor genes, namely succinate dehydrogenase (SDH) and fumarate hydratase, respectively [7, 34, 28, 29, 20]. The encoded enzymes are responsible for conversion of succinate to fumarate and fumarate to malate, respectively, and their functional inactivation leads to abnormal accumulation of succinate that moves freely between the mitochondrion and the cytosol [34]. Increased cytosolic succinate back-feeds the hydroxylation reaction and inhibits PHDs (see the scheme in Fig. 2a).
Apart from the TCA cycle intermediates, ROS have been also proposed to participate in “mitochondrial oxygen sensing” [42]. As SDH plays an additional role in Complex II of the electron transport chain, its inactivation might lead to a buildup of ROS via chemical reaction of the stalled electrons with molecular oxygen [1]. ROS were reported to inhibit PHD activity under normoxic conditions by oxidizing the PHD cofactors ferrous iron and ascorbate [8].
In contrast to the aforementioned studies that focused on individual components of the pathways leading to normoxic stabilization of HIF-1α in the relevant genetic/metabolic context, we wanted to compare the effect of several chemical compounds mimicking hypoxia via inhibition of PHDs on the expression of CA IX in our rat cell lines. DMS, a membrane-permeable derivative of succinate, induced CA IX expression in Rat2 cells, but not in BP6 and H9c2 cells. Similar effect was observed with diazoxide. At 0.1 mmol l−1 concentration, DZO is known to stimulate translocation of PKC-ɛ to mitochondrion followed by opening of mitochondrial KATP channel [18]. However, a much higher concentration (0.5 mmol l−1) reported to inhibit SDH is needed to increase the expression of CA IX, indicating that CA IX induction in Rat2 cells is indeed due to inhibition of SDH.
Differential effects of TCA cycle inhibitors in Rat2 cells vs BP6 and H9c2 cells might be explained by the observation that the latter two cell lines are more sensitive to hypoxia (according to inducibility of CA9 promoter) and possibly less dependent on TCA cycle due to more efficient switch to anaerobic glycolysis. This is in line with much higher lactate production by BP6 cells when compared to Rat2 cells (data not shown) and higher expression of MCT4 (both in BP6 and H9c2) that exports the lactate from cells to extracellular space. Those two cell lines seem to rely more on glycolysis than on TCA cycle for energy production, and therefore, they might not respond to TCA inhibitors.
On the other hand, treatment with DMOG resulted in an increased CA IX level in all three cell lines. Furthermore, considerable induction of CA IX could be observed in all cell types after treatment by TL, which was reported to act as a ROS scavenger when applied for a short time (30 min) at 0.1–5 mmol l−1 concentration [19]. However, this type of treatment was ineffective against CA IX. To achieve CA IX induction, cells were incubated with TL for 48 h at a 2.5–5 mmol l−1 concentration. It appears that in this setting, TL operated by activation of the MAPK pathway as was shown previously in the study of Suy et al. [37]. This explanation would also correspond with earlier data showing that activation of the MAPK pathway leads to transactivation of HIF-1α and to an increased expression of CA IX [33, 21]. Alternatively, TL (at 10 mmol l−1 concentration), described as a compound interfering with the formation of the hydroxyl radicals [3], could directly inhibit HIF-1α hydroxylation. Interestingly, strong induction of CA IX was observed in fibroblasts but not in cardiomyocytes upon combined treatment by DZO and TL. If TL acted as a radical scavenger here, it should have worked against DZO-produced radicals and would not have supported its effect. Nevertheless, based on our data, it seems that the effects of chemical compounds that disrupt oxygen sensing have cell-type-specific, time-related, and dose-dependent effects and that their mechanism of action cannot be simply generalized.
Data from the analysis of other hypoxia-regulated genes add further complexity to evaluation of the effects of hypoxia and hypoxia-mimicking compounds. Each of the examined hypoxia-regulated genes showed different magnitude of induction most probably as a consequence of different molecular mechanisms involved in their regulation. The most steady expression profile was observed with VEGF. Although the regulation of VEGF and CA IX has several common features, there are also clear differences. CA9 promoter contains HIF-binding HRE element at a position close to the transcription initiation site, whereas VEGF possesses HRE element almost 1 kb upstream of the transcription start and utilizes it more as an enhancer element. On the other hand, VEGF mRNA is regulated at the level of posttranscriptional stability, and its translation efficiently proceeds under hypoxia. Finally, posttranslational stability of CA IX is very high, and only 10% of the protein population is shed to extracellular protein, whereas VEGF is not associated with cells but is efficiently secreted [31, 45].
Moreover, each of the three cell lines displays a different expression profile under normoxia, and the effects of the compounds are variable. Therefore, it is important to take these differences into account when drawing principal conclusions from the studies of particular cell models.
Despite all these variations, in this study, we clearly showed that rat CA IX is consistently induced by physiological hypoxia in different cell types and that it can be also induced under normoxia by certain chemical compounds that disrupt oxygen sensing. We also demonstrated that in both fibroblasts and cardiomyocytes, rat CA IX is localized at the cell surface, suggesting that it might play a role in pH control and ion transport across the plasma membrane of hypoxic cells similarly as had been shown for human CA IX [39, 40]. Finally, we also showed that CA IX expression can be induced by physiological hypoxia in rat tissues including the heart, brain, kidney, and stomach. Together, these data approve the relevance of the rat model for further studies of CA IX.
Moreover, hypoxia-induced expression of CA IX in cardiomyocytes might be of particular importance for the study of cardiac pathophysiology, especially in association with different diseases linked to defective supply of oxygen and nutrients. This idea stems from the data showing that chronic moderate hypoxia is cardioprotective against acute hypoxia and reoxygenation injury [6] and that acidosis is an important factor contributing to death of cardiac cells exposed to chronic hypoxia [41]. As chronic moderate hypoxia induces expression of CA IX in cardiomyocytes and CA IX is functionally implicated in pH regulation, it is well conceivable that CA IX can be a part of the cardioprotective machinery. Nevertheless, further studies are needed to verify this assumption.
Abbreviations
- CA IX:
-
carbonic anhydrase IX protein
- CA9 :
-
human carbonic anhydrase 9 gene
- Car9 :
-
rodent carbonic anhydrase 9 gene
- DMOG:
-
dimethyloxalylglycine
- DMS:
-
dimethylester succinate
- DZO:
-
diazoxide
- GLUT-1:
-
glucose transporter 1
- GLUT-4:
-
glucose transporter 4
- HIF:
-
hypoxia-inducible factor
- MCT4:
-
monocarboxylate transporter 4
- iNOS:
-
inducible nitric oxide synthase
- N-TAD:
-
N-terminal transactivation domain
- ODDD:
-
oxygen-dependent degradation domain
- PHD:
-
prolyl hydroxylase
- ROS:
-
radical oxygen species
- TL:
-
tempol
- VEGF:
-
vascular endothelial growth factor
References
Baysal BE (2003) On the association of succinate dehydrogenase mutations with hereditary paraganglioma. Trends Endocrinol Metab 14:453–459
Blancher C, Moore JW, Talks KL, Houlbrook S, Harris AL (2000) Relationship of hypoxia-inducible factor (HIF)-1α and HIF-2α expression to vascular endothelial growth factor induction and hypoxia survival in human breast cancer cell lines. Cancer Res 60:7106–7113
Burlando B, Viarengo A (2005) Ca2+ is mobilized by hydroxyl radical but not by superoxide in RTH-149 cells: the oxidative switching-on of Ca2+ signalling. Cell Calcium 38:507–513
Busija DW, Katakam P, Rajapakse NC, Kis B, Grover G, Domoki F, Bari F (2005) Effects of ATP-sensitive potassium channel activators diazoxide and BMS-191095 on membrane potential and reactive oxygen species production in isolated piglet mitochondria. Brain Res Bull 66:85–90
Casey JR (2006) Why bicarbonate? Biochem Cell Biol 84(6):930–939
Crawford RM, Jovanovic S, Budas GR, Davies AM, Lad H, Wenger RH, Robertson KA, Roy DJ, Ranki HJ, Jovanovic A (2003) Chronic mild hypoxia protects heart-derived H9c2 cells against acute hypoxia/reoxygenation by regulating expression of the SUR2A subunit of the ATP-sensitive K+ channel. J Biol Chem 278:31444–1455
Dalgard CL, Lu H, Mohyeldin A, Verma A (2004) Endogenous 2-oxoacids differentially regulate expression of oxygen sensors. Biochem J 380:419–424
Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, Mechta-Grigoriou F (2004) JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell 118:781–794
Grabmaier K, de Weijert MCA, Verhaegh GW, Schalken JA, Oosterwijk E (2004) Strict regulation of CAIX(G250/MN) by HIF-1alpha in clear cell renal cell carcinoma. Oncogene 23:5624–5631
Gut MO, Parkkila S, Vernerova Z, Rohde E, Zavada J, Hocker M, Pastorek J, Karttunen T, Gibadulinova A, Zavadova Z, Knobeloch KP, Wiedenmann B, Svoboda J, Horak I, Pastorekova S (2002) Gastric hyperplasia in mice with targeted disruption of the carbonic anhydrase gene Car9. Gastroenterology 123:1889–1903
Harris AL (2002) Hypoxia—a key regulatory factor in tumor growth. Nature Cancer Reviews 2:38–47
Hilvo M, Rafajová M, Pastoreková S, Pastorek J, Parkkila S (2004) Expression of carbonic anhydrase IX in mouse tissues. J Histochem Cytochem 52:1313–1322
Hynninen P, Hamalainen JM, Pastorekova S, Pastorek J, Waheed A, Sly WS, Tomas E, Kirkinen P, Parkkila S (2004) Transmembrane carbonic anhydrase isozymes IX and XII in the female mouse reproductive organs. Reprod Biol Endocrinol 2:73
Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG (2001) HIFa targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464–468
Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim AV, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ (2001) Targeting of HIF-alpha to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468–472
Kallio H, Pastorekova S, Pastorek J, Waheed A, Sly WS, Mannisto S, Heikinheimo M, Parkkila S (2006) Expression of carbonic anhydrases IX and XII during mouse embryonic development. BMC Dev Biol 6:22
Kaluz S, Kaluzova M, Chrastina A, Olive PL, Pastorekova S, Pastorek J, Lerman MI, Stanbridge EJ (2002) Lowered oxygen tension induces expression of the hypoxia marker MN/carbonic anhydrase IX in the absence of hypoxia-inducible factor 1α stabilization: a role for phosphatidylinositol 3′-kinase. Cancer Res 62:4469–4477
Kim MY, Kim MJ, Yoon IS, Ahn JH, Lee SH, Baik EJ, Moon CH, Jung YS (2006) Diazoxide acts more as a PKC-ε activator, and indirectly activates the mitochondrial KATP channel conferring cardioprotection against hypoxic injury. Br J Pharmacol 149:1059–70
Kimura S, Zhang GX, Nishiyama A, Shokoji T, Yao L, Fan YY, Rahman M, Abe Y (2005) Mitochondria-derived reactive oxygen species and vascular MAP kinases: comparison of angiotensin II and diazoxide. Hypertension 45:438–44
King A, Selak MA, Gottlieb E (2006) Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 25:4675–4682
Kopacek J, Barathova M, Dequiedt F, Sepelakova J, Kettmann R, Pastorek J, Pastorekova S (2005) MAPK pathway contributes to density- and hypoxia-induced expression of the tumor-associated carbonic anhydrase IX. Biochimica et Biophysica Acta 172:41–49
MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, Watson DG, Gottlieb E (2007) Cell permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol 27:3282–3289
Morgan PE, Pastorekova S, Stuart-Tilley AK, Alper SL, Casey JR (2007) Interactions of transmembrane carbonic anhydrase, CA IX, with bicarbonate transporters. Am J Physiol Cell Physiol 293:738–748
Novakova M, Ela C, Bowen WD, Hasin Y, Eilam Y (1998) Highly selective sigma receptor ligands elevate inositol 1,4,5-triphosphate production in rat cardiac myocytes. Eur J Pharmacol 353:315–327
Pastorekova S, Parkkila S, Parkkila AK, Opavsky R, Zelnik V, Saarnio J, Pastorek J (1997) Carbonic anhydrase IX, MN/CA IX: analysis of stomach complementery DNA sequence and expression in human and rat alimentary tracts. Gastroenterology 112:398–408
Pastorekova S, Pastorek J (2004) Cancer-related carbonic anhydrase isozymes and their inhibition. In: Supuran CT, Scozzafava A, Conway J (eds) Carbonic anhydrase, its inhibitors and activators. CRC, Boca Raton, pp 253–272
Pastorekova S, Parkkila S, Zavada J (2006) Tumor-associated carbonic anhydrases and their clinical significance. Adv Clin Chem 42:167–216
Pollard P, Wortham N, Barcley E, Alam A, Elia G, Manek S, Poulsom R, Tomlinson I (2005) Evidence of increased microvessel density and activation of the hypoxia pathway in tumours from the hereditary leiomyomatosis and renal cell cancer syndrome. J Pathol 205:41–49
Pollard PJ, El-Bahrawy M, Poulsom R, Elia G, Killick P, Kelly G, Hunt T, Jeffery R, Seedhar P, Barwell J, Latif F, Gleeson MJ, Hodgson SV, Stamp GW, Tomlinson IP, Maher ER (2006) Expression of HIF-1alpha, HIF-2alpha (EPAS1), and their target genes in paraganglioma and pheochromocytoma with VHL and SDH mutations. J Clin Endocrinol Metab 91:4593–4598
Potter CPS, Harris AL (2003) Diagnostic, prognostic and therapeutic implications of carbonic anhydrases in cancer. Br J Cancer 89:2–7
Rafajova M, Zatovicova M, Kettmann R, Pastorek J, Pastorekova S (2004) Induction by hypoxia combined with low glucose or low bicarbonate and high posttranslational stability upon reoxygenation contribute to carbonic anhydrase IX expression in cancer cells. Int J Oncol 24:995–1004
Raval RR, Lau KW, Tran MGB, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ (2005) Contrasting properties of hypoxia inducible factor 1 (HIF-1) and HIF-2 in von Hippel–Lindau-associated renal cell carcinoma. Mol Cell Biol 25:5675–5686
Richard DE, Berra E, Gothié E, Roux D, Pouysségur J (1999) p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem 274:32631–32637
Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7:77–85
Semenza GL (2002) Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol 64:993–998
Supuran CT (2004) Carbonic anhydrases: catalytic mechanisms, distribution and physiological roles. In: Supuran CT, Scozzafava A, Conway J (eds) Carbonic anhydrase: its inhibitors and activators. CRC, Boca Raton, pp 1–24
Suy S, Mitchell JB, Ehleiter D, Haimovitz-Friedman A, Kasid U (1998) Nitroxides tempol and tempo induce divergent signal transduction pathways in MDA-MB 231 breast cancer cells. J Biol Chem 273:17871–17878
Svastova E, Zilka N, Zatovicova M, Gibadulinova A, Ciampor F, Pastorek J, Pastorekova S (2003) Carbonic anhydrase IX reduces E-cadherin-mediated adhesion of MDCK cells via interaction with b-catenin. Exp Cell Res 290:332–345
Svastova E, Hulikova A, Rafajova M, Zatovicova M, Gibadulinova A, Casini A, Cecchi A, Scozzafava A, Supuran CT, Pastorek J, Pastorekova S (2004) Hypoxia activates the capacity of tumor-associated carbonic anhydrase IX to acidify extracellular pH. FEBS Lett 577:439–495
Swietach P, Vaughan-Jones RD, Harris AL (2007) Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev 26:299–310
Webster KA, Discher DJ, Kaiser S, Hernandez O, Sato B, Bishopric NH (1999) Hypoxia-activated apoptosis of cardiac myocytes requires reoxygenation or a pH shift and is independent of p53. J Clin Invest 104:239–252
Wenger RH (2005) Mitochondria: oxygen sinks rather than sensors? Med Hypotheses 66:380–383
Wiesener MS, Jurgensen JS, Rosenberger C, Scholze CK, Horstrup JH, Warnecke C, Mandriota S, Bechmann I, Frei UA, Pugh CW, Ratcliffe PJ, Bachmann S, Maxwell PH, Eckardt KU (2003) Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J 17:271–273
Wykoff C, Beasley N, Watson P, Turner L, Pastorek J, Sibtain A, Wilson G, Turley H, Talks K, Maxwell PH, Pugh C, Ratcliffe P, Harris AL (2000) Hypoxia-inducible regulation of tumor-associated carbonic anhydrases. Cancer Res 60:7075–7083
Zatovicova M, Sedlakova O, Svastova E, Ohradanova A, Ciampor F, Arribas J, Pastorek J, Pastorekova S (2005) Ectodomain shedding of the hypoxia-induced carbonic anhydrase IX is a metalloprotease-dependent process regulated by TACE/ADAM17. Br J Cancer 93:1267–1276
Zavada J, Zavadova Z, Pastorekova S, Ciampor F, Pastorek J, Zelnik V (1993) Expression of MaTu-MN protein in human tumor cultures and in clinical specimens. Int J Cancer 54:268–274
Zavada J, Zavadova Z, Pastorek J, Biesova Z, Jezek J, Velek J (2000) Human tumour-associated cell adhesion protein MN/CA IX: identification of M75 epitope and of the region mediating cell adhesion. Br J Cancer 82:1808–1813
Acknowledgement
This work was supported by grants from EU (6FP Integrated project EUROXY, LSCH-CT-2003-502932), from Research and Development Support Agency (Contracts APVV-51-024805 and APVV-51-027404), and from VEGA (2/7126/27).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Holotnakova, T., Ziegelhoffer, A., Ohradanova, A. et al. Induction of carbonic anhydrase IX by hypoxia and chemical disruption of oxygen sensing in rat fibroblasts and cardiomyocytes. Pflugers Arch - Eur J Physiol 456, 323–337 (2008). https://doi.org/10.1007/s00424-007-0400-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-007-0400-6