Abstract
Purpose
Aerobic (AE) and resistance (RE) exercise elicit unique adaptations in skeletal muscle. The purpose here was to compare the post-exercise response of mTOR signaling and select autophagy markers in skeletal muscle to acute AE and RE.
Methods
In a randomized, cross-over design, six untrained men (27 ± 3 years) completed acute AE (40 min cycling, 70% HRmax) and RE (8 sets, 10 repetitions, 65% 1RM). Muscle biopsies were taken at baseline, and at 1 h and 4 h following each exercise. Western blot analyses were performed to examine total and phosphorylated protein levels. Upstream regulator analyses of skeletal muscle transcriptomics were performed to discern the predicted activation states of mTOR and FOXO3.
Results
Compared to AE, acute RE resulted in greater phosphorylation (P < 0.05) of mTORSer2448 at 4 h, S6K1Thr389 at 1 h, and 4E- BP1Thr37/46 during the post-exercise period. However, both AE and RE increased mTORSer2448 and S6K1Thr389 phosphorylation at 4 h (P < 0.05). Upstream regulator analyses revealed the activation state of mTOR was increased for both AE (z score, 2.617) and RE (z score, 2.789). No changes in LC3BI protein were observed following AE or RE (P > 0.05), however, LC3BII protein was decreased after both AE and RE at 1 h and 4 h (P < 0.05). p62 protein content was also decreased at 4 h following AE and RE (P < 0.05).
Conclusion
Both acute AE and RE stimulate mTOR signaling and similarly impact select markers of autophagy. These findings indicate the early adaptive response of untrained human skeletal muscle to divergent exercise modes is not likely mediated through large differences in mTOR signaling or autophagy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Skeletal muscle possesses a remarkable capacity to adapt to various stimuli, and in particular, exercise represents a powerful stimulus for skeletal muscle adaptations. Moreover, it is well understood that different modes of exercise elicit unique molecular responses within skeletal muscle. Notably, chronic aerobic exercise (AE) generally results in increased mitochondrial density thorough mitochondrial biogenesis (Holloszy 1967), ultimately leading to increased fatigue resistance of the skeletal muscle (Holloszy and Coyle 1984). When coupled with increased capillarization, this specific adaptation allows for a prolonged ability to perform muscle contractions, often at an increased intensity (Gollnick 1986). Conversely, chronic resistance exercise (RE) results in a muscle with greater ability to produce force. Increased force output is specifically a result of the accumulation of contractile proteins (Phillips et al. 1997) and is commonly associated with muscle hypertrophy (Moore et al. 2005).
Skeletal muscle adaptation is ultimately governed by the relationship between synthesis and breakdown of proteins. This relationship not only regulates the accumulation of specific proteins in the cell, but also the degradation and removal of damaged and dysfunctional proteins. Together, these anabolic and catabolic processes mediate the cellular adaptations which translate to the improved function of skeletal muscle (Phillips et al. 2009). The synthesis of skeletal muscle proteins is largely regulated by the mammalian/mechanistic target of rapamycin (mTOR) signaling pathway (Drummond et al. 2009; Dickinson et al. 2011), and this pathway is thought to have a primary role in mediating muscle hypertrophy following resistance exercise training (Goodman et al. 2011; Bodine et al. 2001). On the other hand, the response of mTOR signaling to aerobic exercise has received less attention. While in vitro data suggest mTOR to be primarily stimulated by RE-like exercise (Atherton et al. 2005), studies in humans have shown mTOR signaling in skeletal muscle is responsive to AE (namely cycling) (Coffey et al. 2006b; Wang et al. 2011; Wilkinson et al. 2008; Camera et al. 2010). However, direct comparisons of mTOR signaling in skeletal muscle following RE and AE are limited to between-group comparisons in highly active participants (Camera et al. 2010), trained individuals (Coffey et al. 2006b), or in the presence of amino acid feeding (Wilkinson et al. 2008). Importantly, training has been shown to blunt the acute molecular response to the specific exercise (Coffey et al. 2006a, b; Phillips et al. 1999), while amino acid feeding stimulates independent skeletal muscle responses (Reidy and Rasmussen 2016). Consequently, to more fully understand the potential role for mTOR signaling in mediating exercise-mode specific responses, there remains a need to compare the skeletal muscle mTOR signaling response to RE and AE in untrained, exercise naïve individuals.
Acute AE, on the other hand, appears to elicit a greater overall stimulation of “breakdown” processes as compared to RE (Louis et al. 2007; Dickinson et al. 2018). For instance, compared to RE, acute AE has been shown to elicit a greater increase in mRNA markers associated with the ubiquitin proteasome system (Louis et al. 2007), including Forkhead box O3a (FOXO3a), which is a transcriptional regulator known to induce transcription of genes within the two major proteolytic systems in skeletal muscle; the ubiquitin–proteasome and autophagy systems (Mammucari et al. 2007; Sandri et al. 2004; Zhao et al. 2007). In particular, autophagy manages the quality control of cellular organelles and contractile proteins through lysosomal-mediated proteolysis (Dunn 1994). While the acute post-exercise response of the autophagy system in human skeletal muscle remains unclear (Lira et al. 2013; Brandt et al. 2018; Fritzen et al. 2016), autophagy has been highlighted as a key cellular mechanism in skeletal muscle adaptation to chronic AE (Lira et al. 2013). Further, while the specific mechanisms regulating the exercise-induced alterations in autophagy remain to be fully resolved, peroxisome proliferator-activated receptor-gamma coactivator-1 alpha (PGC-1ɑ), which is a key regulator of mitochondrial biogenesis, has been demonstrated to positively regulate autophagy during exercise (Vainshtein and Hood 2016).
We have previously shown in untrained individuals that acute AE and RE elicit unique skeletal muscle transcriptome responses in the immediate hours post exercise (Dickinson et al. 2018). The purpose of this study was to determine the response of mTOR signaling and autophagy markers in skeletal muscle of untrained individuals in the immediate hours following acute AE and RE. We hypothesized that in the untrained state, RE would elicit a greater response in mTOR signaling whereas AE would produce a greater change in select markers of autophagy.
Materials and methods
Participants
Seven healthy males initially volunteered to participate in the study. All participants were recruited from the greater Phoenix, AZ metropolitan area. All of these participants were considered untrained or recreationally active as none of the participants performed regular physical activity (< 2 days/week) at the time of participation. All subjects completed a medical history and physical activity questionnaire along with a blood draw for blood coagulation profile. One subject dropped out prior to any testing for reasons unrelated to the study (scheduling conflict). Six participants completed all experimental trials (mean ± SD, 27 ± 3 years, 179 ± 6 cm, 79 ± 10 kg). Based on means, standard deviations, and effect sizes from our previous work examining changes in mTORSer2448 and S6K1Thr389 phosphorylation following RE (D'Lugos et al. 2018), 4–6 participants were deemed suitable to confidently detect increases in these variables from basal. All participants provided written, informed consent before participation, and all procedures were approved by a University Institutional Review Board (in compliance with the Declaration of Helsinki, as revised in 1983).
Prior to participation in the experimental trials, subjects completed baseline testing of unilateral leg extension 1-repetition maximum (1RM) along with determination of maximal heart rate and peak work rate (W) on a cycle ergometer. 1RM was assessed on two occasions (separated by ~ 1 week) using a knee extension device (Cybex VR3, Medway MA), and the heaviest weight lifted by each leg between the two measurements was considered the participant’s 1RM for each leg. Peak exercise heart rate and work rate were determined on a cycle ergometer (model 828E; Monark Exercise, Vansboro, Sweden) using a ramp protocol (Dickinson et al. 2018). Briefly, participants began cycling at ~ 50 W and work rate was progressively increased by 15 W per minute until volitional fatigue. Heart rate was assessed via telemetry, and maximal heart rate and peak work rate were recorded.
Study design
Details of the study design have been previously presented (Dickinson et al. 2018). Briefly, each participant completed two separate experimental exercise trials that were identical except for the mode of exercise performed (AE or RE). These trials were performed utilizing a randomized, counter-balanced cross-over design with ~ 1 week between each trial (mean ± SD: 9 ± 3 days). Subjects were instructed to abstain from alcohol consumption for 72 h and caffeine for 24 h before each experimental trial.
Experimental trials
For each experimental trial, participants arrived at the laboratory (~ 0700) following a 12-h overnight fast and remained fasted until completion of the trial. Participants were instructed to record their last meal the night before the first experimental exercise trial and to replicate that meal the night before the second experimental exercise trial. Following 30 min of supine rest, and prior to the start of exercise, a basal muscle biopsy was obtained. To minimize the number of invasive procedures for the cross-over study design, the basal biopsy from the first experimental trial represented basal conditions for both experimental exercise trials (no basal biopsy was obtained during the second experimental trial). After the basal biopsy (1st trial) or 45 min of supine rest (2nd trial, to imitate the basal biopsy time frame), subjects performed a 5-min warm-up at a low work rate on a cycle ergometer (model 828E; Monark Exercise) and then completed either acute AE or acute RE (see below). Upon completion of exercise, participants rested supine, engaging in minimal physical activity for the remainder of the trial. Muscle biopsies were then taken at 1 and 4 h relative to the end of the exercise bout. The order of the experimental trials was counter-balanced, such that three participants performed AE during the first experimental trial and three participants performed RE during the first experimental trial. In addition, the use of the dominant leg for the basal biopsy and for the post-exercise biopsies for each exercise mode was also counter-balanced.
Acute exercise
AE consisted of 40 min of cycling on a cycle ergometer (model 828E; Monark Exercise, Vansboro, Sweden) at a prescribed work rate to elicit ~ 75% peak heart rate. Heart rate was continuously monitored (Polar Electro, Lake Success, NY) throughout the exercise bout and work rate and heart rate were recorded in 2-min intervals. RE consisted of isotonic unilateral leg extensions (Cybex VR3), where each subject performed eight sets of ten repetitions at 60–65% unilateral 1RM with each leg, as we have previously utilized (Patel et al. 2017; D'Lugos et al. 2018; Dickinson et al. 2018). Unilateral exercise was chosen to ensure that the leg to be biopsied post-exercise performed the prescribed work. During RE, participants alternated legs such that the leg to be biopsied performed each set first (e.g., 1 leg performed 10 reps and then the other leg performed 10 reps for each set). Participants were given 3 min of rest between each set. The total exercise time for the RE was ~ 40 min.
Muscle biopsies
Muscle biopsy procedures were conducted under strict sterile conditions and were obtained using a 5-mm Bergström needle with suction from the lateral portion of the vastus lateralis following local anesthesia (1% lidocaine, no epinephrine) (D'Lugos et al. 2018; Dickinson et al. 2018; Patel et al. 2017). During both experimental trials, the 4-h biopsy was obtained from the same incision as for the 1-h biopsy; however, the biopsy needle was inclined at a different angle such that the 4-h post-exercise biopsy was taken ~ 5 cm proximal to the 1-h post-exercise biopsy, as previously performed (Dickinson et al. 2018; Volpi et al. 2008). All muscle samples were obtained from a single needle pass for each time point. Muscle tissue was immediately blotted, dissected of connective tissue and adipose tissue and frozen in liquid nitrogen. Muscle tissue was stored at −80 °C until analysis.
These particular post-exercise timepoints were chosen as they represent post-exercise timepoints previously used to examine signaling events (1–5 h post exercise) (D'Lugos et al. 2018; Dickinson et al. 2017b), as well as a timepoint (4 h) that corresponds with changes in gene expression after exercise (e.g., 4–8 h post) (Louis et al. 2007). In addition, these post-exercise timepoints are similar to previous studies examining the response of human skeletal muscle to divergent exercise stimuli (0–4 h post) (Coffey et al. 2006a, b; Camera et al. 2010; Wilkinson et al. 2008).
Analytical techniques
Frozen muscle was weighed (mean ± SD, 38.7 ± 8.0 mg) and homogenized (1:9 wet weight/volume) in an ice-cold homogenization buffer containing 50 mM Tris–HCl, 250 mM mannitol, 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM ethylenediaminetetraacetic acid, 1 mM ethyleneglycotetraacetic acid, 1% Triton X-100, pH 7.4, 1 mM DTT, 1 mM benzamidine, 0.1 mM PMSF, and 5 µg mL−1 soybean trypsin inhibitor. Homogenized samples were then centrifuged at 3400g for 10 min at 4 °C, and the supernatant was collected which represents the cytosolic fraction of proteins (Patel et al. 2017; D'Lugos et al. 2018; Dickinson et al. 2017a).
Immunoblot analyses
Total protein concentrations were determined using the Bradford assay (Pierce Coomassie Plus; Thermo Fisher Scientific, Waltham, MA). The supernatant was diluted (1:1) in a 2X sample buffer mixture containing 125 mM Tris, pH 6.8, 25% glycerol, 2.5% SDS, 2.5% A-mercaptoethanol, and 0.002% bromophenol blue and then boiled for 3 min at 100 °C. Samples were run in duplicate and equal amounts of total protein (40 µg) were loaded into each lane. Samples were separated by electrophoresis (150 V) on a 7.5% or 12% polyacrylamide gel as determined by the size of the target protein (Mini-PROTEAN; BioRad, Hercules, CA). Each gel contained an internal control sample and molecular weight ladder (Precision Plus, BioRad) (Patel et al. 2017; D'Lugos et al. 2018; Dickinson et al. 2017a).
Following electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad) using the Transblot® Turbo™ Transfer System according to manufacturer’s instructions. Blots were then blocked for 1 h in 5% nonfat dry milk and incubated with primary antibody dissolved in nonfat dry milk or bovine serum albumin overnight at 4 °C. Primary antibodies directed against proteins of interest were purchased from Cell Signaling Technology (Beverly, MA) unless otherwise listed: total mTOR (1:1000, no. 2972), mTORSer2448 (1:500, no 2971), total ribosomal protein S6 kinase 1 (S6K1) (1:500, no. 9202), S6K1Thr389 (1:500, no. 9205), total eukaryotic elongation factor 2 (eEF2) (1:2000, no. 2332), eEF2Thr56 (1:5000, no 2331), total eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) (1:1000, no. 9452), 4E-BP1Thr37/46 (1:1000, no. 9459), LC3 (microtubule-associated protein 1 light chain 3) B (1:1000, no. 2775), p62 (1:1000, no. 5114), Forkhead box O3a (FOXO3a) (1:500, no. 2497), PGC-1α (1:1000, Novus Biomedical, no. NBP1-04,676) and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1000, no. 2118). After overnight incubation, blots were incubated with an anti-rabbit IgG HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature. Blots were then washed in TBST (TBS + Tween20), incubated in a chemiluminescent solution (ECL plus, Amersham Biosciences, Piscataway NJ) and exposed with a chemiluminescent imager (ChemiDoc MP, Bio-Rad). All densitometric analyses were performed using Image Lab 6.1.0 software (Bio-Rad). Optical density values for phosphorylated and total proteins were normalized to the internal control. Western blot data are expressed as either phosphorylation divided by total protein or as total protein, and all data are adjusted to fold change of basal. The ratio of LC3BII to LC3BI was also determined. No changes in the density of GAPDH were observed between trials or across time (P > 0.05).
Transcriptome analyses
To supplement our targeted immunoblot analyses, skeletal muscle transcriptomics previously published from these participants [GEO: GSE107934 (Dickinson et al. 2018)] were analyzed using Ingenuity Pathway Analysis (IPA; Qiagen Inc., Hilden, Germany) (Kramer et al. 2014). Specifically, differentially expressed genes (Benjamini–Hochberg adjusted P value ≤ 0.05) at each post-exercise time point were utilized to complete Upstream Regulator Analyses for mTOR and FOXO3 (Kramer et al. 2014; Rundqvist et al. 2019). These analyses were completed to discern the predicted activation state of mTOR and FOXO3 during the post-exercise timeframe. In addition, network analyses were employed to identify differentially expressed genes annotated to “autophagy” in response to each exercise mode.
Statistical analysis
All data were tested for normality through skewness and kurtosis and visual inspection of the normality plots using SPSS. Data for S6K1Thr389 and 4E-BP1Thr37/46 were non-normally distributed, and thus these data were transformed to achieve normal distribution before statistical analyses (raw data presented). A one-way repeated measures analysis of variance (ANOVA) was used to test for differences in GAPDH across time. A two-way repeated measures ANOVA was used to test group by time differences. Pairwise comparisons using the Tukey post hoc test were employed to determine differences within ANOVA. Data analyses were performed using SigmaStat version 12.5 (Systat Software). Significance was set at P ≤ 0.05. Data are presented as raw mean ± SE unless otherwise noted.
Results
Physiological exercise data
Details on the physiological data in response to the acute exercise bouts have been previously detailed (Dickinson et al. 2018). The median intensity for the entire AE bout was 78% of heart rate max, with a range of 75–84% heart rate max for all participants. The median intensity for all sets of RE was 62% 1RM with a range of 52–68% 1RM for all participants.
mTOR signaling response to acute exercise
Representative images related to the assessment of mTOR signaling are presented in Fig. 1. No changes in mTORSer2448 phosphorylation were observed at 1 h for either AE (P = 0.886) or RE (P = 0.329) (Fig. 2A). However, mTORSer2448 phosphorylation was increased at 4 h after RE (P < 0.001) and tended to increase at 4 h after AE (P = 0.066). Further, mTORSer2448 phosphorylation was higher at 4 h in RE compared to AE (P = 0.026). No changes from basal in S6K1Thr389 phosphorylation were observed at 1 h for either AE (P = 0.743) or RE (P = 0.083), however, S6K1Thr389 phosphorylation was higher at 1 h in RE compared to AE (P = 0.020) (Fig. 2B). S6K1Thr389 phosphorylation was increased at 4 h in both AE (P = 0.002) and RE (P = 0.005). 4E-BP1Thr37/46 phosphorylation was unchanged by AE and RE (P = 0.989), however, 4E-BP1Thr37/46 phosphorylation was higher in RE vs. AE during the post-exercise time course (main effect of trial, P = 0.015) (Fig. 2C). eEF2Thr56 phosphorylation was unchanged in response to AE and RE (P = 0.227) and similar between trials (P = 0.578) (Fig. 2D).
Representative images for the assessment of mTOR signaling. All samples from a given subject (basal and both trials) were loaded and analyzed on the same blot in a blinded fashion. All samples were loaded in duplicate and each gel contained a loading standard (STD). Representative images displayed for a given protein target show the data from one participant. mTOR, mammalian/mechanistic target of rapamycin complex 1; S6K1, ribosomal protein S6 kinase 1; 4E-BP1, eukaryotic initiation factor 4E-binding protein 1; eEF2, eukaryotic elongation factor 2; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase
Phosphorylation of mTORSer2448 (A), S6K1Thr389 (B), 4E-BP1Thr37/46 (C), and eEF2Thr56 (D). Subjects completed either 40 min of cycling at 70% heart rate max (AE, aerobic exercise) or 8 sets of 10 repetitions at 65% 1-repetition maximum (RE, resistance exercise). Columns represent mean ± SE (raw data) and individual data are also presented with a given shape/fill representing the same participant. Filled shapes represent individuals that performed RE first while open shapes represent participants that performed AE first. All data are presented as fold change relative to basal levels. Raw data for S6K1Thr389 and 4E-BP1Thr37/46 were transformed to achieve normal distribution for statistical analyses. mTOR, mammalian/mechanistic target of rapamycin complex 1; S6K1, ribosomal protein S6 kinase 1; 4E-BP1, eukaryotic initiation factor 4E-binding protein 1; eEF2, eukaryotic elongation factor 2. n = 6 for both trials. *P < 0.05 versus basal; †P < 0.05 between trials; #P = 0.06 versus basal, ‡P < 0.05 main effect of trial
Utilizing changes in the skeletal muscle transcriptome of these subjects (Dickinson et al. 2018), upstream regulator analysis was performed to discern the activation state of mTOR in response to AE and RE (Table 1). At 1 h, the activation of mTOR was not identified in response to either AE or RE. However, at 4 h, the activation state of mTOR was increased for both AE (z score = 2.617) and RE (z score = 2.789), in which 18 and 32 exercise-responsive target genes were identified, respectively.
Autophagic response to acute exercise
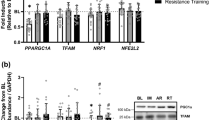
Representative images related to the assessment of autophagy are presented in Fig. 3. No changes in LC3BI protein content were observed in response to either AE or RE (P = 0.169) (Fig. 4A). However, LC3BII protein content was decreased in both AE and RE at 1 h (AE, P < 0.001; RE, P < 0.001) and 4 h post exercise (AE, P = 0.034; RE, P = 0.005) (Fig. 4B). Interestingly, no changes in the LC3BII/I ratio were observed in response to either AE or RE (P = 0.153) and no differences were observed between trials (P = 0.338) (Fig. 4C). No changes in p62 protein content were observed at 1 h for either AE (P = 0.366) or RE (P = 0.122) (Fig. 4D), however, p62 protein content was decreased at 4 h following both AE (P = 0.013) and RE (P = 0.004). No differences in p62 were observed between trials (P = 0.547). No changes in FOXO3a protein content were observed at 1 h for either AE (P = 0.458) or RE (P = 0.301) (Fig. 5A), however, FOXO3a protein content was decreased at 4 h following both AE (P = 0.001) and RE (P = 0.021) (Fig. 5A). No differences in FOXO3a were observed between trials (P = 0.547). PGC-1ɑ protein content was unchanged in response to AE and RE (P > 0.05) and similar between trials (P > 0.05) (Fig. 5B).
Representative images for the assessment of autophagy markers. All samples from a given subject (basal and both trials) were loaded and analyzed on the same blot in a blinded fashion. All samples were loaded in duplicate and each gel contained a loading standard (STD). Representative images displayed for a given protein target show the data from one participant. LC3, microtubule-associated protein 1 light chain 3; p62, sequestosome-1; FOXO3a, Forkhead box O3a; PGC-1ɑ, Peroxisome proliferator-activated receptor-gamma coactivator-1 alpha; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase
Protein content of LC3BI (A), LC3BII (B), LC3BII/I (C), and p62 (D). Subjects completed either 40 min of cycling at 70% heart rate max (AE, aerobic exercise) or 8 sets of 10 repetitions at 65% 1-repetitons maximum (RE, resistance exercise). Columns represent mean ± SE (raw data) and individual data are also presented with a given shape/fill representing the same participant. Filled shapes represent individuals that performed RE first while open shapes represent participants that performed AE first. All data are presented as fold change relative to basal levels. LC3, microtubule-associated protein 1 light chain 3; p62, sequestosome-1. n = 6 for both trials. *P < 0.05 versus basal
Protein content of FOXO3a (A) and PGC-1ɑ (B). Subjects completed either 40 min of cycling at 70% heart rate max (AE, aerobic exercise) or 8 sets of 10 repetitions at 65% 1-repetition maximum (RE, resistance exercise). Columns represent mean ± SE (raw data) and individual data are also presented with a given shape/fill representing the same participant. Filled shapes represent individuals that performed RE first while open shapes represent participants that performed AE first. All data are presented as fold change relative to basal levels. FOXO3a, Forkhead box O3a; PGC-1ɑ, Peroxisome proliferator-activated receptor-gamma coactivator-1 alpha. n = 6 for both trials (n = 5 for PGC-1ɑ due to sample insufficiency of one participant). *P < 0.05 versus basal
The upstream regulator analysis predicted the activation of FOXO3 at 1 h by both AE (z score = 2.387) and RE (z score = 2.575), in which 6 and 7 exercise-responsive target genes were identified, respectively (Table 1). Interestingly, only RE resulted in the activation of FOXO3 at 4 h (z score = 2.638), where 42 exercise-responsive target genes were identified.
Differentially expressed genes annotated to “Autophagy” are displayed in Supplemental Fig. 1. Independent of exercise mode and post-exercise timepoint, a majority of the differentially expressed genes were increased in expression as compared to decreased. The transcriptional response of the autophagy system was modest at 1 h, where AE and RE resulted in the differential expression of 6 and 7 genes, respectively. Comparatively, the autophagy-related gene response was greater at 4 h, especially following RE, which induced the differential expression of 43 autophagy-related genes versus only 17 genes following AE.
Discussion
It is well known that chronic AE and RE foster unique adaptations in skeletal muscle, however, the precise mechanisms mediating these responses are not completely understood. In the current study, we used a counter-balanced, cross-over design in untrained, exercise naïve participants to compare the response of mTOR signaling and select autophagy markers in skeletal muscle to acute AE and RE. These two molecular processes were investigated given their known responsiveness to exercise and their role in regulating exercise-induced muscle growth (Goodman et al. 2011) and adaptation (Lo Verso et al. 2014), respectively. The primary findings from this investigation reveal that both AE and RE are capable of stimulating mTOR signaling in the early post-exercise timeframe, and that only minor differences exist in the mTOR signaling response between exercise modes performed acutely in the untrained state. Similarly, both acute RE and acute AE reduced protein levels of LC3BII and p62 and increased expression of several autophagy-related genes, collectively indicating both AE and RE likely increased autophagy flux, perhaps through degradation of autophagosomes.
Previous research employing an in vitro model of electrical stimulation demonstrated that high-frequency stimulation, meant to mimic RE, preferentially elicited activation of the mTOR pathway as compared to low-frequency stimulation designed to mimic AE (Atherton et al. 2005). Interestingly, we did observe subtle differences between exercise modes in the magnitude of stimulation, with acute RE showing higher phosphorylation of mTORSer2448 at 4 h, S6K1Thr389 at 1 h, and 4E- BP1Thr37/46 during the post-exercise period. It is interesting to speculate that perhaps these differences in phosphorylation status could be related to fiber type recruitment rather than exercise mode. Specifically, we (D'Lugos et al. 2018) and others (Parkington et al. 2003) have identified preferential mTOR activation in type II fibers following RE. Similarly, cycle training has been shown to increase basal mTOR phosphorylation in both type I and type II fibers, but the response is more pronounced in type II fibers (Stuart et al. 2010). Consequently, the subtle differences in mTOR signaling between exercise modes may be related to increased forced requirements during each contraction and greater recruitment of type II fibers during RE.
To our knowledge, this is the first study to employ a cross-over design in untrained, exercise naïve individuals to investigate the early signaling response to divergent exercise modes. While there were subtle differences as discussed above, both acute RE and acute AE increased mTORSer2448 and S6K1Thr389 phosphorylation 4 h after exercise in these participants. These signaling findings at 4 h post exercise were accompanied by IPA analyses of differentially expressed genes indicating mTOR to be active 4 h following each exercise mode. Together, these findings indicate that in untrained individuals both exercise modes stimulate mTOR signaling activity in the early hours after exercise, which may represent a “general” response to what could be considered “unfamiliar” stimuli experience by untrained skeletal muscle (which may also explain the relative variability in some signaling responses). Moreover, we specifically focused our work on exercise naïve individuals as exercise training has been shown to alter (Ogasawara et al. 2013) or blunt (Coffey et al. 2006a, b; Phillips et al. 1999) the acute molecular response of skeletal muscle to exercise, which may impede identification of early adaptive exercise-mode specific molecular responses. Consequently, to what extent similar findings would be observed in response to training requires further investigation. Interestingly, however, our observation of mTOR signaling being stimulated by both exercise modes in untrained individuals is in agreement with previous work consisting of single-leg exercise combined with nutrient ingestion (Wilkinson et al. 2008) and parallel-group designs in highly active individuals (Camera et al. 2010). Moreover, the latter study (Camera et al. 2010) only included a 1-h post-exercise time course, and therefore the findings from the current study indicate similarities in mTOR signaling following RE and AE can be observed for up to 4 h post exercise. Collectively, our findings in untrained individuals, coupled with previous work in highly active participants (Camera et al. 2010), do not provide strong support that large differences in mTOR signaling play a primary role in mediating exercise-mode specific responses between AE and RE. However, mTOR stimulation can lead to a variety of potential cellular responses (Schmelzle and Hall 2000), and thus further research is necessary to uncover to what extent the “downstream” response to mTOR signaling differs following divergent exercise.
In addition to our investigation of mTOR signaling, we also examined the response of select autophagy markers in skeletal muscle. Our interest in specifically examining markers of autophagy in response to each exercise mode is based on (1) previous work demonstrating AE to elicit a greater overall proteolytic transcriptional response compared to RE (Louis et al. 2007), (2) our previous transcriptome analyses indicating AE induced a specific gene cluster with several genes in the FOXO pathway (Dickinson et al. 2018), and (3) suggestions that autophagy may be an important process mediating endurance exercise performance (Lira et al. 2013) and mitochondrial biogenesis, the latter of which may be regulated through PGC-1ɑ (Ju et al. 2016). In contrast to our hypothesis, both acute RE and AE resulted in similar responses in the select autophagy protein makers, despite a lack of change in PGC-1ɑ protein content, during the immediate hours following acute exercise. The lack of change in LC3BI coupled with the reduction in LC3BII and p62 during the post-exercise time course is in agreement with previous exercise-based findings (Fritzen et al. 2016; Dickinson et al. 2017b). Moreover, a change in LC3BII alone can be difficult to interpret as this can occur due to less conversion of LC3BI to LC3BII or through degradation of LC3BII in the lysosome. However, the reduction in LC3BII was accompanied by a reduction in autophagosome membrane protein, p62. Given p62 is degraded in the lysosome during autophagy (Komatsu and Ichimura 2010; Sanchez et al. 2014a), the reduction in both p62 and LC3BII would indicate that both exercise modes likely stimulated increased degradation of autophagosomes (Sanchez et al. 2014b).
The finding that both exercise modes stimulated increased degradation of autophagosomes would also appear to be supported by the drop in FOXO3a protein content, and likely movement of FOXO3a from the cytosol (compartment analyzed) to the nucleus where it serves, in part, as a transcription factor for autophagy-related genes (Sandri et al. 2004). While IPA upstream regulator analysis only identified FOXO3 to be active following RE at 4 h post exercise, we did observe several autophagy-associated genes to be upregulated at 4 h post-exercise in response to both exercise modes, which we interpret to also indicate increased autophagy involvement (Sanchez et al. 2014a). Importantly, this upregulation of autophagy-associated genes at 4 h corresponded to the drop in cytosolic FOXO3a protein content. It is important to note, however, that greater stimulation of autophagy may occur when exercise is performed in the fasted state (Jamart et al. 2013). Therefore, we cannot confirm that the fasted state of the subjects in the current study did not confound exercise-mode specific responses in the selected autophagy markers. Collectively, in the absence of any feeding, our data suggest that lysosomal degradation of autophagosomes may be similarly stimulated in the early/untrained adaptive response of skeletal muscle to both AE and RE.
We acknowledge that the findings from this investigation are confined to the specific exercises examined. While AE, in particular, is not confined to cycling exercise, we specifically chose leg extension RE and cycling AE as these exercises are known to stimulate the skeletal muscle that was biopsied (i.e., vastus lateralis). We also acknowledge that cycling exercise training may promote a muscle hypertrophy response (Harber et al. 2009) that may make identifying differences between AE and RE in mTOR signaling difficult as compared to running. However, it should be noted that running may also stimulate growth, at least at the single muscle fiber level (Harber and Trappe 2008; Harber et al. 2012). We also recognize the impact that nutrient ingestion can have on the signaling pathways and proteins examined. However, the goal of this study was to focus on the independent response of untrained skeletal muscle to each mode of exercise. Future studies are needed to incorporate the impact of nutrient ingestion on the divergent exercise response. Although consistent with group sizes from previous work investigating divergent exercise responses (Coffey et al. 2006b), this study does include a relatively small sample size. However, the use of a cross-over design reduces the overall impact of the smaller sample size. Moreover, to further minimize this potential impact on our conclusions, we chose to also perform upstream regulator analyses for mTOR and FOXO3 utilizing unbiased skeletal muscle transcriptomics to supplement (and strengthen) the immunoblot findings. This study also included only a single basal biopsy. This design was chosen to minimize the number of biopsies, and thus support the cross-over design which also reduces the impact of the single basal biopsy. Lastly, we recognize that the acute adaptive response to exercise lasts beyond 4 h (Louis et al. 2007; Yang et al. 2005). Consequently, a longer time course following exercise is certainly warranted and may elicit different findings.
In summary, our findings indicate that in untrained individuals both acute AE and RE stimulate mTOR signaling and result in similar changes in select autophagy protein markers in skeletal muscle during the early post-exercise period. Moreover, these protein-based findings are supported by transcriptome analyses revealing “activation” of mTOR and increased expression of several autophagy-associated genes following each exercise bout. Our findings provide support that the early adaptive response of human skeletal muscle to divergent exercise modes is not mediated through large differences in mTOR signaling or autophagy flux. Thus, examination of additional molecular pathways, a more comprehensive time course (including chronic training), or perhaps additional transcriptional (Dickinson et al. 2018) or proteomic events are warranted to uncover the precise mechanisms mediating the adaptive response of skeletal muscle to divergent exercise modes.
Availability of data and material
The data used and/or analyzed for this study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Abbreviations
- 1RM:
-
1-Repetition maximum
- 4E-BP1:
-
Eukaryotic initiation factor 4E-binding protein 1
- AE:
-
Aerobic exercise
- ANOVA:
-
Analysis of variance
- eEF2:
-
Eukaryotic elongation factor 2
- FOXO:
-
Forkhead box O
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- LC3:
-
Microtubule-associated protein 1 light chain 3
- mTOR:
-
Mammalian/mechanistic target of rapamycin
- p62:
-
Sequestosome-1
- PGC-1ɑ:
-
Peroxisome proliferator-activated receptor-gamma coactivator-1 alpha
- RE:
-
Resistance exercise
- S6K1:
-
Ribosomal protein S6 kinase 1
References
Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H (2005) Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J 19(7):786–788. https://doi.org/10.1096/fj.04-2179fje
Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD (2001) Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3(11):1014–1019. https://doi.org/10.1038/ncb1101-1014
Brandt N, Gunnarsson TP, Bangsbo J, Pilegaard H (2018) Exercise and exercise training-induced increase in autophagy markers in human skeletal muscle. Physiol Rep 6(7):e13651. https://doi.org/10.14814/phy2.13651
Camera DM, Edge J, Short MJ, Hawley JA, Coffey VG (2010) Early time course of Akt phosphorylation after endurance and resistance exercise. Med Sci Sports Exerc 42(10):1843–1852. https://doi.org/10.1249/MSS.0b013e3181d964e4
Coffey VG, Shield A, Canny BJ, Carey KA, Cameron-Smith D, Hawley JA (2006a) Interaction of contractile activity and training history on mRNA abundance in skeletal muscle from trained athletes. Am J Physiol Endocrinol Metab 290(5):E849-855. https://doi.org/10.1152/ajpendo.00299.2005
Coffey VG, Zhong Z, Shield A, Canny BJ, Chibalin AV, Zierath JR, Hawley JA (2006b) Early signaling responses to divergent exercise stimuli in skeletal muscle from well-trained humans. FASEB J 20(1):190–192. https://doi.org/10.1096/fj.05-4809fje
Dickinson JM, Fry CS, Drummond MJ, Gundermann DM, Walker DK, Glynn EL, Timmerman KL, Dhanani S, Volpi E, Rasmussen BB (2011) Mammalian target of rapamycin complex 1 activation is required for the stimulation of human skeletal muscle protein synthesis by essential amino acids. J Nutr 141(5):856–862. https://doi.org/10.3945/jn.111.139485
Dickinson JM, D’Lugos AC, Mahmood TN, Ormsby JC, Salvo L, Dedmon WL, Patel SH, Katsma MS, Mookadam F, Gonzales RJ, Hale TM, Carroll CC, Angadi SS (2017a) Exercise protects skeletal muscle during chronic doxorubicin administration. Med Sci Sports Exerc 49(12):2394–2403. https://doi.org/10.1249/MSS.0000000000001395
Dickinson JM, Reidy PT, Gundermann DM, Borack MS, Walker DK, D’Lugos AC, Volpi E, Rasmussen BB (2017b) The impact of postexercise essential amino acid ingestion on the ubiquitin proteasome and autophagosomal-lysosomal systems in skeletal muscle of older men. J Appl Physiol 122(3):620–630. https://doi.org/10.1152/japplphysiol.00632.2016
Dickinson JM, D’Lugos AC, Naymik MA, Siniard AL, Wolfe AJ, Curtis DR, Huentelman MJ, Carroll CC (2018) Transcriptome response of human skeletal muscle to divergent exercise stimuli. J Appl Physiol 124(6):1529–1540. https://doi.org/10.1152/japplphysiol.00014.2018
D’Lugos AC, Patel SH, Ormsby JC, Curtis DP, Fry CS, Carroll CC, Dickinson JM (2018) Prior acetaminophen consumption impacts the early adaptive cellular response of human skeletal muscle to resistance exercise. J Appl Physiol 124(4):1012–1024. https://doi.org/10.1152/japplphysiol.00922.2017
Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB (2009) Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol 587(Pt 7):1535–1546. https://doi.org/10.1113/jphysiol.2008.163816
Dunn WA (1994) Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol 4(4):139–143. https://doi.org/10.1016/0962-8924(94)90069-8
Fritzen AM, Madsen AB, Kleinert M, Treebak JT, Lundsgaard AM, Jensen TE, Richter EA, Wojtaszewski J, Kiens B, Frosig C (2016) Regulation of autophagy in human skeletal muscle: effects of exercise, exercise training and insulin stimulation. J Physiol 594(3):745–761. https://doi.org/10.1113/JP271405
Gollnick PD (1986) Metabolic regulation in skeletal muscle: influence of endurance training as exerted by mitochondrial protein concentration. Acta Physiol Scand Suppl 556:53–66
Goodman CA, Frey JW, Mabrey DM, Jacobs BL, Lincoln HC, You JS, Hornberger TA (2011) The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol 589(Pt 22):5485–5501. https://doi.org/10.1113/jphysiol.2011.218255
Harber M, Trappe S (2008) Single muscle fiber contractile properties of young competitive distance runners. J Appl Physiol 105(2):629–636. https://doi.org/10.1152/japplphysiol.00995.2007
Harber MP, Konopka AR, Douglass MD, Minchev K, Kaminsky LA, Trappe TA, Trappe S (2009) Aerobic exercise training improves whole muscle and single myofiber size and function in older women. Am J Physiol Regul Integr Comp Physiol 297(5):R1452–R1459. https://doi.org/10.1152/ajpregu.00354.2009
Harber MP, Konopka AR, Undem MK, Hinkley JM, Minchev K, Kaminsky LA, Trappe TA, Trappe S (2012) Aerobic exercise training induces skeletal muscle hypertrophy and age-dependent adaptations in myofiber function in young and older men. J Appl Physiol 113(9):1495–1504. https://doi.org/10.1152/japplphysiol.00786.2012
Holloszy JO (1967) Biochemical adaptations in muscle. Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J Biol Chem 242(9):2278–2282
Holloszy JO, Coyle EF (1984) Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol Respir Environ Exerc Physiol 56(4):831–838. https://doi.org/10.1152/jappl.1984.56.4.831
Jamart C, Naslain D, Gilson H, Francaux M (2013) Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am J Physiol Endocrinol Metab 305(8):E964–E974. https://doi.org/10.1152/ajpendo.00270.2013
Ju J-s, Jeon S-i, Park J-y, Lee J-y, Lee S-c, Cho K-j, Jeong J-m (2016) Autophagy plays a role in skeletal muscle mitochondrial biogenesis in an endurance exercise-trained condition. J Physiol Sci 66(5):417–430. https://doi.org/10.1007/s12576-016-0440-9
Komatsu M, Ichimura Y (2010) Physiological significance of selective degradation of p62 by autophagy. FEBS Lett 584(7):1374–1378. https://doi.org/10.1016/j.febslet.2010.02.017
Kramer A, Green J, Pollard J Jr, Tugendreich S (2014) Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30(4):523–530. https://doi.org/10.1093/bioinformatics/btt703
Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS, Hoehn KL, Yan Z (2013) Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J 27(10):4184–4193. https://doi.org/10.1096/fj.13-228486
Lo Verso F, Carnio S, Vainshtein A, Sandri M (2014) Autophagy is not required to sustain exercise and PRKAA1/AMPK activity but is important to prevent mitochondrial damage during physical activity. Autophagy 10(11):1883–1894. https://doi.org/10.4161/auto.32154
Louis E, Raue U, Yang Y, Jemiolo B, Trappe S (2007) Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J Appl Physiol 103(5):1744–1751. https://doi.org/10.1152/japplphysiol.00679.2007
Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M (2007) FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6(6):458–471. https://doi.org/10.1016/j.cmet.2007.11.001
Moore DR, Phillips SM, Babraj JA, Smith K, Rennie MJ (2005) Myofibrillar and collagen protein synthesis in human skeletal muscle in young men after maximal shortening and lengthening contractions. Am J Physiol Endocrinol Metab 288(6):E1153-1159. https://doi.org/10.1152/ajpendo.00387.2004
Ogasawara R, Kobayashi K, Tsutaki A, Lee K, Abe T, Fujita S, Nakazato K, Ishii N (2013) mTOR signaling response to resistance exercise is altered by chronic resistance training and detraining in skeletal muscle. J Appl Physiol 114(7):934–940. https://doi.org/10.1152/japplphysiol.01161.2012
Parkington JD, Siebert AP, Lebrasseur NK, Fielding RA (2003) Differential activation of mTOR signaling by contractile activity in skeletal muscle. Am J Physiol Regul Integr Comp Physiol 285(5):R1086–R1090. https://doi.org/10.1152/ajpregu.00324.2003
Patel SH, D’Lugos AC, Eldon ER, Curtis D, Dickinson JM, Carroll CC (2017) Impact of acetaminophen consumption and resistance exercise on extracellular matrix gene expression in human skeletal muscle. Am J Physiol Regul Integr Comp Physiol 313(1):R44–R50. https://doi.org/10.1152/ajpregu.00019.2017
Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR (1997) Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol 273(1 Pt 1):E99-107. https://doi.org/10.1152/ajpendo.1997.273.1.E99
Phillips SM, Tipton KD, Ferrando AA, Wolfe RR (1999) Resistance training reduces the acute exercise-induced increase in muscle protein turnover. Am J Physiol 276(1):E118-124. https://doi.org/10.1152/ajpendo.1999.276.1.E118
Phillips SM, Glover EI, Rennie MJ (2009) Alterations of protein turnover underlying disuse atrophy in human skeletal muscle. J Appl Physiol 107(3):645–654. https://doi.org/10.1152/japplphysiol.00452.2009
Reidy PT, Rasmussen BB (2016) Role of ingested amino acids and protein in the promotion of resistance exercise-induced muscle protein anabolism. J Nutr 146(2):155–183. https://doi.org/10.3945/jn.114.203208
Rundqvist HC, Montelius A, Osterlund T, Norman B, Esbjornsson M, Jansson E (2019) Acute sprint exercise transcriptome in human skeletal muscle. PLoS ONE 14(10):e0223024. https://doi.org/10.1371/journal.pone.0223024
Sanchez AM, Bernardi H, Py G, Candau RB (2014a) Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol 307(8):R956-969. https://doi.org/10.1152/ajpregu.00187.2014
Sanchez AM, Candau RB, Bernardi H (2014b) FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci 71(9):1657–1671. https://doi.org/10.1007/s00018-013-1513-z
Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117(3):399–412. https://doi.org/10.1016/s0092-8674(04)00400-3
Schmelzle T, Hall MN (2000) TOR, a central controller of cell growth. Cell 103(2):253–262. https://doi.org/10.1016/s0092-8674(00)00117-3
Stuart CA, Howell MEA, Baker JD, Dykes RJ, Duffourc MM, Ramsey MW, Stone MH (2010) Cycle training increased GLUT4 and activation of mammalian target of rapamycin in fast twitch muscle fibers. Med Sci Sports Exerc 42(1):96–106. https://doi.org/10.1249/MSS.0b013e3181ad7f36
Vainshtein A, Hood DA (2016) The regulation of autophagy during exercise in skeletal muscle. J Appl Physiol 120(6):664–673. https://doi.org/10.1152/japplphysiol.00550.2015
Volpi E, Chinkes DL, Rasmussen BB (2008) Sequential muscle biopsies during a 6-h tracer infusion do not affect human mixed muscle protein synthesis and muscle phenylalanine kinetics. Am J Physiol Endocrinol Metab 295(4):E959-963. https://doi.org/10.1152/ajpendo.00671.2007
Wang L, Mascher H, Psilander N, Blomstrand E, Sahlin K (2011) Sahlin Resistance exercise enhances the molecular signaling of mitochondrial biogenesis induced by endurance exercise in human skeletal muscle. J Appl Physiol 111(5):1335–1344. https://doi.org/10.1152/japplphysiol.00086.2011
Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, Rennie MJ (2008) Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. J Physiol 586(15):3701–3717. https://doi.org/10.1113/jphysiol.2008.153916
Yang Y, Creer A, Jemiolo B, Trappe S (2005) Time course of myogenic and metabolic gene expression in response to acute exercise in human skeletal muscle. J Appl Physiol 98(5):1745–1752. https://doi.org/10.1152/japplphysiol.01185.2004
Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6(6):472–483. https://doi.org/10.1016/j.cmet.2007.11.004
Acknowledgements
We thank the participants for their time. Funding provided, in part, by R01 DK109948 (JMH). ACD is supported by a National Heart, Lung and Blood Institute Grant (T32HL134621).
Funding
R01 DK109948 (JMH). ACD is supported by a National Heart, Lung and Blood Institute Grant (T32HL134621).
Author information
Authors and Affiliations
Contributions
CEM, ACD, CCC, and JMD conceived and designed the research; CEM, ACD, KS, JMH, SSA, CCC, and JMD conducted research; CEM, ACD, and JMD, analyzed data and performed statistical analysis; CEM, ACD, and JMD, wrote the manuscript and have primary responsibility for final content; all authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare there are no conflicts or competing interests.
Ethics approval
All study procedures were conducted in accordance with ethical standards and under protocols approved by the Midwestern University Institutional Review Board, and in compliance with the Declaration of Helsinki, as revised in 1983.
Consent to participate
Written, informed consent was obtained from all participants prior to study participation.
Consent for publication
The consent form signed by all participants acknowledged publication of study data.
Additional information
Communicated by Philip D. Chilibeck.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mazo, C.E., D’Lugos, A.C., Sweeney, K.R. et al. The effects of acute aerobic and resistance exercise on mTOR signaling and autophagy markers in untrained human skeletal muscle. Eur J Appl Physiol 121, 2913–2924 (2021). https://doi.org/10.1007/s00421-021-04758-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00421-021-04758-6