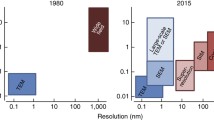
Abstract
Development of new fluorescent probes and fluorescence microscopes has led to new ways to study cell biology. With the emergence of specialized microscopy units at most universities and research centers, the use of these techniques is well within reach for a broad research community. A major breakthrough in fluorescence microscopy in biology is the ability to follow specific targets on or in living cells, revealing dynamic localization and/or function of target molecules. One of the inherent limitations of fluorescence microscopy is the resolution. Several efforts are undertaken to overcome this limit. The traditional and most well-known way to achieve higher resolution imaging is by electron microscopy. Moreover, electron microscopy reveals organelles, membranes, macromolecules, and thus aids in the understanding of cellular complexity and localization of molecules of interest in relation to other structures. With the new probe development, a solid bridge between fluorescence microscopy and electron microscopy is being built, even leading to correlative imaging. This connection provides several benefits, both scientifically as well as practically. Here, I summarize recent developments in bridging microscopy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent advancements in fluorescence microscopy have opened the way to study protein (co)localization, interaction and function in live cells. These developments have been boosted by the molecular cloning, development and implementation of a jelly-fish-derived green fluorescent protein. Visualizing proteins attached at specific sites in living cells and organisms are now widely applied in biosensors that can change color [based on fluorescence energy resonance transfer (FRET)], for studying protein dynamics using fluorescence recovery after photobleaching (FRAP), dynamic (co)localization studies and many other assays (reviewed in Giepmans et al. 2006). Although these achievements help to study and understand biology in live cells, the localization of proteins at high resolution and at the ultrastructural level can only be studied by electron microscopy (EM). Fluorescent microscopy and EM are highly additive. Combining EM and non-fluorescent light microscopy is done for decades, with molecular identification initially restricted to a single protein that could be visualized by a dark, electron-dense precipitate (for a detailed overview, see Sosinsky et al. 2007). Scientists have long sought to combine fluorescent and electron microscopic techniques, but methodological differences hampered a general and efficient use on a single sample. The good news is that these problems are now successfully being tackled, and a bridge has been built. By combining both imaging techniques, (sub)cellular processes in living cells can now be studied with follow-up at higher resolution at the ultrastructural level, making correlative microscopy a new powerful research tool.
Bridging microscopy not only allows to study or manipulate protein localization or function in live cells prior to deadly fixation and EM analysis, but also aids in detection of rare events at low magnification at the light microscope, followed by further EM-analysis of a selected area. Specimen size is limited for EM and preparation is often laborious, making a prescreen at the light level very rewarding. Such a “google earth” approach might also help to orient and navigate within the tissue. Correlative microscopy is the ultimate combination of LM and EM allowing a particular cell to be studied from the micron to the nanometer scale while maintaining spatial orientation. Here, I discuss our contributions as well as the latest developments in building and bridging probes for live-cell and correlated microscopy, with emphasis on fluorescent proteins, the biarsenical-tetracysteine system and Quantum dots (QDs) and their application that revealed new insight into Golgi neogenesis.
Protein identification: targeting and labeling
Key in identification of specific proteins is the specific targeting of a specific label (Fig. 1). The general and most widespread targeting methods are immuno-based or are using genetically encoded tags, or are a combination of both. Whole-cell immunolabeling of endogenous proteins has the disadvantage of damaging the ultrastructure due to the use of detergents to deliver immunoreagents inside cells. Moreover, both pre-embedding and postembedding immunolabeling require use of relative mild fixatives to preserve epitopes. This dramatic impact on the ultrastructure goes unnoticed by light microscopic examination. Expressing exogenous genetically tagged proteins avoids the need of permeabilizing the cells prior to staining but requires modification of the protein of interest (for a detailed discussion about the pros and cons of the probes and techniques highlighted here, see Giepmans et al. 2006). In the combinatorial approaches, fluorescent proteins can be immunolabeled with gold for EM detection. Vice versa, enzymes such as peroxidases used for EM detection can be immunolabeled with a fluorescent group.
Probes that allow relative straight-forward switching between the two forms of microscopy have led to a broader implementation of correlated microscopy. These probes can either be combinatorial or have the intrinsic properties that allow detection at both fluorescence and EM (Sosinsky et al. 2007). A combinatorial probe typically consists of a separate fluorophore, and an electron-dense particle/initiator for deposition of electron-dense material can be conjugated to an antibody to allow correlated microscopy. An example of probes that have the intrinsic properties to be visible at both fluorescence and EM are QDs. These are fluorescent nanocrystals with an electron-dense core, directly visible in the electron microscope, and well-suited for multiprotein labeling at both the light microscope and electron microscope levels (see below).
A third method for correlated microscopy is by local generation of diaminobenzadine (DAB) polymers at the fluorophore by so-called photoconversion. In working with fluorescence in living cells, one should avoid phototoxicity resulting from illumination of fluorophores that produce high concentrations of reactive oxygen species. However, these fluorophores are well suited for photoconversion of diaminobenzidine into polymers that are osmiophilic and readably detectable at the EM (reviewed in Sosinsky et al. 2007). One of the earliest examples of photoconversion to detect proteins was through immunotargetting the small organic dye eosin (Deerinck et al. 1994). Nowadays, photoconversion can be applied using genetically encoded tagging of proteins as well.
Fluorescent proteins: a fruitful development
Fluorescence microscopy is usually limited to study a few spectrally different probes simultaneously, thus also to a limited amount of proteins. Much effort has been undertaken to broaden the availability of fluorescent proteins with different, optimized characteristics—although it should be noted that simultaneous optimization of several key characteristics remains a challenge (Shaner et al. 2004). Mostly used are the Aequorea victoria green fluorescent protein-derivatives (GFP, YFP and CFP for green, yellow and cyan fluorescent protein, respectively). Improved Discosoma sp. red fluorescent protein-derivatives, including the mFruits, such as mCherry (Shaner et al. 2004; Chudakov et al. 2005) are nowadays also widely implemented in live-cell microscopy studies. The broadened availability of fluorescent proteins now allows “comparing apples and oranges” simultaneously. Efforts of many researchers to improve spectral properties, bleaching resistance, folding, etc., of fluorescent proteins are still ongoing (Mishin et al. 2008; Shaner et al. 2008 and references therein).
Besides the widespread application of fluorescent proteins to study protein (co)localization, the fluorescent proteins are very suitable to study protein dynamics and function in living cells (reviewed in Lippincott-Schwartz and Patterson 2003; Giepmans et al. 2006; Chudakov et al. 2005). Using FRAP, the diffusion rate of proteins can be determined in subcellular compartments. Initiated by the engineering of a photoactivatable GFP, now a generation of photoswitchable proteins (i.e., these will change green to red upon specific illumination) have been made that not only allow to study recovery as in FRAP, but also allow tracking of the converted protein. In addition, these photoswitchable proteins can be used to increase spatial resolution by activation of a single protein in a diffraction-limited space followed by imaging of red fluorescence until bleaching, and subsequent localization of the center of the fluorescent spot. Iterative rounds of photoactivation, imaging, bleaching and localizing lead to the localization of the proteins with subnanometer accuracy in the specimen. This fluorescent image can even be projected on a subsequent acquired EM-image (so called photoactivation localization microscopy or “PALM”; Betzig et al. 2006; see also Heintzmann and Ficz 2006 for an overview of several other fluorescent microscopic techniques that break the classical resolution limit in light microscopy).
Other spectral switches are engineered fluorescent protein-based biosensors, which revolutionized the studies of protein function in living cells. Diverse events can be reported by these optical spies, ranging from promoter activity of genes of interest to the activity of certain signaling pathways. Several of these biosensors are based on a change of fluorescence (or Förster) resonance energy transfer (FRET) initiated by a conformational change of a specific probe that contains two fluorophores leading to an altered emission spectrum of a probe (Jares-Erijman and Jovin 2003). CFP/YFP, or combinations of improved derivatives, is the most commonly used FRET-pair. The development of new fluorescent proteins has led to novel efficient and spectral distinct FRET pairs to study protein–protein interaction and create novel FRET-based biosensors (Goedhart et al. 2007; van der Krogt et al. 2008; Piljic and Schultz 2008). Analysis of multiple functional parameters in a single cell nowadays can be achieved by simultaneous FRET using multiple sensors that can be either spatially resolved (by specific subcellular targeting) and/or spectrally resolved by using distinct FRET pairs (Piljic and Schultz 2008). Since FRET is limited to ~7 nm, it can also be used to detect protein–protein proximity at the molecular level (reviewed in Chudakov et al. 2005; Giepmans et al. 2006).
Correlated light and EM with fluorescent proteins is mostly applied using immuno-detection of GFP at the electron microscope. Inefficient photoconversion of DAB into polymers by GFP compared to other fluorophores hampers the broad application of more direct detection of GFP at the ultrastructural level. However, three studies now showed successful visualization of fluorescent proteins at the light and electron microscopic levels using fluorescent protein-mediated photoconversion of DAB (Monosov et al. 1996; Grabenbauer et al. 2005; Meisslitzer-Ruppitsch et al. 2008). A benefit of genetically encoded photoconversion is the specific targeting of the probe by direct fusion to the protein of interest and the introduction into the cells by means of DNA transfection. This allows high quality preservation of the ultrastructure and does not need cellular delivery of antibodies and destruction of membranes. Parallel to the exploration of the possibilities using genetically encoded fluorescent proteins, Roger Tsien and his research group developed a small genetically encoded tag that can be visualized or modulated upon binding of engineered specific ligands, the biarsenical-tetracysteine system.
The biarsenical-tetracysteine system: versatile applications
Numerous approaches are being developed that allow imaging of proteins depending on epitope tagging (reviewed in Chen and Ting 2005; O’Hare et al. 2007) and/or antibody–fluorogen combinations (Szent-Gyorgyi et al. 2008). Here, I discuss the biarsenical-tetracysteine system, which is based on a small genetically encoded tag (core sequence Cys-Cys-Pro-Gly-Cys-Cys) that can bind cell-permeable biarsenical derivatives of fluorescein (such as FlAsH or ReAsH), which then become fluorescent. Since the introduction of the system (Griffin et al. 1998), several rounds of optimization of the amino-acid sequence flanking the cysteines have lead to an increase of the affinity of the biarsenicals for the peptide tags (Martin et al. 2005). The tetracysteine-biarsenical system is very versatile: it can be used to study localization at the light microscopic level of the tagged protein of interest (including in live cells). Moreover, the tetracysteine–ReAsH complex can be used to photoconvert DAB in osmiophilic polymers for EM (Fig. 4). The combination of a genetically encoded targeting with a cell-permeant labeling step, which does not require permeabilization and allows stringent fixation, makes this system unique in retaining sample preservation that is so important for ultrastructural studies.
The development of biarsenicals with different characteristics allows proteins of interest to be targeted for different purposes. The subcellular turnover of proteins can be determined using a fluorescent pulse-chase experiment (Gaietta et al. 2002; Fig. 2): the tetracysteine-labeled protein of interest can first be labeled with, e.g., ReAsH, which is then removed from the medium. After a chase that allows new protein to be synthesized, a spectrally distinct biarsenical is then added (e.g., FlAsH) that will only label the new protein. By specific photoconversion of the bound ReAsH, the pulse chase can even be analyzed at the EM level (Gaietta et al. 2002).
Pulse-chase in the microscope. Four amino-acid substitutions in the gap junction protein conexin43 create a binding site for tetracysteines. This minor protein modification allows determination of gap junction turnover. Sequence of labeling: proteins in transfected cells are labeled with FlAsH (green); excess label is washed away. After a 6-h chase, the new protein is labeled with ReAsH (red), revealing that gap junctions grow at the edge of the plaque. This finding is consistent with another study, where FRAP was used to study the gap junction turnover (see Gaietta et al. 2002; Lauf et al. 2002 for further details). [Image: BNGG at the National Center for Microscopy and Imaging Research (NCMIR), San Diego, CA, USA]
The system also allows protein-inactivation in living cells by inducing the production of reactive oxygen species through illumination of tetracysteine-bound ReAsH, resulting in destruction of the protein of interest (reviewed in Giepmans et al. 2006). The engineering of another biarsenical that changes fluorescent intensity allows to determine calcium-concentrations at tetracysteine-tagged proteins (Tour et al. 2007). In addition, the tetracysteine flanking sequences contribute to the versatility of the system. In the screen to detect high-affinity binding sequences for ReAsH (Martin et al. 2005), a peculiar sequence turned out that, in combination with GFP, results in the induction of protein aggregation of the tetracysteine–GFP–ReAsH complex. When proteins of interest carry the specific tag, they will be dragged into the aggregate. Release of interaction can then be initiated by bleaching the ReAsH. While this is an obvious unwanted phenomenon when determining protein localization, Brent Martin and colleagues used this observation to study binding characteristics of macromolecular complexes (Martin et al. 2007). The tetracysteine-biarsenical system thus allows multiple studies depending on the binding sequence, biarsenical and technique that helps to study diverse processes, including dynamic light microscopic imaging of protein location, function modulation and high resolution-correlated EM.
Quantum dot nanocrystals: single molecule detection
Much attention had been given to the optical properties of QDs since their first application in cell biology (Bruchez et al. 1998; Chan and Nie 1998). QDs consist of a core crystal (like CdSe and CdTe) with a ZnS shell. Water solubility of QDs has been reached by development of a passivating layer: polymers to which targeting molecules can be coupled. Since QDs are very bright, they relatively easily allow single molecule detection. Moreover, the electron-dense crystal is directly visible by EM, and because of the different sizes and shapes, three different QDs can be used simultaneously to label three distinct proteins at the EM level, while the spectral resolution would allow to use even more QDs simultaneously for light microscopy (Nisman et al. 2004; Giepmans et al. 2005; Deerinck et al. 2007; Fig. 3). Being an immune-affinity-based method, QD labeling is typically suitable to detect endogenous proteins, in contrast to the earlier mentioned genetic-based labeling approaches. Optimization and first analysis can be performed at the light microscopy level prior to more laborious preparation for EM or correlated microscopic analysis. Moreover, we showed the beneficial use of QDs for pre-embedding electron microscopic labeling, a technique now allowing labeling of multiple proteins and detection in EM-tomograms (manuscript submitted). Benefits of QDs over gold-labeling are the fluorescent properties and higher penetration efficiency in cells and tissue. Note that the first generation of commercially available QDs penetrate less than traditional fluorophores. Recently, Alice Ting and coworkers reported their successful development of QDs that were downsized by half but with the same core size. The smaller passivating layer and use of a monovalent streptavidine as targeting device resulted in more efficient labeling of synapses compared to the first generation, but small fluorophores still remained slightly more efficient (Howarth et al. 2008). For EM studies, it will be important to address if these QDs penetrate better in cells and tissue, making them more generally applicable than the first generation of QDs for correlated microscopy applications. The potency of some of the probes discussed here is illustrated in their application to study the Golgi apparatus during the cell cycle.
Detection of proteins by Quantum dots. Top Composite of six cultured fibroblasts (false-colored) from six different dishes immuno-labeled for a-tubulin and detected with Quantum dots analyzed by confocal microscopy: QD525 (blue), QD565 (green), QD585 (yellow), QD605 (orange), QD655 (red) and QD705 (purple). Bottom EM-analysis of a similar experiment. Note the specific labeling of microtubules, but not of the actin fibers. [Image: T.J. Deerinck and BNGG at NCMIR]
Correlative microscopic studies: birth of twins during Golgi neogenesis
Several studies on the Golgi apparatus during mitosis revealed that the Golgi transforms in a so-called Golgi-haze, consisting of ~50 nm vesicles. Whether the Golgi apparatus fuses with endoplasmatic reticulum components at this stage remains controversial. Therefore, we set out to study Golgi structure in living cells at high spatiotemporal resolution followed by EM examination (Gaietta et al. 2006). Central in our study is a combinatorial probe consisting of a part of α-mannosidase II (ensuring Golgi-targeting), GFP (for live-cell imaging) and a tetracysteine-tag (for ReAsH labeling, photoconversion and EM examination). Given the proximity of GFP and ReAsH, we were able to increase specificity of photoconversion: excitation of GFP results in FRET to the adjacent tetracysteine labeled with ReAsH. While we initially focused our research on the Golgi haze, surprisingly, we found a striking phenomenon following the disappearance of the haze and reconstitution of the Golgi apparatus: instead of the expected appearance of a single Golgi in each of the daughter cells, Golgi biogenesis in mitotic cells starts by formation of four clusters, two in each cell. The smallest cluster forms at the midbody and gradually migrates, along tracks, towards the other perinuclear cluster and one Golgi apparatus per cell is then being reconstituted (Gaietta et al. 2006; Fig. 4). EM examination of these “Golgi twins” revealed the typical Golgi-like structure in both the perinuclear compartment and the twin near the midbody. The labeled Golgi-structures were found in close proximity with microtubules. These observations prompted us to dynamically image microtubules, using mCherry-α-tubulin, and the Golgi apparatus, using the probe described above, simultaneously. The reconstitution of the smaller twins at the midbody was even more obvious by using dual fluorescence that highlights the prominent enrichement of microtubules at this subcellular structure (see also Fig. 4). Moreover, microtubules appear to dictate the route of travel of the smaller Golgi twins to reconstitute a single Golgi apparatus (Gaietta et al. 2006). This study exemplifies how pioneering the applications of novel probes for correlated microscopy aids in disclosure of dynamic biological processes in high spatiotemporal detail. Future studies need to answer whether the smaller Golgi twins are involved in subcellular delivery of proteins, which may potentially aid in cytokinesis, and what the driving force is for formation of the smaller twins at the midbody and subsequent journey to the perinuclear region.
Golgi twins revealed by LM/EM. Main picture Proliferating population of HeLa cells, of which some cells express Golgi-targeted GFP-tetracysteine (green) fixed and stained for the Golgi protein Giantin (blue) and microtubules (green). Note the different morphologies of the Golgi apparatus depending on cell cycle phase. Inset Sequence of time lapse of the same marker (GFP: green; tetracysteine-labeled with ReAsH red, depicted on the transmission image). The cells were fixed when the Golgi twins were formed and subsequently processed and analyzed by electron microscopy (bottom-right). See text and (Gaietta et al. 2006) for further details. [Image: G.M. Gaietta, T.J. Deerinck and BNGG at NCMIR]
Concluding remarks
Exaggeratedly stated, with fluorescence microscopy, you almost see nothing, i.e., only your fluorescent signal(s); with EM, you see everything, i.e., organelles, macromolecules and membranes. Bridging dynamic imaging in living cells and ultrastructural examination of proteins of interest opens a wide variety of possibilities to study protein localization, dynamics, activity and function at high spatiotemporal resolution inside cells. The parallel development of genetic-encoded tags for live-cell imaging, i.e., the fluorescent tags and the tetracysteine-system, as well as fluorescent nanocrystals that have to be targeted by affinity, i.e., QDs, are now finding their way into labs as standard tools to help to uncover molecular mechanisms in biology. The combination of microscopy not only helps to develop high-throughput, quantitative EM, but also aids in easier implementation of EM by researchers not familiar with the technique who can optimize labeling by light microscopic analysis. The novel probes and techniques form a whole new and powerful toolbox brightening the future for scientists who study molecular mechanisms that underlie cell behavior in health and diseases.
References
Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645
Bruchez M Jr, Moronne M, Gin P, Weiss S, Alivisatos AP (1998) Semiconductor nanocrystals as fluorescent biological labels. Science 281:2013–2016
Chan WC, Nie S (1998) Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 281:2016–2018
Chen I, Ting AY (2005) Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol 16:35–40
Chudakov DM, Lukyanov S, Lukyanov KA (2005) Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol 23:605–613
Deerinck TJ, Martone ME, Lev-Ram V, Green DP, Tsien RY, Spector DL, Huang S, Ellisman MH (1994) Fluorescence photooxidation with eosin: a method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol 126:901–910
Deerinck TJ, Giepmans BN, Smarr BL, Martone ME, Ellisman MH (2007) Light and electron microscopic localization of multiple proteins using quantum dots. Methods Mol Biol 374:43–53
Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH (2002) Multicolor and electron microscopic imaging of connexin trafficking. Science 296:503–507
Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, Tsien RY, Ellisman MH (2006) Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc Natl Acad Sci USA 103:17777–17782
Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH (2005) Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat Methods 2:743–749
Giepmans BN, Adams SR, Ellisman MH, Tsien RY (2006) The fluorescent toolbox for assessing protein location and function. Science 312:217–224
Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW Jr (2007) Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS ONE 2:e1011
Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T (2005) Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods 2:857–862
Griffin BA, Adams SR, Tsien RY (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281:269–272
Heintzmann R, Ficz G (2006) Breaking the resolution limit in light microscopy. Brief Funct Genomic Proteomic 5:289–301
Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY (2008) Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat Methods 5:397–399
Jares-Erijman EA, Jovin TM (2003) FRET imaging. Nat Biotechnol 21:1387–1395
Lauf U, Giepmans BN, Lopez P, Braconnot S, Chen SC, Falk MM (2002) Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells. Proc Natl Acad Sci USA 99:10446–10451
Lippincott-Schwartz J, Patterson GH (2003) Development and use of fluorescent protein markers in living cells. Science 300:87–91
Martin BR, Giepmans BN, Adams SR, Tsien RY (2005) Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol 23:1308–1314
Martin BR, Deerinck TJ, Ellisman MH, Taylor SS, Tsien RY (2007) Isoform-specific PKA dynamics revealed by dye-triggered aggregation and DAKAP1alpha-mediated localization in living cells. Chem Biol 14:1031–1042
Meisslitzer-Ruppitsch C, Vetterlein M, Stangl H, Maier S, Neumuller J, Freissmuth M, Pavelka M, Ellinger A (2008) Electron microscopic visualization of fluorescent signals in cellular compartments and organelles by means of DAB-photoconversion. Histochem Cell Biol. 8 May 2008 (Epub ahead of print)
Mishin AS, Subach FV, Yampolsky IV, King W, Lukyanov KA, Verkhusha VV (2008) The first mutant of the Aequorea victoria green fluorescent protein that forms a red chromophore. Biochemistry 47:4666–4673
Monosov EZ, Wenzel TJ, Luers GH, Heyman JA, Subramani S (1996) Labeling of peroxisomes with green fluorescent protein in living P pastoris cells. J Histochem Cytochem 44:581–589
Nisman R, Dellaire G, Ren Y, Li R, Bazett-Jones DP (2004) Application of quantum dots as probes for correlative fluorescence, conventional, and energy-filtered transmission electron microscopy. J Histochem Cytochem 52:13–18
O’Hare HM, Johnsson K, Gautier A (2007) Chemical probes shed light on protein function. Curr Opin Struct Biol 17:488–494
Piljic A, Schultz C (2008) Simultaneous recording of multiple cellular events by FRET. ACS Chem Biol 3:156–160
Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotechnol 22:1567–1572
Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY (2008) Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5(6):545–551
Sosinsky GE, Giepmans BN, Deerinck TJ, Gaietta GM, Ellisman MH (2007) Markers for correlated light and electron microscopy. Methods Cell Biol 79:575–591
Szent-Gyorgyi C, Schmidt BF, Creeger Y, Fisher GW, Zakel KL, Adler S, Fitzpatrick JA, Woolford CA, Yan Q, Vasilev KV et al (2008) Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol 26:235–240
Tour O, Adams SR, Kerr RA, Meijer RM, Sejnowski TJ, Tsien RW, Tsien RY (2007) Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat Chem Biol 3:423–431
van der Krogt GN, Ogink J, Ponsioen B, Jalink K (2008) A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS ONE 3:e1916
Acknowledgments
I thank Lisette van Esseveldt for editorial assistance and Mark Ellisman (National Center for Microscopy and Imaging Research, UCSD), Roger Tsien (UCSD) and their lab members (special thanks to Stephen Adams, Tom Deerinck, Guido Gaietta and Brent Martin) for support and collaborations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Giepmans, B.N.G. Bridging fluorescence microscopy and electron microscopy. Histochem Cell Biol 130, 211–217 (2008). https://doi.org/10.1007/s00418-008-0460-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-008-0460-5