Abstract
It has recently been shown in mice that the plasma membrane Ca2+-ATPase isoform 4 (PMCA4) is essential for sperm fertilization capacity. We analyzed whether sperm PMCA4 is formed in the rat during spermatogenesis or is synthesized in the epididymis and transferred onto sperm during sperm maturation. We could show that PMCA4 is conserved in sperm from testis to epididymis. In testis, PMCA4 mRNA was restricted to spermatogonia and early spermatocytes, while the PMCA4 protein was detected in spermatogonia, late spermatocytes, spermatids and in epididymal sperm. In epididymis PMCA4 mRNA was localized in basolateral plasma membranes of epithelial cells of the caput, corpus and cauda epididymidis. In contrast, the protein was only detectable in the epithelial cells of the caput, indicating that PMCA4 mRNA is only translated into protein in caput epithelium. In the epididymal corpus and cauda, PMCA4 mRNA and protein, respectively, was localized and in peritubular cells. Furthermore, we detected an identical distribution of PMCA4a and b splice variants in rat testis, epididymal corpus and cauda. In the caput epididymidis, where PMCA4 is located in the epithelium splice variant 4b was more prominent. Further experiments have to clarify the functional importance of the differences in the PMCA4 distribution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Like all eukaryotic cells, sperm use Ca2+ signals to elicit physiological responses. In mammalian spermatozoa, Ca2+ signals are considered as prime regulators of sperm motility, capacitation and the acrosome reaction (Babcock and Pfeiffer 1987; Darszon et al. 1999; O’Toole et al. 2000; Publicover and Barratt 1999; Ren et al. 2001). Ca2+ ions entering the sperm cytoplasm from extracellular space or from intracellular stores must be returned to the extracellular milieu or the intracellular storage organelles to guarantee a Ca2+ ion concentration gradient between the extracellular space and the cytoplasm. Therefore, efficient systems must exist to remove the Ca2+ ions from the cytosol. In the plasma membrane, this function is performed by two membrane proteins, Na+/Ca2+ exchanger and the plasma membrane Ca2+-ATPase (PMCA). Furthermore, a Ca2+ pump is responsible for the import into the endo/sarcoplasmic reticulum (SERCA). As has recently been shown in mouse sperm, the Ca2+-ATPase pump of the plasma membrane performs the major task of Ca2+ clearance to achieve a low resting Ca2+ concentration, whereas the contribution of the Na+/Ca2+ exchanger, mitochondrial uniporter and the SERCA pump to this effect is low or nil (Wennemuth et al. 2003).
The PMCAs belong to a family of P-type primary ion transport ATPases, which form a phosphorylated intermediate (an aspartyl phosphate) during the reaction cycle. The amount of PMCA in the plasma membrane is low and never exceeds 0.1–0.3% of total membrane protein. Till date, four isoforms (PMCA1-4) have been identified, each encoded by a separate gene (Carafoli 1992; Carafoli and Brini 2000; Guerini 1998; Stauffer et al. 1993). The sequence differences are found mostly in the C- and N-terminal regions of the protein. Furthermore, by alternative splicing at least two different sites, more than 20 PMCA variants can be generated. As the PMCA isoforms are characterized by variable regulatory and kinetic properties, they are obviously optimized for the different functional tasks in different tissues. PMCA1 and PMCA4 are expressed in most tissues, suggesting that they are necessary in housekeeping functions. PMCA2 and PMCA3 are restricted to a limited number of tissues, suggesting tissue-specific functions (Strehler and Treiman 2004; Strehler and Zacharias 2001). Studies of knockout mice carrying PMCA1, PMCA2 or PMCA4 null mutations point to the in vivo function of these isoforms. Loss of both the copies of the gene encoding PMCA1 causes embryonic lethality. Heterozygous PMCA1 mutants exhibit no apparent disease phenotype. These results are consistent with an essential housekeeping function of PMCA1 (Okunade et al. 2004; Prasad et al. 2007, 2004). In contrast, PMCA2 null mice show deafness and balance disorders (Kozel et al. 1998; Street et al. 1998; Takahashi and Kitamura 1999). Homozygous mice with a targeted gene deletion of PMCA4 are infertile due to severely impaired sperm motility. Null mice had normal spermatogenesis and mating behavior. Sperm taken from these animals that had not undergone capacitation exhibited normal motility, but could not achieve hyperactivated motility needed to traverse the female genital tract. Therefore, a pivotal role of PMCA4 on the regulation of sperm function and intracellular Ca2+ levels in sperm was claimed (Okunade et al. 2004; Schuh et al. 2004; Withers et al. 2006).
Sperm leaving the testis are morphologically differentiated, but functionally immature; they can neither swim nor fertilize an egg. During their transit through the epididymis, spermatozoa obtain their fertilization capacity. One main function of the epididymis is sperm maturation. Thereby, many biochemical characteristics are modified, like changes in phospholipid and cholesterol composition of the plasma membrane, modification of plasma membrane protein composition and nuclear condensation of the sperm (Cooper 1995, 1996; Jones 1989, 1998). Furthermore, it is hypothesized that epididymis-derived proteins, e.g. some GPI-anchored proteins, can be transferred to the sperm plasma membrane. The vehicles of transfer are presumably the so-called epididymosomes, which are secreted in an apocrine mode by the epididymal epithelium (Eickhoff et al. 2001; Frenette and Sullivan 2001; Kirchhoff and Hale 1996; Sullivan et al. 2005, 2007). All together this transfer improves the ability of sperm motility, zona pellucida binding property and fusion with the egg plasma membrane (Cooper 1995, 1996, 2007; Jones 1989, 1998; Jones et al. 2007; Sullivan et al. 2005).
As PMCA4 plays an important role in sperm fertilization capacity, the aim of the present study was to elucidate whether PMCA4 is generated already during spermatogenesis or is synthesized in diverse parts of the epididymis and transferred onto sperm during sperm maturation. Therefore, we studied the gene and protein expression of PMCA4 in both rat spermatogenic cells as well in the epididymis and in epididymal sperm.
Materials and methods
Collection of rat tissue and epididymal sperm
Male mature Wistar rats (56 days old, 250–300 g) were anesthetized with forene® and killed by cervical dislocation. A total number of 65 rats were used. Sperm were released from rat caput and cauda epididymidis by incising the duct several times with a sharp razor blade and swirling the tissue in 250 mM sucrose solution containing 5 mM Tris–HCl buffer pH 7.4, 1 mM EDTA. Released sperm were centrifuged for 10 min at 2,500×g at 4°C. Rat brain, testis and epididymis (caput, corpus, and cauda) were taken from the animal and were immediately frozen in liquid nitrogen.
Gene expression of PMCA4 in rat testis and epididymis
Total RNA isolation and reverse transcriptase-PCR
Total RNA from rat brain, epididymis (caput, corpus, and cauda) and testis were extracted with TriFAST Reagent (Peqlab, Erlangen, Germany) according to the manufacturer’s protocol. The organs of three rats were used for RNA isolation. RNA was treated with DNase I (RNase-free, Roche Mannheim, Germany). The concentration of total RNA was measured spectrophotometrically at 260 nm. One microgram of total RNA was reverse-transcribed to a single-stranded cDNA using M-MLV H-reverse transcriptase (200.000 U/ml, Promega Madison, WI), Oligo-dT15 (Promega, Madison, WI) and 0.5 mM dNTPs (Promega, Madison, WI) in a final volume of 20 μl for 90 min at 37°C.
The cDNA fragments of PMCA4 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were amplified in a total volume of 50 μl, by addition of 50 pmol of reverse and forward primers, 2 mM MgCl2, 5 μl 10× PCR buffer, 0.25 U PlatinumTaq DNA polymerase (Invitrogen, de Schelp, The Netherlands), 10 mM nucleotide mix (Promega, Madison, WI), to 1–2 μl of the obtained reverse transcriptase (RT) reaction mixture on a MJ Research PTC-200 Peltier Thermal cycler (Biozym, Hessisch Oldendorf, Germany). The following oligonucleotide primers for PMCA4 and GAPDH were purchased from MWG, (Ebersberg, Germany). The primer sequences were as follows: GAPDH: forward primer 5′-CAT TGT TGC CAT CAA CGA CC-3′, reverse primer 5′-TCA CAC CCA TCA CAA ACA TG-3′, (320 bp, bp111–430; Tso et al. 1985). PMCA4-iso: forward primer 5′-AAC CAG GTC TCC ATC CAC AG-3′; reverse primer 5′-CGG CAT TGT TAT TCG TGT TG-3′ (175 bp, bp 3,706–3,880, gi:54312087; Keeton and Shull 1995). PMCA4-spv: forward primer 5′-CAC GTT TGT GCT GAT GCA ACT-3′; reverse primer 5′-TGA AAC AGT TTC AGC ATC CG-3′ (793 bp, bp 3,062–3,855, gi:54312087; Keeton and Shull 1995). For GAPDH, 25 cycles consisting of a denaturation period of 30 s at 94°C, an annealing period of 30 s at 60°C, and an elongation period of 40 s at 72°C were performed. For PMCA4-iso, 36 cycles performing a denaturation period of 30 s at 94°C, an annealing period of 60 s at 58°C, and an elongation period of 60 s at 72°C were carried out. For PMCA4-spv, 33 cycles composed of a denaturation period of 30 s at 94°C, an annealing period of 20 s at 59°C, and an elongation period of 30 s at 72°C were performed. The first denaturation phase was extended to 1 min 30 s at 98°C and the last elongation phase was extended to 10 min. As negative controls, the master-mixture without addition of cDNA and RNA without RT were used.
Cloning of PCR products
The resulting PCR products were cloned in a pCR®II vector using the TA Cloning Kit Dual Promotor (Invitrogen, de Schelp, Netherlands) and transformed into E.coli strain inv-alpha F´. Plasmid preparation was performed using the Plasmid-Kit from Quiagen (Hilden, Germany). The presence of cloned insert was checked by digestion with EcoRI (Amersham, Freiburg, Germany) by gel electrophoretic analysis. Furthermore, the plasmids were sequenced by MWG (Ebersberg, Germany) and the sequences received were aligned using BLAST at NCBI.
Labeling of riboprobes by in vitro transcription
For in vitro transcription, the template PMCA4 cDNAs (793 bp fragment and 619 bp fragment, primer PMCA4-spv) were linearized by digestion with BamHI and XhoI. Subsequently, the samples were treated with alkaline phosphatase and proteinase K (Roche Diagnostics, Mannheim, Germany), as described in the instructions, and were purified by phenol/chloroform-extraction and ethanol-precipitation, as described previously (Sambrook et al. 1989). The RNA labeling with digoxigenin (DIG) was performed with the RNA-labeling kit (Roche Diagnostics, Mannheim, Germany) following the manufacturer’s instructions.
In situ hybridization
Three adult rats (56 days, 250–300 g) were perfused by vascular perfusion with Bouin’s fixative in PBS (pH 7.4). Testis and epididymis were removed and subsequently postfixed in the same solution for another 6 h. Tissues were dehydrated in ethanol, transferred into methyl benzoate and benzene and embedded in paraffin. Sections were cut at 5–6 μm thickness, mounted on superfrost plus glass slides (Sigma, München, Germany) and processed for in situ hybridization after removal of paraffin in xylene and ethanol. In situ hybridization was performed, as previously described in detail (Wilhelm et al. 1999). Briefly, sections were treated with 0.1 M HCl for 10 min and incubated with proteinase K (15 μg/ml) for 30 min at 37°C to increase probe accessibility. After incubation with 4% paraformaldehyde in PBS (pH 7.4) for 5 min, sections were rinsed twice in PBS. Thereafter, sections were acetylated and prehybridized (50% deionized formamide, 50 mM Tris–HCl-buffer, pH 7.5 containing, 25 mM EDTA, 20 mM NaCl, 250 μg/ml t-RNA and 2.5× Denhardt’s solution) for 2h at 45°C. Hybridization was performed overnight at 45°C in hybridization buffer, consisting of prehybridization buffer plus 10% dextran sulphate and DIG-labelled probe (5 ng/μl). Both the anti-sense and sense probe of the 793 bp fragment (splice variant a) and 619 bp fragment (splice variant b) were used. Unbound probe was removed by sequential washing, as previously described (Wilhelm et al. 1999). The hybridized probe was localized using an anti-DIG alkaline phosphatase-conjugated Fab-fragment (diluted 1:500 overnight, Roche Diagnostics, Mannheim, Germany). Bound alkaline phosphatase was visualized by an enzyme-catalysed color reaction utilizing 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium salt (NBT; Promega, Madison, WI) as chromogenic substrates.
Immunohistochemistry of PMCA and PMCA4 in rat testis and epididymis
Deparaffinized sections of rat testis and epididymis were rinsed two times in PBS, blocked in 5% dried non-fat milk powder for 30 min and were incubated at 4°C over night with the pan-PMCA antibody (5F10, monoclonal, Dianova, Hamburg, Germany, 1:2000) or isoform specific PMCA4 antibody (JA9, monoclonal, Dianova, Hamburg, Germany, 1:1000). Bound antibody was detected using the CSA-Kit (Dako, Hamburg) according to the manufacturer’s instructions. Sections were counterstained with hematoxilin.
Identification of PMCA4 in rat epididymal sperm
Immunofluorescence studies on sperm smears
Immunofluorescence studies on sperm smears were performed as previously described (Okunade et al. 2004). Sperm released from caput, corpus or cauda epididymidis were dropped on slides and allowed to settle for 30 min. Adherent sperm were fixed overnight in 4% formalin in PBS at 4°C and washed four times in PBS. Sperm were permeabilized with 0.1% SDS in PBS for 15 min at room temperature and washed several times in PBS. Nonspecific binding sites were blocked with blocking medium (2% goat serum in 1% BSA, 0.1% gelatine in PBS) for 1 h. Slides were incubated with primary antibody (5F10, JA9) diluted 1:50 in blocking medium overnight at 4°C. After washing the slides four times with blocking medium at room temperature, antibodies were detected with goat anti mouse IgG labelled with Cy3 (Dianova, Hamburg, 1:200) for 1 h. After washing three times with PBS, slides were mounted on VecaMount (Vector Laboratories) and nuclei were fluorescence-labeled with DAPI.
Preparation of microsomal membranes
Microsomal membranes of rat sperm were prepared according to Sikdar et al. (1991). Epididymal sperm of 30 rats were pooled for one preparation of microsomal membranes. The preparation of membranes was repeated twice. Sperm originating either from the epididymal caput or cauda as well as rat brain were homogenized in homogenization buffer containing 25 mM imidazole pH 7.5, 1 mM EDTA, 1 mM DTT, 250 mM sucrose and the protease inhibitor Complete™ (1 tablet/50 ml, Roche Diagnostics, Mannheim, Germany) using a Potter-Elvejem tissue homogenizer. The homogenate was centrifuged for 10 min at 600×g at 4°C, the supernatant was collected and the pellet was resuspended in 25 mM imidazole buffer and was centrifuged again at 600×g. This step was repeated and the pooled supernatants were spun down at 12,000×g and 4°C for 10 min. The microsomal membranes were pelleted for 1 h at 100,000×g at 4°C. The pellet was resuspended in a small volume of homogenization buffer and the protein concentration was determined as previously described (Bradford 1976).
Western blot
The sperm and brain membrane pellets (positive control) were then resuspended in SDS-gel sample buffer without 2-mercaptoethanol and boiled for 5 min. After centrifugation at 10,000×g for 5 min, the supernatant was removed, and boiled in the presence of 5% 2-mercaptoethanol for 5 min. Sperm and brain proteins were separated on 4–12% Bis–Tris SDS gels with MES-running buffer (NuPAGE, Invitrogen, Paisley, UK) according to the manufacturer’s instructions and were transferred onto nitrocellulose membranes (Hybond, Amersham, Freiburg, Germany). Prior to the incubation with antibodies, non-specific binding was blocked with 5% dried non-fat milk powder in TBS. Blots were incubated with the pan-PMCA antibody (5F10, monoclonal, Dianova, Hamburg, Germany) or isoform specific PMCA4 antibody (JA9, monoclonal, Dianova, Hamburg, Germany), both at a dilution of 1:1000 at room temperature over night. Subsequently, blots were incubated with ECL-anti-mouse IgG horseradish-peroxidase (POD) antibody (1:2000, Amersham, Freiburg, Germany). To visualize the peroxidase reaction, the enhanced chemiluminescence detection (ECL, Amersham, Freiburg, Germany) was used according to manufacturer’s instructions.
Ca2+-ATPase activity assay
Mg2+-dependent- and Mg2+-independent Ca2+-ATPase activity assays were performed using microsomal membranes of sperm from the caput or cauda epididymidis according to Sikdar et al. (1991). For assay of Mg2+-dependent activity, 5 μg sperm protein and 3 mM ATP were added to the reaction buffer containing 54.5 mM histidine hydrochloride, 2 mM EGTA, 0.2 mM DTE, 50 mM sucrose, 1 mM MgCl2, 4 mM CaCl2 in a total volume of 250 μl. After 30 min at 37°C, the reaction was stopped by adding 70 μl of 30% ice-cold trichloric acid (TCA) and the sample was subsequently diluted with 550 μl of distilled water. To determine the amount of released inorganic phosphate (Pi), 100 μl of 1.75% ammonium molybdate and 100 μl of 2% ascorbic acid were added. After 10 min of incubation at room temperature the complexed Pi was measured colorimetrically at 820 nm. The activity of the Mg2+-independent Ca2+-ATPase was measured in a buffer system containing 50 mM imidazole, 2 mM EGTA, 0.2 mM DTE, 50 mM sucrose, 4 mM CaCl2, as described above. In both cases Ca2+-ATPase activity was defined as the difference of activity in the presence and absence of Ca2+ ions.
Results
Gene expression of PMCA4 in rat testis and epididymis
In order to analyze the stage of spermatogenesis and the cell types of the epididymis where PMCA4 is expressed, in situ hybridization on paraffin sections of rat testis and rat epididymis were performed. The experiments gave evidence of a substantial expression of PMCA4 RNA in spermatogonia and primary spermatocytes but not in later spermatogenic stages such as secondary spermatocytes or spermatids (Fig. 1a). Strong expression was found in the epithelium in the principal cells of the caput, corpus and cauda epididymidis, but not in clear cells of the cauda epididymidis. The stroma was negative, while in the peritubular cells of the corpus and cauda epididymidis a weak signal was visible (Fig. 1b–d).
In situ hybridization on paraffin-embedded tissues: in rat testis strong expression of PMCA4-RNA was visible in spermatogonia and primary spermatocytes using the DIG-labeled PMCA4 anti-sense probes. In contrast, no expression was detected in later spermatogenic stages and in Sertoli cells (a). In the epididymal caput, strong mRNA expression is detected in all epithelial cells and peritubular cells. The stroma shows no PMCA4-RNA expression (b). PMCA4 expression was also shown in epithelial cells in addition to a weak signal in peritubular cells of the corpus epididymidis (c). In the cauda epididymidis, a strong mRNA expression was shown for epithelial cells excluding the clear cells. A faint reaction was detected in peritubular cells (d). Both the PMCA4 anti-sense probe of the 793 and 619 bp PCR-fragment showed identical results. The figures displayed here represent the in situ hybridization using the 793 bp PCR-fragment probe. In negative controls, the DIG-labeled PMCA4 sense probe failed to label all tissues (inserts). Sg Spermatogonia, Sc spermatocytes, arrows sertoli cells, E epithelial cell, C clear cells, arrow head peritubular cell. Bar 10 μm
To investigate the expression of PMCA4 and the PMCA4 splice variants, a and b, RT-PCR was performed using cDNA of rat testis as well as of caput, corpus and cauda epididymidis as templates. Rat brain cDNA served as a positive control. We demonstrated strong expression of PMCA4 in all organs using the PMCA4-iso primer, which recognizes all PMCA4 splice variants (Fig. 2). A combination of PCR analysis of alternative splicing patterns at splice site C and sequence analysis of the PCR products using the PMCA4-spv primer showed the distribution of PMCA4a and PMCA4b splice variant. Sequence analysis of the 793 bp PCR product corresponded to the PMCA4 cDNA sequence from bp 3,062 to 3,855 representing the PMCA4a splice variant (Schuh et al. 2003). The sequence analysis of the 619 bp PCR product matched with the cDNA sequence of PMCA4 from bp 3,062 to 3,478 and bp 3,652 to 3,855, the part from 3,479 to 3,651 was missing (Schuh et al. 2003). This sequence stands for PMCA4b. Our RT-PCR results illustrate a more or less identical distribution of PMCA4a and b in rat testis and epididymal caput and cauda, while the expression in epididymal corpus in general is weaker. In the epididymal caput, PMCA4b is more prominent than PMCA4a (Fig. 2).
RT-PCR analysis of rat testis (lane 2), epididymal caput (lane 3), corpus (lane 4) and cauda (lane 5) RNA using primers specific for PMCA4. Primer PMCA4-spv is overlapping splice site C. The PCR product of 794 bp represents PMCA4a and the 619 bp the PMCA4b. Primer PMCA4-iso corresponds to both the splice variants. Rat brain RNA (lane 1) served as positive control. RT-PCR of GAPDH shows RNA integrity in all organs. The RT-PCR was repeated three times, showing equal results. The PCR displayed in the figure, shows one representative experiment
Immunohistochemistry of PMCA and PMCA4 in rat testis and epididymis
To analyze the localization of PMCA4 protein in rat spermatogenic cells and epididymis, immunohistochemical studies were performed on paraffin embedded sections of rat testis and epididymis. For general localization of PMCA in the tissues, the commercial pan-antibody 5F10 was used. Thereby, PMCA was detectable in the plasma membranes of spermatogonia, spermatocytes and spermatids. The existence in spermatocytes was restricted to specific late spermatogenic stages. In addition, the cytoplasmic droplet of spermatozoa was strongly labeled. Furthermore, the peritubular cells were marked while the stroma in-between the seminiferous tubules was free of label (Fig. 3a–c; Table 1). In the caput and corpus epididymidis, the basolateral plasma membranes of the epithelial cells (principal cells) and the peritubular cells were stained. In the cauda epididymidis, the basolateral plasma membranes of the epithelial cells (principal cells) and the peritubular cells were marked, too. Clear cells appearing dominantly in the cauda epididymidis were unstained. In all the segments, the stroma was negative for PMCA (Fig. 4a–c; Table 1).
Immunohistochemistry in paraffin-embedded rat testis using monoclonal antibodies against PMCA (5F10; a–c) and PMCA4 (JA9; d–f), respectively: in rat testis, both overall PMCA immunoreactivity and specific PMCA4 immunoreactivity were detected stage-specific. PMCA and PMCA4 were localized in spermatogonia, late spermatocytes and spermatids, while the signal of PMCA4 was weaker than the overall PMCA signal. The cytoplasmic droplet was intensively marked. Sertoli cells were free of label. Sections were counterstained with hematoxilin. Sg Spermatogonia, Sc spermatocytes, St spermatids, Str round spermatids, Ste elongated spermatids, arrow head cytoplasmic droplet, arrow sertoli cell. Bar 10 μm
Immunohistochemistry in paraffin-embedded rat epididymis using monoclonal antibodies against PMCA (5F10; a–c), and PMCA4 (JA9; d–f): in the epididymal caput, overall PMCA immunoreactivity was shown in the basolateral plasma membranes of epithelial cells and in peritubular cells (a). PMCA4 was visible in the basolateral plasma membranes of the epithelial cells, too but was not localized in peritubular cells (d). In the corpus epididymidis, PMCA was detected in the basolateral membranes of epithelial and peritubular cells (b). In contrast, PMCA4 was only detected in peritubular cells (e). In the cauda epididymidis the basolateral membranes of epithelial cells, apart from clear cells and the peritubular cells were positive for PMCA (c). In contrast, PMCA4 was only detectable in peritubular cells of the cauda epididymidis (f). Sections were counterstained with haematoxilin. E Epithelial cell, C clear cells, arrow head peritubular cell. Bar 10 μm
In order to specify the localization of PMCA4 in more detail, a specific, commercial antibody against PMCA4 was used. In rat testis, the same stages of spermatogenesis showed PMCA4 in the plasma membrane, but the signal was diminished compared to the overall PMCA signal (Fig. 3d–f; Table 1). In caput epididymidis PMCA4 was localized in the basolateral plasma membranes of epithelial cells, while peritubular cells were negative. In contrast, PMCA4 was not detectable in the epithelial cells (principal cells) of the corpus and cauda epididymidis and in the clear cells of the cauda epididymidis. A pronounced signal was only demonstrated in the peritubular cells of the corpus and cauda epididymidis. The stroma of all the three epididymal parts was negative for PMCA4. (Fig. 4d–f, Table 1).
Identification of PMCA4 in rat epididymal sperm
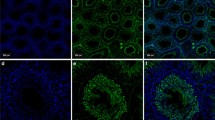
In order to elucidate whether or not epididymal sperm exhibit PMCA4, immunofluorescence studies, Western blot analysis and activity assays were carried out. Immunostaining of sperm using the pan antibody 5F10 and the PMCA4 specific antibody JA9 showed an intense fluorescence signal in the principal piece of the sperm tail of sperm originated from all parts of the epididymis. Furthermore, a more or less faint signal, which was more prominent using the pan PMCA antibody, was detected in the middle piece of the sperm tail (Fig. 5).
Subcellular localization of PMCA and PMCA4 in rat epididymal sperm. Sperm originating from caput (a, d) corpus (b, e) and cauda (c, f) epididymidis were analyzed using the pan PMCA antibody 5F10 (a–c) and the PMCA4-specific antibody JA9 (d–f) visualized with a Cy3 labeled secondary antibody (red). Nuclei were fluorescence-labeled using DAPI. PMCA and PMCA4 immunoreactivity was found predominately in the principal piece of the flagellum in all parts of the epididymis. In the mid piece a weak signal was only detected with the pan PMCA antibody. PP Principal piece, MP mid piece. Bar 10 μm
Performing Western blot studies, we detected immunoreactive bands of about 130 kDa in epididymal sperm using the pan PMCA antibody 5F10 as well as the isoform 4 specific antibody (JA9). Rat brain served as positive control (Fig. 6). Furthermore, we have found Ca2+-ATPase activity in sperm microsomal membranes. Both Mg2+-independent and -dependent Ca2+-ATPase activities were biochemically analyzed. Mg2+-independent activity was twice as high as Mg2+-dependent Ca2+-ATPase activity. Interestingly, a slightly higher activity was observed for Mg2+-independent Ca2+-ATPase activity in caput sperm, while the activity of Mg2+-dependent Ca2+-ATPase activity was somewhat higher in cauda sperm. The differences measured were not significant (Fig. 7).
Western blot analyses of rat epididymal caput (lane 1, 15 μg) and cauda sperm (lane 2, 15 μg). Microsomal membranes of rat brain (lane C, 15 μg) served as positive control. Proteins were probed with PMCA-pan antibody (clone 5F10) and isoform specific antibody against PMCA4 (JA9). A pronounced immunoreaction was shown at 130 kDa with both antibodies in epididymal caput and cauda sperm, respectively
Activity of Mg2+-independent and -dependent Ca2+-ATPase of microsomal membranes of rat epididymal caput (column 1) and cauda sperm (column 2). Vertical bars show SEM (n = 3). Ca2+-ATPase activity differences between epididymal caput and cauda sperm were not significant neither for Mg2+-independent nor for -dependent Ca2+-ATPase. Mg2+-independent Ca2+-ATPase activity was twice as high as Mg2+-dependent Ca2+-ATPase activity
Discussion
Functions of sperm PMCA
Capacitation, motility and acrosomal reaction are essential functions of the spermatozoon in the process of fertilization. Several steps of these functions are triggered by calcium dependent mechanisms, requiring pumping systems to remove excess intracellular calcium (Babcock and Pfeiffer 1987; Darszon et al. 1999; O’Toole et al. 2000; Publicover and Barratt 1999; Ren et al. 2001). It is known that PMCA performs the major task of Ca2+clearance in mouse spermatozoa (Wennemuth et al. 2003). Four different isoforms (PMCA 1-4) have been identified and multiple splice variants of these isoforms have been described (Carafoli et al. 1996; Guerini et al. 1998; Strehler and Zacharias 2001). Recently, PMCA4 has been reported to be important for male fertility. It has been shown in homozygous mice with a targeted gene deletion of isoform 4 of PMCA that these animals are infertile due to severely impaired sperm motility. The knock out mice had normal spermatogenesis and mating behavior. Sperm taken from these animals that had not undergone capacitation, exhibited normal motility, but could not achieve hyperactivated motility needed to traverse the female genital tract (Okunade et al. 2004; Schuh et al. 2004).
Expression and distribution of PMCA4 in testicular spermatogenic and epididymal sperm cells
In order to investigate the expression and distribution of PMCA4 in particular in rat germ cells stages, in situ hybridization and immunohistochemistry on rat testis sections were performed. In addition, rat epididymal spermatozoa were characterized by immunofluorescence technique, Western blot analysis and activity assays. Our results clearly point out that the mRNA expression of PMCA4 is evident in the basal compartment beneath the blood-testis barrier of the germinal epithelium in spermatogonia and in primary spermatocytes but not in secondary spermatocytes and spermatids. In contrast, the PMCA4 protein was localized in all stages of spermatogenesis, in spermatogonia, in singular specific stages of spermatocytes and in spermatids. The discrepancies between PMCA4 mRNA expression and PMCA4 protein localization indicate the different functional activities in the spermatogenic cells related to PMCA protein synthesis. While mRNA expression is restricted to premeiotic spermatogonia and early spermatocytes, the protein translation is expanded to later stages in spermatogenesis. Performing immunofluorescence studies, PMCA4 was detected in the principal piece of rat epididymal sperm originating from all parts of the epididymis. This is the same distribution, which was shown in mouse sperm (Okunade et al. 2004) and is located in direct vicinity to the Catsper Ca2+ channel, responsible for the Ca2+ influx, which is required for hyperactivated sperm motility (Ren et al. 2001). The PMCA occurrence in the middle piece must predominantly arise from another PMCA isoform than isoform 4. Furthermore, performing Western bloting analyses, we detected bands of about 130 kD using both the pan-PMCA antibody and the PMCA4 isoform specific antibody in epididymal sperm originating from the epididymal caput and cauda, respectively. The four PMCA isoforms range from 129 to 139 kDa in size, while the molecular weight of PMCA4b is 133 kDa and of PMCA4a is 129 kDa, respectively (Carafoli 1992; Caride et al. 1996; Strehler and Zacharias 2001). The PMCA4 positive bands lower in size in cauda and caput sperm might result from degradation of the protein (personal communication by A. Fileteo, Rochester, MN). The pan antibody (clone 5F10) used in this study, recognizes an epitope located between amino acids 724 and 783 of the human PMCA (Filoteo et al. 1997). This epitope is highly conserved in all isoforms, and, therefore, reacts with all isoforms. The isoform specific antibody (JA9) used has been mapped to amino acids 51–75 of PMCA4 ATPase. This sequence is common to both PMCA4a and 4b ATPase splice variants (Caride et al. 1996; Filoteo et al. 1997). To prove the functional activity of the protein Ca2+-ATPase activity assays were performed. Mg2+-dependent Ca2+-ATPase- and Mg2+-independent Ca2+-ATPase activity have been identified in both the epididymal sperm fractions. Activities in caput and cauda sperm were more or less similar, while the Mg2+-independent Ca2+-ATPase activity was twice has high as Mg2+-dependent Ca2+-ATPase activity. Both, Mg2+-dependent Ca2+-ATPase- and Mg2+-independent Ca2+-ATPase activity have also been shown in goat (Sengupta et al. 2007; Sikdar et al. 1991) and bull spermatozoa (Sanchez-Luengo et al. 2004; Triphan et al., 2007) as well as in microsomal membranes of rat testis (NagDas et al. 1988). Sen and co workers (NagDas et al. 1988; Sikdar et al. 1991) showed similar results with a Mg2+-independent activity, higher than Mg2+-dependent activity in both goat spermatozoa and rat testis microsomal membranes. Whether or not Mg2+-independent- and Mg2+-dependent Ca2+-ATPase are two different enzymes or identical molecules having the same or different catalytic sites has not yet been elucidated. Sen and co-workers favor the model of representing one enzyme with different catalytic sites (Sikdar et al. 1993).
Distribution of PMCA4 in rat epididymis
The main function of the epididymis besides resorption of the fluid derived from the seminiferous tubules and sperm transport is sperm maturation and storage (Cooper 1996). This sperm maturation process encompasses changes in phospholipid and cholesterol composition of the plasma membrane, modification of plasma membrane protein composition and nuclear condensation (Cooper 1995, 1996; Jones 1989, 1998). Furthermore, a transfer of proteins originating from the epididymal epithelium onto the spermatozoa is presumed (Kirchhoff and Hale 1996; Sullivan et al. 2005). In order to localize the PMCA4-mRNA and -protein in rat epididymis, in situ hybridization and immunohistochemistry studies were performed. In the caput epididymidis, the mRNA was detected in epithelial cells, and the PMCA4 protein was localized in the basolateral plasma membranes of principal and basal cells, respectively. The peritubular cells of the epididymal caput region show general PMCA immunoreactivity, but neither PMCA4 mRNA nor PMCA4 protein could be detected, indicating the presence of different isoforms in the peritubular cells of the epididymal caput. In contrast, although the mRNA of PMCA4 is expressed in principal cells of the corpus and cauda epididymidis, the PMCA4 protein was absent in all epithelial cells. One interpretation could be that the RNA is not translated into protein in the epididymal corpus or cauda epithelium. Nevertheless, PMCA in general was detectable in the basolateral membranes of principal cells pointing again to a different isoform pattern in principal cells of the corpus and cauda. Clear cells show neither PMCA4 mRNA expression nor PMCA nor PMCA4 immunoreactivity. The stroma in between the epididymal duct was negative in all parts. Main function of the cauda epididymidis is storage of spermatozoa, while the caput and corpus epididymidis are more involved in sperm maturation (Cooper 1995, 1996). Possibly, these different functions require peculiar conditions in the Ca2+ balance, and, therefore, unequal distribution of PMCA4 and other isoforms. A concentrated basolateral distribution of PMCA4 in general was also shown for other epithelial cells involved in resorption, e.g. in the intestines and distal tubules of the kidney (Friedman and Gesek 1995; Hoenderop et al. 2002; Larsson and Nemere 2002). In secretory epithelial cells such as pancreatic acinar cells, salivary gland cells and in epithelial cells of the rat-coagulating gland, PMCA is largely concentrated in the apical plasma membrane (Belan et al. 1996; Lee et al. 1997; Post et al. 2002). The principal cells of the epididymis combine both resorptive and secretory functions (Cooper 1999), therefore, the localization of PMCA in the plasma membrane in the caput epididymidis is conclusive; it indicates that PMCA4 contributes substantially to vectorial, transcellular Ca2+ transport from the apical to the basolateral compartment.
Each of the four PMCA genes can undergo alternative splicing, which may occur at two different sites. As the PMCA isoforms are characterized by different regulatory and kinetic properties, the splice variants are obviously optimized for the different functional tasks in variable tissues. In order to obtain more information on PMCA4 function in the reproductive organs, we studied which PMCA4 splice variant is expressed in sperm and their pathways. Splicing site A is located N-terminally yielding the splice variants x and y, while splicing site C is located C-terminally generating the splice variants a and b in PMCA4 (Keeton and Shull 1995). The functional effect of splicing site A is scarcely discussed, while the functional consequence of splice site C has been characterized in more detail. The presence of splice variants at this site affects the regulation of PMCA by calmodulin and phosphorylation as well as differential interaction with the PDZ domain containing anchoring and signaling proteins (Strehler and Zacharias 2001; Withers et al. 2006). In our analyses we could show some differences in the distribution of splice variant 4a and 4b in rat testis and epididymal corpus and cauda. Interestingly, splice variant 4b is more prominent especially in epididymal caput, which is the part of the epididymis where PMCA4 is localized in the epithelium. PMCA4b has the lower basal activity, but a higher stimulatory dependence on calmodulin compared to PMCA4a. PMCA4a has a lower affinity for calcium (Enyedi et al. 1994; Strehler and Zacharias 2001). Furthermore, the last four COOH-terminal residues of PMCA4b conform to the minimal consensus sequence for binding to the PDZ protein interaction domain, while PMCA4a has an aberrant C-terminus and is, therefore, unable to interact with PDZ-domains (Kim et al. 1998). PDZ proteins, the potential binding partner for the splice variant b, have the ability of linking membrane channels, receptors and transporters to the cytoskeleton. As both the splice variants are present in testis and epididymis, an optimized function of PMCA4 in the tissues is obvious.
Conclusions
The focus of the present study was to analyze whether or not PMCA4 is formed already during spermatogenesis or is differently synthesized in the various parts of the epididymis during sperm maturation. Therefore, the distribution of PMCA4 in different germ cell stages and epididymis was analyzed. We clearly show that PMCA4 is conserved in sperm from testes to epididymis. While mRNA expression of PMCA4 is restricted to spermatogonia and early spermatocytes, the protein is translated in spermatogonia, singular specific stages of spermatocytes and more developed germ cell stages and is present in sperm down to the epididymal cauda. In rat epididymal epithelium PMCA4 is restricted to the epididymal caput, where it is located in the basolateral plasma membranes of the epithelium. The epithelium from corpus and cauda epididymis exhibits no PMCA4 protein anymore. Interestingly, this corresponds to the distribution of the splice variants. We could show an identical distribution of PMCA4a and 4b splice variants in testis, epididymal corpus and cauda, while PMCA4b is more prominent in the epididymal caput. Further experiments have to clarify the functional significance of PMCA4 in sperm in general and the different PMCA4 splice variants in the reproductive tissues.
Reference
Babcock DF, Pfeiffer DR (1987) Independent elevation of cytosolic [Ca2+] and pH of mammalian sperm by voltage-dependent and pH-sensitive mechanisms. J Biol Chem 262:15041–15047
Belan PV, Gerasimenko OV, Tepikin AV, Petersen OH (1996) Localization of Ca2+ extrusion sites in pancreatic acinar cells. J Biol Chem 271:7615–7619
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal biochem 72:248–254
Carafoli E (1992) The Ca2+ pump of the plasma membrane. J Biol Chem 267:2115–2118
Carafoli E, Brini M (2000) Calcium pumps: structural basis for and mechanism of calcium transmembrane transport. Curr Opin Chem Biol 4:152–161
Carafoli E, Garcia-Martin E, Guerini D (1996) The plasma membrane calcium pump: recent developments and future perspectives. Experientia 52:1091–1100
Caride AJ, Filoteo AG, Enyedi A, Verma AK, Penniston JT (1996) Detection of isoform 4 of the plasma membrane calcium pump in human tissues by using isoform-specific monoclonal antibodies. Biochem J 316(Pt 1):353–359
Cooper TG (1995) Role of the epididymis in mediating changes in the male gamete during maturation. Adv Exp Med Biol 377:87–101
Cooper TG (1996) Epididymis and sperm function. Andrologia 28(Suppl 1):57–59
Cooper TG (ed) (1999) Epididymis. Academic Press, San Diego
Cooper TG (2007) Sperm maturation in the epididymis: a new look at an old problem. Asian J Androl 9:533–539
Darszon A, Labarca P, Nishigaki T, Espinosa F (1999) Ion channels in sperm physiology. Physiol Rev 79:481–510
Eickhoff R, Wilhelm B, Renneberg H, Wennemuth G, Bacher M, Linder D, Bucala R, Seitz J, Meinhardt A (2001) Purification and characterization of macrophage migration inhibitory factor as a secretory protein from rat epididymis: evidences for alternative release and transfer to spermatozoa. Mol Med 7:27–35
Enyedi A, Verma AK, Heim R, Adamo HP, Filoteo AG, Strehler EE, Penniston JT (1994) The Ca2+ affinity of the plasma membrane Ca2+ pump is controlled by alternative splicing. J Biol Chem 269:41–43
Filoteo AG, Elwess NL, Enyedi A, Caride A, Aung HH, Penniston JT (1997) Plasma membrane Ca2+ pump in rat brain. Patterns of alternative splices seen by isoform-specific antibodies. J Biol Chem 272:23741–23747
Frenette G, Sullivan R (2001) Prostasome-like particles are involved in the transfer of P25b from the bovine epididymal fluid to the sperm surface. Mol Reprod Dev 59:115–121
Friedman PA, Gesek FA (1995) Cellular calcium transport in renal epithelia: measurement, mechanisms, and regulation. Physiol Rev 75:429–471
Guerini D (1998) The significance of the isoforms of plasma membrane calcium ATPase. Cell Tissue Res 292:191–197
Guerini D, Garcia-Martin E, Zecca A, Guidi F, Carafoli E (1998) The calcium pump of the plasma membrane: membrane targeting, calcium binding sites, tissue-specific isoform expression. Acta Physiol Scand Suppl 643:265–273
Hoenderop JG, Nilius B, Bindels RJ (2002) Molecular mechanism of active Ca2+ reabsorption in the distal nephron. Annu Rev Physiol 64:529–549
Jones R (1989) Membrane remodelling during sperm maturation in the epididymis. Oxf Rev Reprod Biol 11:285–337
Jones R (1998) Plasma membrane structure and remodelling during sperm maturation in the epididymis. J Reprod Fertil Suppl 53:73–84
Jones RC, Dacheux JL, Nixon B, Ecroyd HW (2007) Role of the epididymis in sperm competition. Asian J Androl 9:493–499
Keeton TP, Shull GE (1995) Primary structure of rat plasma membrane Ca(2+)-ATPase isoform 4 and analysis of alternative splicing patterns at splice site A. Biochem J 306(Pt 3):779–785
Kim E, DeMarco SJ, Marfatia SM, Chishti AH, Sheng M, Strehler EE (1998) Plasma membrane Ca2+ ATPase isoform 4b binds to membrane-associated guanylate kinase (MAGUK) proteins via their PDZ (PSD-95/Dlg/ZO-1) domains. J Biol Chem 273:1591–1595
Kirchhoff C, Hale G (1996) Cell-to-cell transfer of glycosylphosphatidylinositol-anchored membrane proteins during sperm maturation. Mol Hum Reprod 2:177–184
Kozel PJ, Friedman RA, Erway LC, Yamoah EN, Liu LH, Riddle T, Duffy JJ, Doetschman T, Miller ML, Cardell EL, Shull GE (1998) Balance and hearing deficits in mice with a null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2. J Biol Chem 273:18693–18696
Larsson D, Nemere I (2002) Vectorial transcellular calcium transport in intestine: integration of current models. J Biomed Biotechnol 2:117–119
Lee MG, Xu X, Zeng W, Diaz J, Kuo TH, Wuytack F, Racymaekers L, Muallem S (1997) Polarized expression of Ca2+ pumps in pancreatic and salivary gland cells. Role in initiation and propagation of [Ca2+]i waves. J Biol Chem 272:15771–15776
NagDas SK, Mukherjee S, Mazumder B, Sen PC (1988) Identification and characterization of a Mg2+-dependent and an independent Ca+2-ATPase in microsomal membranes of rat testis. Mol Cell Biochem 79:161–169
O’Toole CM, Arnoult C, Darszon A, Steinhardt RA, Florman HM (2000) Ca(2+) entry through store-operated channels in mouse sperm is initiated by egg ZP3 and drives the acrosome reaction. Mol Biol Cell 11:1571–1584
Okunade GW, Miller ML, Pyne GJ, Sutliff RL, O’Connor KT, Neumann JC, Andringa A, Miller DA, Prasad V, Doetschman T, Paul RJ, Shull GE (2004) Targeted ablation of plasma membrane Ca2+-ATPase (PMCA) 1 and 4 indicates a major housekeeping function for PMCA1 and a critical role in hyperactivated sperm motility and male fertility for PMCA4. J Biol Chem 279:33742–33750
Post H, Wiche R, Sen PC, Hoffbauer G, Albrecht M, Seitz J, Aumüller G, Wilhelm B (2002) Identification of a plasma membrane Ca2+-ATPase in epithelial cells and aposomes of the rat coagulating gland. Prostate 52:159–166
Prasad V, Okunade GW, Miller ML, Shull GE (2004) Phenotypes of SERCA and PMCA knockout mice. Biochem Biophys Res Commun 322:1192–1203
Prasad V, Okunade G, Liu L, Paul RJ, Shull GE (2007) Distinct phenotypes among plasma membrane Ca2+-ATPase knockout mice. Ann N Y Acad Sci 1099:276–286
Publicover SJ, Barratt CL (1999) Voltage-operated Ca2+ channels and the acrosome reaction: which channels are present and what do they do? Hum Reprod 14:873–879
Ren D, Navarro B, Perez G, Jackson AC, Hsu S, Shi Q, Tilly JL, Clapham DE (2001) A sperm ion channel required for sperm motility and male fertility. Nature 413:603–609
Sambrook J, Fritsch EF, Maniatis T (eds) (1989) Molecular cloning: laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor
Sanchez-Luengo S, Aumuller G, Albrecht M, Sen PC, Rohm K, Wilhelm B (2004) Interaction of PDC-109, the major secretory protein from bull seminal vesicles, with bovine sperm membrane Ca2+-ATPase. J Androl 25:234–244
Schuh K, Uldrijan S, Gambaryan S, Roethlein N, Neyses L (2003) Interaction of the plasma membrane Ca2+ pump 4b/CI with the Ca2+/calmodulin-dependent membrane-associated kinase CASK. J Biol Chem 278:9778–9783
Schuh K, Cartwright EJ, Jankevics E, Bundschu K, Liebermann J, Williams JC, Armesilla AL, Emerson M, Oceandy D, Knobeloch KP, Neyses L (2004) Plasma membrane Ca2+ ATPase 4 is required for sperm motility and male fertility. J Biol Chem 279:28220–28226
Sengupta T, Ghoshal S, Sen PC (2007) Stimulation of Mg2+-independent form of Ca2+-ATPase by a low molecular mass protein purified from goat testes cytosol. Comp Biochem Physiol 146:131–138
Sikdar R, Ganguly U, Pal P, Mazumder B, Sen PC (1991) Biochemical characterization of a calcium ion stimulated-ATPase from goat spermatozoa. Mol Cell Biochem 103:121–130
Sikdar R, Ganguly U, Chandra GA, Sen PC (1993) Calcium uptake and Ca2+-ATPase activity in goat spermatozoa membrane vesicles do not require Mg2+. J Biosci 18:73–82
Stauffer TP, Hilfiker H, Carafoli E, Strehler EE (1993) Quantitative analysis of alternative splicing options of human plasma membrane calcium pump genes. J Biol Chem 268:25993–26003
Street VA, McKee-Johnson JW, Fonseca RC, Tempel BL, Noben-Trauth K (1998) Mutations in a plasma membrane Ca2+-ATPase gene cause deafness in deafwaddler mice. Nat Genet 19:390–394
Strehler EE, Treiman M (2004) Calcium pumps of plasma membrane and cell interior. Curr Mol Med 4:323–335
Strehler EE, Zacharias DA (2001) Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev 81:21–50
Sullivan R, Saez F, Girouard J, Frenette G (2005) Role of exosomes in sperm maturation during the transit along the male reproductive tract. Blood Cells Mol Dis 35:1–10
Sullivan R, Frenette G, Girouard J (2007) Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian J Androl 9:483–491
Takahashi K, Kitamura K (1999) A point mutation in a plasma membrane Ca(2+)-ATPase gene causes deafness in Wriggle Mouse Sagami. Biochem Biophys Res Commun 261:773–778
Triphan J, Aumüller G, Brandenburger T, Wilhelm B (2007) Localization and regulation of plasma membrane Ca(2+)-ATPase in bovine spermatozoa. Eur J Cell Biol 86:265–273
Tso JY, Sun XH, Kao TH, Reece KS, Wu R (1985) Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res 13:2485–2502
Wennemuth G, Babcock DF, Hille B (2003) Calcium clearance mechanisms of mouse sperm. J Gen Physiol 122:115–128
Wilhelm B, Meinhardt A, Renneberg H, Linder D, Gabius HJ, Aumuller G, Seitz J (1999) Serum albumin as a potential carrier for the apocrine secretion of proteins in the rat coagulating gland. Eur J Cell Biol 78:256–264
Withers S, Cartwright EJ, Neyses L (2006) Sperm phenotype of mice carrying a gene deletion for the plasma membrane calcium/calmodulin dependent ATPase 4. Mol Cell Endocrinol 250:93–97
Acknowledgments
The technical assistance of Michael Dreher, Anne Henkeler, Gudrun Hoffbauer and Claudia Keppler is gratefully acknowledged. The grant sponsor for this study is P. E. Kempkes-Foundation Marburg; Grant number: 12/04
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wilhelm, B., Brandenburger, T., Post, H. et al. Expression and localization of PMCA4 in rat testis and epididymis. Histochem Cell Biol 129, 331–343 (2008). https://doi.org/10.1007/s00418-007-0362-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-007-0362-y