Abstract
Alpha smooth muscle actin (α-SMA) was recently shown to be present in mouse subcutaneous tissue fibroblasts in the absence of tissue injury. In this study, we used a combination of immunohistochemistry and correlative confocal scanning laser and electron microscopy to investigate the structural organization of α-SMA in relation to the nucleus. Furthermore, we explored colocalization analysis as a method for quantifying the amount of α-SMA in close approximation to the nucleic acid marker, 4′,6-diamidino-2-phenyl-indole, dihydrochloride. Our findings indicate the presence of α-SMA within nuclear invaginations in close proximity to the nuclear membrane, but not in the nucleoplasm. Although the function of these α-SMA-rich nuclear invaginations is at present unknown, the morphology of these structures suggests their possible involvement in cellular and nuclear mechanotransduction as well as nuclear transport.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In a previous study, we determined that mouse subcutaneous tissue fibroblasts contained non-filamentous alpha smooth muscle actin (α-SMA). The distribution of α-SMA within these fibroblasts was dynamically responsive to changes in mechanical tissue forces. In contrast to cultured fibroblasts, however, α-SMA did not form distinct stress fibers and did not colocalize with phalloidin, indicating that it was not in filamentous or F-actin form (Langevin et al. 2006). In addition, α-SMA was observed in close proximity to the nucleus, raising the question of whether it was present within the nucleus itself. The existence of actin in the nucleus has long been a matter of debate, made difficult by its complex geometry and the presence of invaginations containing cytoplasm within the nuclear domain. At the time of our previous study, a more detailed examination of the intracellular distribution of α-SMA and its relationship to the nucleus was not possible at the electron microscopy level using the immunogold technique. Putatively, the low concentration of α-SMA in fibroblasts within mouse subcutaneous tissue in the absence of prior tissue injury, coupled with the potential detrimental effect of tissue preparatory methods on actin antigenicity, had yielded inconclusive results. To remedy this, we applied the recently developed quantum dot (Q-dot) labeling technology (Giepmans et al. 2005, 2006; Nisman et al. 2004) in a correlative technique combining confocal scanning laser and transmission electron microscopy on the same tissue sample. Q-dots are highly stable semiconductor nanocrystals (Arya et al. 2005) that are insensitive to laser exposure unlike fluorescent dyes, simplifying viewing and image capture by confocal microscopy. Moreover, Q-dots are electron dense, allowing specific viewing by electron microscopy of α-SMA distribution in relation to nuclear ultrastructure. In this study, this technique allowed us to specifically examine the relationship of α-SMA to nuclear invaginations. The distribution of α-SMA within the cell and its relationship to the nucleus may have important implications for our understanding of mechanical signal transduction from the cell periphery to the nucleus.
Materials and methods
Subcutaneous tissue sample preparation
Subcutaneous connective tissue was harvested immediately after death from the abdomen of C57BL/6 male mice (19–21 g). An 8 cm × 3 cm flap containing dermis, subcutaneous muscle, and subcutaneous tissue was dissected away from the abdominal wall musculature and excised. Tissue samples were incubated in a relaxed state for 30 min in HEPES physiological buffer, then fixed in 3% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for light microscopy. After fixation of whole tissue flaps, 1 cm × 1 cm subcutaneous tissue samples were dissected from the dermis while applying minimal traction on the tissue and mounted on Fisherbrand Superfrost/Plus (Fisher Scientific, Pittsburgh, PA, USA) glass microscope slides and rinsed three times for 10 min in PBS/1.0% bovine serum albumim (BSA)/0.1% Triton X-100.
Immunohistochemistry
Following tissue fixation and sample preparation, immunohistochemistry was performed for the detection of α-SMA using indirect immunofluorescence with a primary mouse monoclonal anti-α-SMA antibody from Sigma (St Louis, MO, USA) at a dilution of 1:100 in PBS/1.0% BSA and an Alexa 488-conjugated secondary goat anti-mouse antibody from Invitrogen Corporation (Carlsbad, CA, USA) at a dilution of 1:200 in PBS/1.0% BSA as previously described (Langevin et al. 2006). Following secondary antibody staining, samples were rinsed in PBS/1.0% BSA for 10 min then incubated with 4′,6-diamidino-2-phenyl-indole, dihydrochloride (DAPI) at a dilution of 1:1,000 in PBS/1.0% BSA for 5 min at room temperature (RT) to reveal cell nuclei. After two 10-min rinses in PBS/1.0% BSA, samples were overlaid with a #1.5 glass coverslip (Fisher Scientific, Pittsburgh, PA, USA) using 50% glycerol in PBS with 1% N-propylgallate as a mounting medium.
Histochemistry
Phalloidin, a specific stain for polymerized actin, was used to visualize subcutaneous tissue fibroblasts with confocal microscopy. After tissue fixation and sample preparation, samples were stained with Texas Red conjugated phalloidin (4 U/ml; Molecular Probes, Eugene, OR, USA) at a dilution of 1:25 in PBS/1.0% BSA for 40 min at 4°C.
Confocal scanning laser microscopy
Samples from a total of seven mice were imaged with a Zeiss LSM 510 META confocal scanning laser microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY, USA). For quantitative colocalization only, four to six fields per animal containing an average of two cells per field were imaged using a PlanApochromat 100× (NA = 1.4) oil immersion lens, yielding a total of 76 cells. All images were obtained using the following settings, ensuring that the most accurate colocalization data could be acquired: zoom of 2, box size of 1,024 × 1,024 pixels, scan speed of 7, and average line scan of 2. For each image, an average of 15 z-sections were captured with an optical section thickness of 0.7 μm for all laser lines and a 0.33-μm inter-image interval. Image stacks were imported into two different image analysis software packages where colocalization between α-SMA and DAPI was assessed using both the Zeiss LSM 510 and Volocity (Improvision, Lexington, MA, USA) programs. In this paper, we display colocalization images using the colocalization module in the LSM software package, but only voxel-based algorithms (Volocity software) were used for quantification of colocalization. In these calculations, colocalization is defined as the detection of signal at the same voxel location (defined as the three-dimensional equivalent of a pixel, the smallest discreet spatial component of a digital volume; volumes within Volocity consist of voxels, each with an associated color and/or intensity value) in each of two channels [DAPI (X) and α-SMA (Y)]. The image was cropped by thresholding the DAPI channel; to account for different staining levels between samples, a threshold of average intensity + 1.5σ was used in order to select the entire nucleus, the “fill holes” option was chosen, and objects less than 1 μm3 in size were excluded to reduce noise. Both X and Y channels were displayed as a scatterplot representing the intensity values of the two labels. For both channels, a maximum threshold value of 255 and a minimum threshold value of 50 were entered to reject low intensity pixels. Next, a region of interest (ROI) that selected the colocalized pixels, as defined by their color, was drawn directly on the scatterplot and colocalization statistics were generated for the selected ROI and computed based on their voxel intensity. The Volocity software colocalization macro utilizes the mathematical algorithms described by Manders et al. (1993). We specifically used colocalization coefficient, M(y), as our outcome measurement where M(y) is the fraction of actin (y) colocalized with DAPI, and represented mathematically as \( {{\sum\limits_i {y_{{i,{\text{coloc}}}} } }} \mathord{\left/ {\vphantom {{{\sum\limits_i {y_{{i,{\text{coloc}}}} } }} {{\sum\limits_i {y_{i} } }}}} \right. \kern-\nulldelimiterspace} {{\sum\limits_i {y_{i} } }}. \) This correlation coefficient (values are between 0 and 1) provides the most biologically relevant assessment of colocalization (Costes et al. 2004; Manders et al. 1993), where a value of 0 indicates an absence of colocalization between the two channels, and a value of 1 indicates complete colocalization.
Correlative confocal and electron microscopy
Confocal and electron microscopy were performed sequentially on the same tissue sample for the detection of α-SMA using a primary mouse monoclonal anti-α-SMA antibody at a dilution of 1:100, followed by a goat anti-mouse IgG conjugated to 655 nm Q-dots from Molecular Probes at a dilution of 1:200. Samples were prepared for primary incubation with the α-SMA antibody as described above, and after overnight primary antibody incubation, samples were rinsed twice for 5 min in PBS/1.0% BSA, followed by an overnight incubation at 4°C with the goat anti-mouse 655-Q-dots in PBS/0.1% Triton X-100. Control samples were incubated in the absence of the primary antibody. After incubation, samples were rinsed twice for 15 min in PBS/1.0% BSA, and stained with DAPI at a dilution of 1:1,000 for 5 min at RT. Finally, after two 15-min rinses in PBS/1.0% BSA, samples were coverslipped as above and imaged for confocal light microscopy with a Zeiss LSM 510 META confocal scanning laser microscope (Carl Zeiss MicroImaging Inc., Thornwood, NY, USA) using a PlanApochromat 63× (NA = 1.4) oil immersion lens and 488 nm laser excitation with a 650 long-pass emission filter. Immediately following confocal microscopy imaging, coverslips were removed from the slides and the tissue was further processed for electron microscopy. Briefly, the tissue was fixed for 30 min at 4°C in Karnovsky’s fixative (2.5% glutaraldehyde, 1.0% PFA in 0.1 M Millonig’s phosphate buffer, pH 7.2). After rinsing in Millonig’s buffer, the tissue was post-fixed in 1% osmium tetroxide for 1 h at 4°C and en bloc stained in 2% aqueous uranyl acetate overnight at 4°C. The tissue was dehydrated through graded ethanols and embedded in Spurr’s epoxy resin. Semithin sections (1 μm) were cut with glass knives on a Reichert ultracut microtome, stained with methylene blue—azure II, and evaluated for areas of interest. Ultrathin sections (60–80 nm) were cut with a diamond knife, retrieved onto 200 mesh nickel thinbar grids and examined with a JEOL 1210 Transmission Electron Microscopy (JEOL USA Inc., Peabody, MA, USA) operating at 80 kV.
Results
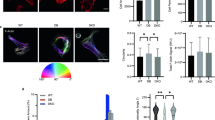
In mouse subcutaneous tissue, cytoplasmic α-SMA immunoreactivity was mostly diffuse and granular without clearly demarcated stress fibers and was not in F-actin form (indicated by lack of phalloidin colocalization) which was consistent with our previous report (Fig. 1). In addition to its presence in the cytoplasm, α-SMA staining also was observed within the nuclear domain, defined in this paper as the nucleoplasm plus a boundary layer of adjacent cytoplasm within 500 nm of the outer nuclear membrane. Figure 2 shows serial optical sections (0.33 μm interimage interval) through a subcutaneous tissue fibroblast. Several distinct α-SMA-containing structures are seen in cross section through the center of the nucleus (Fig. 2 middle row). The structures are predominantly oriented parallel to the long axis of the cell and appear continuous with the pattern of cytoplasmic α-SMA. In a given optical section (taken parallel to the plane of the tissue), the dimensions of these actin structures ranged from 2 to 12 μm in length and 0.2 to 1 μm in width, while their estimated “thickness” (measured across optical sections) ranged from 2 to 5 μm. In comparison, the nuclei of mouse subcutaneous tissue fibroblasts typically measured approximately 7 μm × 12 μm in the plane of the tissue and had a thickness of 4–7 μm (perpendicular to the tissue). Therefore, the observed α-SMA structures in these cells did not appear to be thin filaments, but rather resembled flat “sheets” extending through a large portion of the thickness of the nucleus and oriented predominantly perpendicular to the tissue plane. Figure 3 shows several branching α-SMA-containing structures that span the nucleus and appear continuous with cytoplasmic α-SMA (indicated by arrowhead), suggesting that these structures are deep, branching nuclear clefts or invaginations. When light microcopy images were analyzed using the LSM software pixel-based method with an intensity threshold of 75 for both α-SMA and DAPI, we observed that α-SMA staining colocalized quite extensively with DAPI predominately at the nuclear periphery including the nuclear clefts (Fig. 4), raising the question of whether α-SMA could be present within the nucleus itself. Among the 76 cells examined using the Volocity voxel-based method with a threshold intensity of 50 for both α-SMA and DAPI, mean ± SD colocalization coefficient M(y) of α-SMA and DAPI was 36 ± 0.1% (figure not shown).
Immunohistochemical staining of mouse subcutaneous tissue with anti-α-SMA antibody (green), counterstained with phalloidin (red) and DAPI (blue) and imaged with confocal laser scanning microscopy using a PlanApochromat 63× (NA = 1.4) oil immersion lens. The image shown is a projection of 12 optical sections representing a total thickness of 4 μm. Scale bar 20 μm
Serial confocal microscopy images showing a mouse subcutaneous tissue fibroblast stained for α-SMA (green) and DAPI (blue). The middle row of optical sections shows α-SMA positive staining in channel-like structures through the center of the nucleus. Optical sections represent a 0.33-μm interimage interval. Scale bar 5 μm
a and b Immunohistochemical staining of mouse subcutaneous tissue fibroblast with α-SMA (green), counterstained with DAPI (blue) and imaged with confocal laser scanning microscopy. Images depict one optical section imaged in the plane of tissue. Colocalization analysis was applied to this image in the LSM program, where white dots are overlayed on the image indicating colocalization of α-SMA and DAPI. Scale bars 2 μm
To further investigate the relationship of α-SMA to the nucleus, as well as additionally characterize its pattern within the cytoplasm, we performed correlative confocal fluorescence and transmission electron microscopy using Q-dots. Figures 5 and 6 show α-SMA in the same tissue sample, imaged first with laser confocal microscopy (Fig. 5) and then with electron microscopy (Fig. 6a, b). Abundant α-SMA staining was seen with electron microscopy both in the cytoplasm and within nuclear invaginations filled with cytoplasmic content (Figs. 6, 7, 8). Most Q-dots within the invaginations appeared to be in close proximity to the outer nuclear membrane. We saw no evidence that α-SMA was present within the nucleoplasm. Note that the Q-dot immunostaining for α-SMA was not found along filament bundles, but rather at discrete locations along the nuclear invaginations, corroborating the absence of phalloidin staining within these structures.
a Immunohistochemical staining of mouse subcutaneous tissue showing the distribution of α-SMA using 655 nm Q-dots (pseudocolored green) in relation to DAPI (blue) in fibroblasts. The image shown was captured using confocal laser scanning microscopy with a PlanApochromat 100× (NA = 1.4) oil immersion lens without a zoom and is a projection of 20 optical sections representing a total thickness of 7 μm. b Control tissue incubated with primary antibody omitted. Scale bar 10 μm
a and b Correlative electron microscopy of mouse subcutaneous tissue fibroblast stained with α-SMA antibody followed by 655 nm Q-dots conjugated to a secondary antibody and previously imaged by confocal microscopy. a Note the intense Q-dot labeling in the cytoplasm (arrowheads), while mitochondria (m) and nucleus (n) are devoid of label. b The Q-dot label (arrowheads) in a nuclear invagination is concentrated at the periphery of the nuclear envelope. Scale bars 200 nm
a and b Correlative electron microscopy of mouse subcutaneous tissue fibroblast stained with α-SMA antibody followed by 655 nm Q-dots conjugated to a secondary antibody and previously imaged by confocal microscopy. This micrograph shows the morphology of nuclear invaginations at low magnification (a), while the region boxed is shown at higher magnification in (b). Note that the Q-dots (arrowheads) are mostly associated with the periphery of the nuclear envelope, while the nucleoplasm itself (n) is devoid of label. Scale bars 1 μm (a); 200 nm (b)
a and b Correlative electron microscopy of mouse subcutaneous tissue fibroblast stained with α-SMA antibody followed by 655 nm Q-dots conjugated to a secondary antibody and previously imaged by confocal microscopy. a In this image a nucleus (n) is almost completely bisected by a nuclear invagination, illustrating the continuity of the invagination with the cytoplasm. The boxed region is shown at higher magnification in (b), where the Q-dots (arrowheads) are seen to be associated with the periphery of the nuclear envelope. Scale bars 500 nm (a); 200 nm (b)
Discussion
We report that, in fibroblasts within whole mouse subcutaneous tissue, α-SMA was located within deep nuclear invaginations containing cytoplasm. Q-dots were frequently seen in close proximity to the outer nuclear membrane, but not in the nucleus itself. Nuclear invaginations, folds or clefts containing cytoplasm are known to be related to the state of cellular differentiation and are thought to play a role in nucleocytoplasmic transport by bringing cytoplasmic space closer to the interior of the nucleus (Bourgeois et al. 1979; Fricker et al. 1997; Johnson et al. 2003; Lui et al. 1998). Furthermore, an association between invaginations and nucleoli has been reported, further suggesting their role in transport activities (Dupuy-Coin et al. 1986). Our finding that α-SMA is (1) abundant in nuclear invaginations, (2) in close proximity to the outer nuclear membrane, and (3) continuous with cytoplasmic actin suggest that nuclear invaginations may play a role in nuclear mechanotransduction in addition to nuclear transport. Direct transmission of forces through the cytoplasmic and nuclear cytoskeletons via changes in cell and nuclear shape have been proposed as a source of specific coupling between tissue mechanical forces and the genome (Hu et al. 2005; Ingber 2006; Maxwell and Hendzel 2001; Thomas et al. 2002). Recent evidence suggests that such force transduction may be mediated via nuclear membrane protein complexes including nesprins, Sun, and emerins (Crisp et al. 2006) which bind actin on the cytoplasmic side and lamins on the nuclear side of the nuclear membrane.
The presence of monomeric cytoplasmic actin within nuclear invaginations was previously suggested by a study using fluorescence recovery after photobleaching in cultured cells expressing fluorescent actin (Johnson et al. 2003). Our study of mouse fibroblasts in whole tissue is, to our knowledge, the first confirmation of the presence of cytoplasmic non-filamentous α-SMA within nuclear invaginations at the electron microscopy level. In a previous investigation (Langevin et al. 2006) we were unable to detect the presence of α-SMA in mouse subcutaneous tissue fibroblasts by immunoelectron microscopy using the immunogold technique. These negative results may have been due to the relative paucity of antigenic sites for α-SMA available in a thin section (60–80 nm thick) from Lowicryl K4M embedded tissues. The specimen surface relief of a Lowicryl section is of the order of 2–6 nm (Kellenberger et al. 1987), and these authors have calculated in their very extensive study that dispersed cellular proteins need to be present in a concentration of 10–100 μM in order to obtain satisfactory post-embedding immunoelectron microscopic results. The situation is likely more dire for compact proteins (such as cytoskeletal elements) where issues such as steric hindrance and non-randomness of orientation will certainly affect immunolabeling efficiency, prohibiting a theoretical calculation of required concentrations (Kellenberger et al. 1987). Since pre-embedding immunolabeling (as illustrated in the present manuscript by the Q-dot staining) exposes several orders of magnitude greater antigenic sites than the 2–6-nm specimen surface of a Lowicryl thin section, this may indeed explain our current success in detecting α-SMA antigenicity at the electron microscopic level.
The results of this study also illustrate the complexity of establishing the precise location of a given structural protein in relation to the nucleus as well as the superior power of electron microscopy, compared with confocal fluorescent microscopy, to resolve detailed cellular structure. In this study, our initial confocal microscopy results indicating colocalization of α-SMA and DAPI suggested that α-SMA may have been located in the nucleus itself. We pursued this finding using a rigorous colocalization analysis method, quantifying the amount of α-SMA associated with DAPI. We used established tissue fixation and processing techniques, followed by conventional fluorescence staining protocols and high-resolution confocal scanning laser microscopy where images were acquired to satisfy the Nyquist oversampling theorem, making them amenable for subsequent colocalization analyses. The colocalization analysis itself was performed on a voxel-by-voxel basis in Improvision Volocity software, and amongst all of its statistical parameters calculated, we selected the colocalization coefficient M(y), which describes the amount of α-SMA colocalized with DAPI, seemingly the most relevant biological parameter. In addition, we used several different image display techniques including orthogonal views and three-dimensional rendering to examine the structure occupied by α-SMA within the nucleus. In using all of these protocols, we detected extensive colocalization of α-SMA with DAPI. This false positive result, apparently indicating the presence of α-SMA within the nucleoplasm, was most likely due to lack of sufficient resolution in the plane orthogonal to the tissue section relative to the size of the nuclear invaginations, or to inherent uncertainty in setting the appropriate intensity colocalization threshold. Because raw, unprocessed confocal images were analyzed in this study, it is also possible that, although we exercised due care in the image parameter settings, the resolution and signal to noise ratio of the colocalization analysis could have been improved if the images had been subjected to medium filtering or deconvolution processing (Landmann and Marbet 2004). In this case, the Q-dot technique at the electron microscopy level allowed us to resolve this issue.
In summary, mouse subcutaneous tissue fibroblasts contain α-SMA that is distributed throughout the cytoplasm as well as in deep nuclear invaginations containing cytoplasm. Within these invaginations, α-SMA was in close approximation to the outer nuclear membrane. Although the function of this actin distribution is at present unknown, we suggest their possible involvement in cellular and nuclear mechanotransduction. Further studies will investigate the effect of tissue stretch on the distribution of α-SMA within the cell.
References
Arya H, Kaul Z, Wadhwa R, Taira K, Hirano T, Kaul SC (2005) Quantum dots in bio-imaging: revolution by the small. Biochem Biophys Res Commun 329:1173–1177
Bourgeois CA, Hemon D, Bouteille M (1979) Structural relationship between the nucleolus and the nuclear envelope. J Ultrastruct Res 68:328–340
Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S (2004) Automatic and quantitative measurement of protein–protein colocalization in live cells. Biophys J 86:3993–4003
Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D (2006) Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 172:41–53
Dupuy-Coin AM, Moens P, Bouteille M (1986) Three-dimensional analysis of given cell structures: nucleolus, nucleoskeleton and nuclear inclusions. Methods Achiev Exp Pathol 12:1–25
Fricker M, Hollinshead M, White N, Vaux D (1997) Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol 136:531–544
Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH (2005) Correlated light and electron microscopic imaging of multiple endogenous proteins using quantum dots. Nat Methods 2:743–749
Giepmans BN, Adams SR, Ellisman MH, Tsien RY (2006) The fluorescent toolbox for assessing protein location and function. Science 312:217–224
Hu S, Chen J, Butler JP, Wang N (2005) Prestress mediates force propagation into the nucleus. Biochem Biophys Res Commun 329:423–428
Ingber DE (2006) Cellular mechanotransduction: putting all the pieces together again. FASEB J 20:811–827
Johnson N, Krebs M, Boudreau R, Giorgi G, LeGros M, Larabell C (2003) Actin-filled nuclear invaginations indicate degree of cell de-differentiation. Differentiation 71:414–424
Kellenberger E, Durrenberger M, Villiger W, Carlemalm E, Wurtz M (1987) The efficiency of immunolabel on Lowicryl sections compared to theoretical predictions. J Histochem Cytochem 35:959–969
Landmann L, Marbet P (2004) Colocalization analysis yields superior results after image restoration. Microsc Res Tech 64:103–112
Langevin HM, Storch KN, Cipolla MJ, White SL, Buttolph TR, Taatjes DJ (2006) Fibroblast spreading induced by connective tissue stretch involves intracellular redistribution of alpha- and beta-actin. Histochem Cell Biol 125:487–495
Lui PP, Kong SK, Kwok TT, Lee CY (1998) The nucleus of HeLa cell contains tubular structures for Ca2+ signalling. Biochem Biophys Res Commun 247:88–93
Manders EMM, Verbeek FJ, Aten JA (1993) Measurement of colocalization of objects in dual-color confocal images. J Microsc 169:375–382
Maxwell CA, Hendzel MJ (2001) The integration of tissue structure and nuclear function. Biochem Cell Biol 79:267–274
Nisman R, Dellaire G, Ren Y, Li R, Bazett-Jones DP (2004) Application of quantum dots as probes for correlative fluorescence, conventional, and energy-filtered transmission electron microscopy. J Histochem Cytochem 52:13–18
Thomas CH, Collier JH, Sfeir CS, Healy KE (2002) Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA 99:1972–1977
Acknowledgments
The authors thank Dr Michael J. Hendzel and Alan K. Howe for helpful discussions. This work was funded by the NIH Center for Complementary and Alternative Medicine. Research Grant RO1-AT01121 and by NIH Grant P20 RR16435 from the COBRE Program of the National Center for Research Resources. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Complementary and Alternative Medicine, National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Storch, K.N., Taatjes, D.J., Bouffard, N.A. et al. Alpha smooth muscle actin distribution in cytoplasm and nuclear invaginations of connective tissue fibroblasts. Histochem Cell Biol 127, 523–530 (2007). https://doi.org/10.1007/s00418-007-0275-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-007-0275-9