Abstract
The synthesis of primary bile acids is confined to the hepatocytes. This study aimed to evaluate the expression pattern within the liver architecture of the rate-limiting enzyme of the neutral pathway, cholesterol 7α-hydroxylase (Cyp7a1), and sterol 12α-hydroxylase (Cyp8b1), the enzyme necessary for the synthesis of cholic acid. Specific Cyp8b1 and Cyp7a1 peptide antiserums were used for immunohistochemical staining of livers from wild type and Cyp8b1 null mice, the latter instead expressing β-galactosidase (β-Gal) as a replacement reporter gene. Cyp8b1 was mainly expressed in the hepatocytes in a zonal pattern surrounding the central vein while the areas surrounding the portal zones showed much lower levels. The zonation was maintained in cholic acid-depleted mice using β-Gal as a reporter protein. Cyp7a1 expression in wild type mice also showed a zonal distribution pattern, although less distinct, with a maximal expression within a 1–2 cell thick layer of hepatocytes surrounding the central vein. In Cyp8b1 null mice, a more intense staining was obtained, in accordance with the higher expression level of Cyp7a1, although the overall expression pattern was maintained. Our results in mice indicate possible differences in the regulation of the cellular zonation of Cyp7a1 and Cyp8b1. Also, cholic acid affects the setpoint of Cyp7a1 expression but not its zonal distribution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bile acids are important in the absorptive process of dietary lipids in the small intestine. As a corollary the conversion of precursor cholesterol into bile acids also represents a major atheroprotective pathway leading to fecal elimination of excess cholesterol. The complete synthesis of bile acids only occurs in the liver, and includes several enzymatic steps involving hepatic P-450 cytochromes such as cholesterol 7α-hydroxylase (CYP7A1), sterol 12α-hydroxylase (CYP8B1), sterol 27-hydroxylase (CYP27A1), and oxysterol 7α-hydroxylase (CYP7B1). Among these, CYP7A1 and CYP27A1 are considered rate limiting for the respective neutral and acidic pathways of bile acid synthesis (Eggertsen et al. 1996; Chiang 1998; Björkhem and Eggertsen 2001).
In human bile the two major primary bile acids are cholic acid (CA) and chenodeoxycholic acid (CDCA), present in a molar ratio of approximately 2:1. Cyp8b1 is the only P-450 cytochrome identified that can 12α-hydroxylate substrates flowing through both the neutral and acidic pathways (Andersson et al. 1998, 1999). No compensatory P-450 seems to exist since in Cyp8b1 null mice, with deletion of the entire coding region of the gene, the synthesis of CA was completely abolished (Li-Hawkins et al. 2002). As a consequence of the loss of CA, the mice decreased their intestinal cholesterol absorption, increased the hepatic de novo cholesterol synthesis, and were protected from hyperlipidemia and gallstone formation (Murphy et al. 2005; Wang et al. 2006a, b). Apart from its role in enhancing micellar formation in the small intestine, CA also performs a regulatory role within the bile acid synthesis in the mouse as a physiological ligand to the farnesoid X receptor (FXR), exerting regulation of many genes like short heterodimerizing partner (SHP) resulting in an increase of its mRNA. CA is important for the regulation of Cyp7a1 expression since it appears to be one of its physiological suppressants, operating via the FXR-SHP axis and the c-Jun N-terminal kinase pathway, leading to a tonic inhibition of Cyp7a1 mRNA, protein and activity (Gupta et al. 2001; Li-Hawkins et al. 2002).
In healthy humans, gallstone patients and patients treated with cholestyramine, the expression of CYP8B1 was found to be homogenously distributed within the liver lobuli showing no sign of zonation (Wang et al. 2005). However, species differences may occur since a zonal distribution was reported for rat Cyp7a1 and Cyp27a1, both proteins mainly expressed in hepatocytes surrounding the central vein (Brassil et al. 1995; Twisk et al. 1995; Massimi et al. 1998). In addition, some P-450 cytochromes like CYP7B1 and CYP27A1 are also expressed in Kupffer cells or cholangiocytes. The present study was performed to investigate whether loss of the tonic suppressive effect of CA affects the pattern of expression of Cyp8b1 and Cyp7a1 within the hepatic lobuli in mice and whether the pattern of expression differs between these proteins.
Materials and methods
Chemicals and reagents
Enhanced chemoluminescence reagents and Hydrogen C nitrocellulose membranes were obtained from Amersham-Pharmacia, Uppsala, Sweden. Rabbit anti-goat IgG conjugated with horseradish peroxidase (HRP) and acrylamide-bisacrylamide 37.5:1 were from BioRad Laboratories, Hercules, CA, USA. For the immunohistochemical studies a specific goat antiserum against Cyp8b1 was obtained from Santa Cruz Biotechnology, Inc. (P-18: sc-23515). The antiserum was raised against a peptide localized to an internal region of the protein. For the Cyp7a1 immunohistochemistry, a specific affinity purified rabbit antiserum was used, raised against the five carboxy-terminal amino acid residues of the protein (Lundåsen et al. 2003).
Animals and tissue preparation
Male Cyp8b1 null mice on a mixed 129Sv/Ev; C57Bl/6 background as previously described and wild type littermates were used (Li-Hawkins et al. 2002). In addition, Balb/cBy mice were utilized.
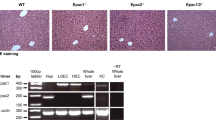
In order to continue the cellular studies of Cyp8b1 gene expression in vivo, the Cyp8b1 null mice were further modified. As reported, the entire coding sequence of the Cyp8b1 gene (Gåfvels et al. 1999) was replaced with a modified lacZ gene (Cat. No 6044-1, Clontech, Palo Alto, California, USA) inserted in the correct orientation downstream to the Cyp8b1 transcriptional start site (Gåfvels et al. 1999). To eliminate potential transcriptional interference by the oppositely oriented 1.4 kb neomycin resistance gene (Artelt et al. 1991; Xu et al. 2001) the latter was flanked by a pair of loxP sites, allowing its removal at the one-cell stage from the progeny of the mixed genotype 129Sv/Ev;C57Bl/6 mice. This was achieved by mating male knockout mice with females transgenically overexpressing cre recombinase, driven by the HPRT promoter (Fig. 1a). The neomycin resistance gene deletion was verified by a multiplex PCR approach, using the oligonucleotide CTTCCTCTATCGCCTGAAGCC as forward primer for the wild-type allele, the oligonucleotide ACACAGGCATAGAGTGTCTGC as a forward primer for the targeted allele, and one common 3′ reverse primer, TGAGCTGACCACATGTGTTCC. The wild type and Cyp8b1 null mice with deletions of the neomycin resistance gene each generated a single PCR product of 209 and 532 base pairs, respectively. Two heterozygous males and one heterozygous female (Fig. 1b) were mated to create one group homozygous for the wild type allele to be used as controls and another with a floxed β-galactosidase reporter gene where the neomycin resistance gene was deleted on both chromosomes (GRS mice). Male mice aged 3–5 months were used in the study.
a Strategy for creating the β-galactosidase reporter mouse strain (GRS). The coding region (CR/DA) of the intronless Cyp8b1 gene was deleted and substituted by homologous recombination in ES cells with the β-galactosidase gene (β-GAL) placed in correct orientation immediately downstream to the Cyp8b1 gene transcription start site (filled square) in tandem with the oppositely oriented neomycin resistance gene (NEO) to create the initial 129Sv/Ev;C57/Bl6 mixed genotype knockout mice. To create the GRS reporter mouse the neomycin resistance gene flanked with loxP sites (filled triangle) was floxed at the one-cell stage by mating male Cyp8b1 null mice carrying the neomycin resistance gene with cre recombinase overexpressing females. b PCR to demonstrate the heterozygous deletion of the neomycin resistance gene in the floxed 129; C57/Bl6 mixed genotype knockout mice. The 532 bp product signifies the floxed allele and the 209 bp product the wild type allele. To obtain homozygous wild type or GRS mice two males (lane 3 and 4) and one female (lane 8) carrying the floxed allele were mated
To study the regulation of gene expression, Cyp8b1 wild type, Cyp8b1 knockout mice and GRS mice were fed a normal chow (R36 powder, Lactamin, Sweden) or a chow supplemented with 0.5% (w/w) sodium cholate in 10% (w/w) peanut oil ad libitum for 1 week.
Immunohistochemical detection of Cyp8b1 and Cyp7a1 expression
The mice were deeply anaesthetized by an intraperitoneal injection of pentobarbital, whereafter the animals were fixed by transcardial perfusion with PBS followed by 4% (w/v) freshly prepared paraformaldehyde in PBS. Dissected organs were postfixed overnight at 4°C in the same fixative. After trimming, the organs were dehydrated and embedded in paraffin according to standard procedures. The procedures were approved by the local committee for ethical standards of animal experiments at Karolinska Institutet, Stockholm.
Paraffin sections (5 μm thickness) were rehydrated and microwaved for 10 min in 10 mM citrate buffer, pH 6.0. After rinsing in Tris–HCl NaCl buffer (TBS), endogenous peroxidase activity was quenched with 3% (v/v) hydrogen peroxide in methanol. Sections were then pretreated with 4% (v/v) normal rabbit serum in TBS. Endogenous biotin binding was quenched by preincubation using an avidin–biotin blocking kit (SP-2001, Vector Laboratories Inc., Buringgame, CA, USA). For blocking experiments the Cyp8b1 antibodies were first preincubated with the corresponding peptide (Santa Cruz Biotechnology Inc., cat. no. Sc_23515p) in Tris–HCl buffer and normal rabbit serum (4%, v/v) overnight at 4°C. Cyp8b1 antibodies were used in a final dilution of 1:50, while the Cyp7a1 antibodies were used at a concentration of 2 μg/ml; incubation with the primary antibodies was performed overnight at 4°C. Binding of the primary antibodies was visualized by sequential incubation with biotin labeled secondary antibodies followed by ABC-complex_HRP. The peroxidase activity was developed with DAB/H2O2. After rinsing, sections were lightly counterstained with hematoxylin, dehydrated, and permanently cover slipped. The cellular expression of β-galactosidase activity was localized by X-gal histochemistry, performed on 10 μm frozen sections that had been fixed in buffered 4% (w/v) paraformaldehyde and 0.5% (w/v) glutaraldehyde as previously described (Robertson et al. 2003). As controls, fixed frozen sections from wild type mice were used.
Expression of Cyp8b1 and the corresponding β-galactosidase reporter in mouse liver
Total RNA from snap frozen mouse liver tissue was used to investigate the hepatic expression of Cyp8b1 and β-galactosidase mRNA by Northern blotting. Relative expression was quantified by phosphorimaging (FujiBas-1800) using GAPDH mRNA as a reference. β-galactosidase mRNA was also measured by real-time PCR (Applied Biosciences) using GAPDH as a reference gene. The oligonucleotides CCTACAGAGATTTAAAGCTCTAAGATTCC and CTCTAGATGGCATTTCTTCTGAGC were used as 5′ and 3′ flanking PCR primers, together with a FAM/Dark-labeled dabcylated oligonucleotide probe with the sequence AAGGCATTCCACCACTGCTCCCATT. To assure specificity, the 5′ flanking PCR primer was designed to span a splice site positioned in the 5′ untranslated region of the lacZ gene. Expression of the β-galactosidase protein in the livers of wild type and GRS mice was determined by immunoblotting using a monoclonal anti-β-galactosidase antibody from Promega (Madison, WI, USA cat. No. Z3781). As a control, purified Escherichia coli β-galactosidase (Sigma, St. Louis, MO, USA, cat. No. G-6008) was used.
Statistics
Values are given as mean ± SEM. Statistical analysis was performed with Student’s t test. P < 0.05 was considered statistically significant.
Results
Hepatic expression pattern of Cyp8b1 in wild-type mice
Immunohistochemical detection of Cyp8b1 expression in inbred Balb/cBy mouse liver revealed a zonal distribution pattern. As shown in Fig. 2a, b (upper panel), the areas surrounding the portal triads (P) were negative, but hepatocytes closer to the central veins (CV) showed a granular cytoplasmic reaction, indicating expression of Cyp8b1. By preincubating the antibodies with the corresponding peptide used for raising them in goats, all staining was lost (Fig. 2e). The distribution of the Cyp8b1 protein in livers of wild type mice displayed a similar zonal distribution as in the Balb/cBy mouse livers, although with somewhat less intensity (Fig. 2c). As an additional control of the specificity, liver sections prepared from Cyp8b1 null mice were incubated. Since no staining was seen in the hepatocytes (Fig. 2d) this verified the specificity of the antibodies used here. However, some sinusoidal cells still showed a weak positive reaction (also seen in Fig. 2b), perhaps indicating that an antigenic epitope on an unknown nontarget molecule may also be reacting with the Cyp8b1 antiserum.
Distribution of Cyp8b1 in mouse liver. In a and b, the staining patterns in Balb/cByJ mice is illustrated. Note clear-cut differences between hepatocytes located around central veins (CV) and periportal zones (P). In c the distribution of Cyp8b1 in liver from wildtype 129Sv/Ev;C57Bl/6 is shown. d shows that only scattered positive cells are present in Cyp8b1 null mice. In e the corresponding peptide was used to block the Cyp8b1 antibody, resulting in a completely negative staining
Hepatic expression pattern of the reporter β-galactosidase in cholic acid-depleted mice
To test whether the replacement of Cyp8b1 with the β-galactosidase marker affected the regulation pattern of the Cyp8b1 gene, groups of both genotypes were fed ordinary chow diet or a diet supplemented with 0.5% (w/w) sodium cholate. Liver tissue from wild type and GRS mice was investigated for the expression of Cyp8b1 and β-galactosidase mRNA. As expected, GRS mice showed no signs of Cyp8b1 mRNA. Instead, a ≈ 4 kb transcript corresponding to β-galactosidase was detected (Fig. 3a). It was confirmed by Western blotting that GRS but not wild type mice expressed β-galactosidase protein (Fig. 3b). Northern blotting also showed a significant 5–7-fold suppression of the Cyp8b1 mRNA in wild type mice by sodium cholate feeding (Fig. 4a, b). A similar suppression of β-galactosidase mRNA expression was recorded in GRS mice by real-time PCR (Fig. 4c). No β-galactosidase mRNA was detected in wild type mice. Our results indicated that the reporter gene was regulated by cholic acid feeding in a manner similar to the pattern seen for the original Cyp8b1 gene. Histochemical localization of the β-galactosidase gene expression in GRS mice showed positive staining in the presence of X-gal, largely confined to the hepatocytes (Fig. 5). In agreement with the pattern for Cyp8b1 expression, as shown in Fig. 2, the pattern of β-galactosidase expression was also uneven and stronger around the central vein of the liver acini, while the periportal fields stained much weaker. X-gal staining of liver sections from wild type mice showed practically no background activity (data not shown).
Expression of Cyp8b1 and β-galactosidase mRNA in mouse liver. a Levels of Cyp8B1 and β-galactosidase mRNA in wild type (+/+), heterozygous and GRS mice (−/−). b Expression of β-galactosidase protein in livers from GRS mice but not in wild type mice. As a control β-galactosidase protein purified from E. coli was used
Regulation of Cyp8b1 and β-galactosidase mRNA by cholate feeding. a Regulation of Cyp8b1 mRNA by cholic acid feeding in wild type mice (+/+) and absence of expression of Cyp8b1 expression in GRS (−/−) mice. Sixteen individual +/+ or GRS (−/−) mice were fed normal chow or a chow supplemented with 0.5% (w/w) sodium cholate for 1 week. b Northern blot and phosphor imaging quantitation of the relative expression of Cyp8b1 and β-galactosidase mRNA in wild type and GRS mice fed normal chow or a chow supplemented with 0.5% (w/w) sodium cholate. *P < 0.05 versus wild type; nd not detected. c Real-time PCR quantitation of the hepatic levels of β-galactosidase mRNA in GRS mice fed a normal chow (control, black bar) or a chow containing 0.5% sodium cholate (CA, grey bar). *P < 0.05 versus wild type
Cellular expression of β-galactosidase in mouse liver. Fixed sections from livers of GRS mice were incubated with X-gal and the cell-specific expression pattern of β-galactosidase was indirectly assessed. In accord with Cyp8b1 expression (Fig. 2) the reporter expression was more intense in the region surrounding the central vein (CV) and decreasing towards the periportal area (P). Magnification ×250
Effects of cholic acid depletion on the expression and distribution of Cyp7a1
In Cyp8b1 wild type mice, the hepatic Cyp7a1 expression, as detected by immunohistochemistry, showed an even distribution in most part of the lobules, with an increased expression in 1–2 cell layers immediately surrounding the central veins (Fig. 6a, b, d, e). In Cyp8b1 null mice, a markedly increased staining reaction was seen, as expected from our studies on the mRNA and protein levels (Li-Hawkins et al. 2002). The pattern of Cyp7a1 expression seen in Cyp8b1 null mice was identical to the expression pattern seen in the Cyp8b1 wild type mice, indicating a generally enhanced expression. Feeding mice with 0.5% cholic acid for 1 week resulted in a suppression of staining for Cyp7a1 (Fig. 6c, f) in both wild type and Cyp8b1 null mice. However, scattered cells throughout the liver parenchyma, especially in wild type mice, appeared to maintain a low level of expression of Cyp7a1. In Cyp8b1 null mice fed cholic acid, a distinct zonation previously described in mice fed control diet was preserved in spite of the overall suppression of Cyp7a1 staining (Fig. 6f).
Distribution of Cyp7a1 expression in wild type (a–c) and Cyp8b1 null (d–f) animals. In a and d the distribution of Cyp7a1 is shown to be rather uniform throughout the liver parenchyma except from 1 to 2 layers of hepatocytes surrounding the central vein (CV). Note a generally higher intensity of Cyp7a1 staining in livers of Cyp8b1 null mice (d, e) but no difference in the pattern of distribution as compared to wild type mice (a, b). Feeding of 0.5% sodium cholate (CA) in the diet for 1 week (c, f) reduced staining both in wild type (wt) and in Cyp8b1 null mice but some staining remained in scattered cells throughout the liver parenchyma as well as cells close to the central vein in livers of Cyp8b1 null mice (f)
Discussion
Our results indicated that the major expression of Cyp8b1 and Cyp7a1 is localized within the hepatocytes in a zonal pattern surrounding the central vein. Cyp8b1 expression showed a more distinct zonation, the hepatocytes closer to the portal area being negative. Two other cytochrome P-450 enzymes active in the bile acid synthesis, Cyp7a1 and Cyp27a1, have also been described to have the same expression pattern within the rat liver lobulus (Twisk et al. 1995). The pattern of Cyp8b1 expression appears to differ among species since, in the human liver, Cyp8b1 was expressed in a homogenous pattern (Wang et al. 2005). The regulatory mechanisms underlying the differences in zonation pattern for Cyp8b1 between the species are not known but may be related to differences in regulatory bile acids along the portocentral axis and the specific regional distribution of regulatory nuclear receptors expressed in the liver (Bookout et al. 2006). Generally, thyroid hormones are candidate regulatory hormones for basal as well as diurnal variation of zonal expression given the distribution of thyroid hormone receptors in mouse liver in general (Zandieh-Doulabi et al. 2003) and more specifically TRβ-mediated modulation of Cyp7a1 expression (Johansson et al. 2005). Differences in Cyp8b1 zonation could also relate to interspecies differences in regulation of gene transcription as such, as it was observed that the DNA sequence of the Cyp8b1 promoter was poorly conserved between mouse and man (Gåfvels et al. 1999). Differences in regulation of gene transcription between rodent species and man relating to presence or absence of certain discrete trans-acing motifs have already been described, since CYP7A1 gene transcription is responsive to liver X receptor (LXR) agonism in mouse and rats but not in man (Chen et al. 2002; Goodwin et al. 2003).
Considering the distribution pattern of Cyp8b1 and Cyp7a1 described herein, one might wonder how large a fraction of the mouse hepatocytes that generally are actively synthesizing bile acids. The bulk of bile acids, representing about 95% of the total amount of secreted bile acids, is reabsorbed into periportal hepatocytes and quantitatively re-excreted into the bile canaliculi within the very same area (Groothuis et al. 1982; Jones et al. 1980). Since high bile acid levels might suppress the synthetic machinery in the portal areas this indicates that the total amount of newly synthesized bile acids would be localized in a compartmentalized manner to the perivenous areas where the bile acid concentrations may be lower. Whether an intracellular gradient along the portocentral axis contributes to the differential expression pattern in rodents remains open for discussion, but the mouse Cyp8b1 would also fit into this pattern since a local increase of bile acid concentrations in the periportal areas hampers the Cyp8b1 gene transcription (Vlahcevic et al. 2000). This is in contrast to the pattern of Cyp7a1 expression, which appears more even implying a less marked compartmentalization of the initial, rate-limiting step of the neutral pathway of bile acid synthesis.
Both Cyp8b1 and Cyp7a1 expression are sensitive to tonic inhibition by cholate (Strawitz et al. 1993; Twisk et al. 1993; Li-Hawkins et al. 2002) but slight differences in sensitivity may lead to individual hepatocellular expression patterns within liver lobuli, and furthermore in man, chenodexycholic acid and not cholic acid is considered to be the primary bile acid responsible for FXR-mediated feedback inhibition of these genes. In rats, interruption of the enterohepatic circulation by administration of cholestyramine induced production of Cyp7a1 and Cyp27a1 mRNA in larger areas of the liver acinus (Twisk et al. 1995), but no such changes were found for the Cyp7a1 expression in our Cyp8b1 knockout mice. The full set of genes necessary for de novo synthesis of bile acids in rodents, especially of CA, would thus be expressed mainly in perivenous hepatocytes, where the cellular concentrations of bile acids are lower. However, synthesis of bile acids via the alternative pathway could nevertheless occur in hepatocytes not expressing Cyp8b1 or Cyp7a1 if oxysterols produced in other cell types like, e.g. Kupffer cells are used (Björkhem and Eggertsen 2001). Another factor affecting the expression pattern of Cyp7a1 and Cyp8b1 may be circadian variations directed by various nuclear receptors (Berkowitz et al. 1995; Yang et al. 2006). Mechanisms of regulation may also be different during embryogenesis and early life, as Cyp7a1 shows an even expression pattern within the liver lobules during these periods (Massimi et al. 1998).
Zonation of expression has been reported for other genes expressed in the liver-like glucose-6-phosphatase, succinate dehydrogenase, albumin, NADPH dehydrogenase, and glutamate dehydrogenase (Racine-Samson et al. 1996). In the rat, immunohistochemical mapping of several P-450 cytochromes showed that a majority were primarily localized around the central vein while a few displayed other patterns of expression (Oinonen and Lindros 1998). However, an even distribution of hepatocyte proteins within the lobuli does occur, as is the case for the rat bile salt export pump (ABCB11; BSEP) (Van Waarde et al. 2002). Expression patterns for transcription factors are particularly interesting as they may affect expression levels of many proteins in the bile acid synthetic chains. Thus, mRNA quantities for rat HNF4a, C/EBPα and Tcf2 showed slightly higher expression in perivenous than in periportal hepatocytes (Lindros et al. 1997). It is not known whether the FXR-SHP or FGF15 and FGF receptor 4, constituting novel regulatory proteins for CYP8B1 and CYP7A1 expression, may somehow direct the zonal distribution seen in rodents (Inagaki et al. 2005).
References
Andersson U, Eggertsen G, Björkhem I (1998) Rabbit liver contains one major sterol 12alpha-hydroxylase with broad substrate specificity. Biochim Biophys Acta 1389:150–154
Andersson U, Yang Y, Björkhem I, Einarsson C, Eggertsen G, Gåfvels M (1999) Thyroid hormone suppresses hepatic sterol 12alpha-hydroxylase (CYP8B1) activity and messenger ribonucleic acid in rat liver: failure to define known thyroid hormone response elements in the gene. Biochim Biophys Acta 1438:167–174
Artelt P, Grannemann R, Stocking C, Friel J, Bartsch J, Hauser H (1991) The prokaryotic neomycin-resistance-encoding gene acts as a transcriptional silencer in eukaryotic cells. Gene 99:249–254
Berkowitz CM, Shen CS, Bilir BM, Guibert E, Gumucio JJ (1995) Different hepatocytes express the cholesterol 7 alpha-hydroxylase gene during its circadian modulation in vivo. Hepatology 21:1658–1667
Björkhem I, Eggertsen G (2001) Genes involved in initial steps of bile acid synthesis. Curr Opin Lipidol 12:97–103
Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ (2006) Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126:789–799
Brassil PJ, Edwards RJ, Davies DS (1995) Expression and distribution of cholesterol 7α-hydroxylase in rat liver. Biochem Pharmacol 50:311–316
Chen JY, Levy-Wilson B, Goodart S, Cooper AD (2002) Mice expressing the human CYP7A1 gene in the mouse CYP7A1 knock-out background lack induction of CYP7A1 expression by cholesterol feeding and have increased hypercholesterolemia when fed a high fat diet. J Biol Chem 277:42588–42595
Chiang JY (1998) Regulation of bile acid synthesis. Front Biosci 3:176–193
Eggertsen G, Olin M, Andersson U, Ishida H, Kubota S, Hellman U, Okuda KI, Björkhem I (1996) Molecular cloning and expression of rabbit sterol 12alpha-hydroxylase. J Biol Chem 271:32269–32275
Gåfvels M, Olin M, Chowdhary BP, Raudsepp T, Andersson U, Persson B, Jansson M, Björkhem I, Eggertsen G (1999) Structure and chromosomal assignment of the sterol 12alpha-hydroxylase gene (CYP8B1) in human and mouse: eukaryotic cytochrome P-450 gene devoid of introns. Genomics 56:184–196
Goodwin B, Watson MA, Kim H, Miao J, Kemper JK, Kliewer SA (2003) Differential regulation of rat and human CYP7A1 by the nuclear oxysterol receptor liver X receptor-α. Mol Endocrinol 17:386–394
Groothuis GM, Hardonk MJ, Keulemans KP, Nieuwenhuis P, Meijer DK (1982) Autoradiographic and kinetic demonstration of acinar heterogeneity of taurocholate transport. Am J Physiol 243:G455–G462
Gupta S, Stravitz RT, Dent P, Hylemon PB (2001) Down-regulation of cholesterol 7α-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem 276:15816–15822
Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA (2005) Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2:217–225
Johansson L, Rudling M, Scanlan TS, Lundåsen T, Webb P, Baxter J, Angelin B, Parini P (2005) Selective thyroid receptor modulation by GC-1 reduces serum lipids and stimulates steps of reverse cholesterol transport in euthyroid mice. Proc Natl Acad Sci USA 102 (29):10297–10302.
Jones AL, Hradek GT, Renston RH, Wong KY, Karlaganis G, Paumgartner G (1980) Autoradiographic evidence for hepatic lobular concentration gradient of bile acid derivatives. Am J Physiol 238:G233–G237
Li-Hawkins J, Gåfvels M, Olin M, Lund EG, Andersson U, Schuster G, Björkhem I, Russell DW, Eggertsen G (2002) Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest 110:1191–1200
Lindros KO, Oinonen T, Issakainen J, Nagy P, Thorgeirsson SS (1997) Zonal distribution of transcripts of four hepatic transcription factors in the mature rat liver. Cell Biol Toxicol 13:257–262
Lundåsen T, Liao W, Angelin B, Rudling M (2003) Leptin induces the hepatic high density lipoprotein receptor scavenger receptor B type I (SR-BI) but not cholesterol 7alpha-hydroxylase (Cyp7a1) in leptin-deficient (ob/ob) mice. J Biol Chem 278:43224–43228
Massimi M, Lear SR, Huling SL, Jones AL, Erickson SK (1998) Cholesterol 7alpha-hydroxylase (CYP7A): patterns of messenger RNA expression during rat liver development. Hepatology 28:1064–1072
Murphy C, Parini P, Wang J, Björkhem I, Eggertsen G, Gåfvels M (2005) Cholic acid as a key regulator of cholesterol synthesis, intestinal absorption and hepatic storage in mice. Biochim Biophys Acta 1735:167–175
Oinonen T, Lindros KO (1998) Zonation of hepatic cytochrome P-450 expression and regulation. Biochem J 329(Pt 1):17–35
Racine-Samson L, Scoazec JY, D’errico A, Fiorentino M, Christa L, Moreau A, Roda C, Grigioni WF, Feldman G (1996) The metabolic organization of the adult human liver: a comparative study of normal, fibrotic, and cirrhotic liver tissue. Hepatology 24:104–113
Robertson GR, Field J, Goodwin B, Bierach S, Tran M, Lehnert A, Liddle C (2003) Transgenic mouse models of human CYP3A4 gene regulation. Mol Pharmacol 64:42–50
Strawitz RT, Hylemon PB, Heuman DM, Hagey LR, Schteingart CD, Ton-Nu H-T, Hofmann AF, Vlahcevic ZR (1993) Transcriptional regulation of cholesterol 7α-hydroxylase mRNA by conjugated bile acids in primary cultures of rat hepatocytes. J Biol Chem 268:13987–13993
Twisk J, Lehmann EM, Princen HMG (1993) Differential feedback regulation of cholesterol 7α-hydroxylase mRNA and transcriptional activity by bile acids in primary monolayer cultures of rat hepatocytes. Biochem J 290:685–691
Twisk J, Hoekman MF, Mager WH, Moorman WF, De Boer PA, Scheja L, Princen HM, Gebhardt R (1995) Heterogeneous expression of cholesterol 7 alpha-hydroxylase and sterol 27-hydroxylase genes in the rat liver lobulus. J Clin Invest 95:1235–1243
Van Waarde WM, Verkade HJ, Wolters H, Havinga R, Baller J, Bloks V, Muller M, Sauer PJ, Kuipers F (2002) Differential effects of streptozotocin-induced diabetes on expression of hepatic ABC-transporters in rats. Gastroenterology 122:1842–1852
Vlahcevic ZR, Eggertsen G, Björkhem I, Hylemon PB, Redford K, Pandak WM (2000) Regulation of sterol 12alpha-hydroxylase and cholic acid biosynthesis in the rat. Gastroenterology 118:599–607
Wang J, Greene S, Eriksson LC, Rozell B, Reihnér E, Einarsson C, Eggertsen G, Gafvels M (2005) Human sterol 12alpha-hydroxylase (CYP8B1) is mainly expressed in hepatocytes in a homogenous pattern. Histochem Cell Biol 123:441–446
Wang J, Einarsson C, Murphy C, Parini P, Björkhem I, Gåfvels M, Eggertsen G (2006a) Studies on LXR-and FXR-mediated effects on cholesterol homeostasis in normal and cholic acid depleted mice. J Lipid Res 47(2):421–430
Wang J, Einarsson C, Murphy C, Rudling M, Björkhem I, Gåfvels M, Eggertsen G (2006b) Critical role of cholic acid for development of hypercholesterolemia and gallstones in diabetic mice. Biochem Biophys Res Commun 342(4):1382–1388
Xu X, Li C, Garrett-Beal L, Larson D, Wynshaw-Boris A, Deng CX (2001) Direct removal in the mouse of a floxed neo gene from a three-loxP conditional knockout allele by two novel approaches. Genesis 30:1–6
Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM (2006) Nuclear receptor expression links the circadian clock to metabolism. Cell 126:801–810
Zandieh-Doulabi B, Dop E, Schneiders M, Shiphorst M P-T, Mansen A, Vennström B, Dijkstra CD, Bakker O, Wiersinga WM (2003) Zonal expression of the thyroid hormone receptor α isoforms in rodent liver. J Endocrinol 179:379–385
Acknowledgments
This study was supported by grants from the The Swedish Research Council, The Swedish Heart and Lung Foundation, The Swedish Society for Medicine, Wallenberg Consortium North and the Julin Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, J., Olin, M., Rozell, B. et al. Differential hepatocellular zonation pattern of cholesterol 7α-hydroxylase (Cyp7a1) and sterol 12α-hydroxylase (Cyp8b1) in the mouse. Histochem Cell Biol 127, 253–261 (2007). https://doi.org/10.1007/s00418-006-0239-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-006-0239-5