Abstract
Elf5 belongs to the ets family of transcription factors and was cloned by homology in the DNA binding domain to the related, epithelial-specific ets factor, Elf3. Elf5 mRNA is expressed highly in normal tissue rich in secretory epithelial cells, including mammary gland, lung, kidney, prostate, salivary gland and stomach. The function of Elf5 and the cell types in which it is expressed remain uncharacterised. The presence of Elf5 mRNA in normal tissues, but absence in cancer tissues, may suggest a role for Elf5 in differentiation and development. We have generated a rabbit antiserum directed against a peptide in the Elf5 DNA-binding domain that is conserved between murine and human sequences. The antiserum specifically detects human and murine Elf5 proteins on western blots and shows specific staining on paraffin-embedded sections obtained from tissues including mammary gland, kidney, salivary gland and stomach. Epithelia from the bladder lining, lung and prostate did not stain for the presence of Elf5, though these organs express Elf5 mRNA. We show for the first time that Elf5 is primarily expressed in epithelial cells and is likely to be an epithelial-specific protein. The antiserum should prove useful in further analysis of the expression and function of Elf5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Elf5/Ese-2 gene is a member of the ets family of winged-turn-helix transcription factors that recognise the consensus sequence GGA(A/T) found in the promoters and enhancers of many genes. Although ets family members bind to a similar core DNA recognition sequence, GGAA/T, the differences in the sequences flanking the core appear to contribute towards ets factor binding specificity and, hence, function (Oettgen et al. 1999; Wang et al. 1992; Wasylyk et al. 1992). Most ets transcription factors are expressed either ubiquitously or restricted to haemopoietic cells. In haemopoietic cells, ets genes such as the archetypal family member, Ets-1, are crucial for maintaining a normal differentiated cell phenotype. Ets factors integrate extracellular signals at the transcriptional level, resulting in the activation of a large set of target genes (Wasylyk et al. 1993). Ets-1 and Ets-2 are also important in fundamental processes such as development and apoptosis (Wolvetang et al. 2003; Xu et al. 2002). In addition, ets factors can promote or repress transcription depending on their interactions with other proteins. Therefore, these genes could have distinct functions.
Members of the epithelial-specific ets (ESE) subfamily, Elf5, Elf3/Ese-1/Ert/Esx/Jen, Ehf/Ese-3 and Pdef/Pse, are expressed only in organ systems rich in epithelium (Chang et al. 1997; Kas et al. 2000; Kleinbaum et al. 1999; Nozawa et al. 2000; Tymms et al. 1997; Wasylyk et al. 1993; Yamada et al. 2000; Zhou et al. 1998). Human and mouse Elf5 cDNAs were isolated by our laboratory from human and mouse lung cDNA libraries, respectively, through homology cloning based on the DNA binding domain sequence of Elf3 (Zhou et al. 1998), and independently identified as Ese-2 (Oettgen et al. 1999).
The different expression patterns of Elf5, Elf3, Ese-3 and Pse, initially established using RNA transcript analyses on whole human and/or mouse organ lysates, may also contribute to their different functions. For example, in the human, Pse is expressed only in prostatic epithelium and regulates Psa gene expression (Chen et al. 2002; Oettgen et al. 2000). Elf3, which transactivates epithelial-specific target genes such as Keratin-4, Sprr2A, Endo-A and Her2, is expressed in a wide variety of simple and stratified epithelial tissues with highest expression in the gastrointestinal epithelium and involuting mammary gland (Brembeck et al. 2000; Chang et al. 1997; Neve et al. 1998; Oettgen et al. 1997; Tymms et al. 1997). Indeed, our laboratory has shown that Elf3 has a non-redundant role in the development of the gastrointestinal tract (Ng et al. 2002). Elf5 mRNA is present in a more restricted set of tissues encompassing the salivary and mammary glands, lung, kidney, bladder, stomach and prostate (Oettgen et al. 1999; Zhou et al. 1998). Ese-3 mRNA is most highly expressed in many of the same tissues as Elf5 but is also expressed at lower levels in additional organs. Therefore, despite the highly conserved DNA-binding domains and overlapping spatial and temporal expression patterns of the ESE subfamily, their functions in vivo are apparently distinct.
In the ESE subfamily, protein expression analysis using immunohistochemistry has been reported only for human Pse, establishing the restriction of this factor to the prostatic glandular epithelium (Nozawa et al. 2000). In this study, to perform immunohistochemical localisation studies and determine whether Elf5 is expressed in an epithelial-restricted manner, we have generated a specific anti-peptide antibody. We report that the Elf5 protein is expressed in the epithelial cells of exocrine glands.
Materials and methods
Generation of Elf5 peptide antibody
A 12 amino acid peptide, identical in mouse and human Elf5, was identified in the Elf5 DNA-binding domain using the MacVector (Accelrys, SanDiego, CA, USA) software program that analyses hydrophilicity, flexibility and antigenicity of protein sequences. The peptide, CGILEWEDREQG, conjugated to the carrier diptheria toxoid via its cysteine residue and a maleimidocaproyl-N-hydroxysuccinimide linker, was synthesised and RP-HPLC purified to a purity of >70% by Mimetope Chiron (Melbourne, Australia). Rabbits were immunised initially with 250 nmol peptide in Freund’s complete adjuvant, followed by three booster immunisations of 250 nmol peptide in Freund’s incomplete adjuvant. Four weeks after the final immunisation the rabbits were killed by exsanguination.
Purification of immunoglobulin
Rabbit sera were affinity-purified on a protein G Sepharose column (Amersham Pharmacia Biotech, Piscataway, NJ, USA), using 0.1 M glycine pH 2.5 for elution, as described in Harlow and Lane (1988).
Western blotting
Recombinant proteins containing 6×Histidine tag at their C-termini were produced in E. coli using the PQE30 vector (Qiagen, Valencia, CA, USA) and purified according to the manufacturer’s protocol. Protein was quantified by the Bradford method (Bradford 1976). Samples containing 200 ng recombinant protein and molecular weight standards (BioRad Laboratories) were subjected to SDS-PAGE on 12% gels under reducing conditions followed by transfer to Immobilon-P membranes (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Purified anti-Elf5 IgG was used at 1 μg/ml in blocking solution (5% non-fat milk in TRIS-buffered saline/0.1% Tween). Secondary goat anti-rabbit IgG conjugated to horseradish peroxidase (Dako, Carpinteria, CA, USA) was used at 1:1,000 dilution in blocking solution. Chemiluminescence was detected using the SuperSignal reagent (Pierce Endogen, Rockford, IL, USA). Blots were exposed to Kodak X-OMAT AR film for up to 2 min and developed.
Silver-staining polyacrylamide gels
Polyacrylamide gels were silver-stained using the BioRad Laboratories kit according to the manufacturer’s protocol.
Immunohistochemistry
Murine organs were dissected from C57Bl6×SV129 J mixed background mice, rinsed in cold phosphate-buffered saline (PBS) and fixed overnight at 4°C in 10% neutral-buffered formalin. The fixative was removed by replacing with 70% ethanol. Organs were processed and embedded in paraffin wax and cut in 5-μm sections onto Superfrost Plus slides (Menzel-Glaser, Braunschweig, Germany). Immunohistochemical detection was performed using the TSA-Indirect kit (NEN Life Sciences, Boston, MA, USA) according to the manufacturer’s instructions. Antigen retrieval was performed using 0.2% trypsin in 0.4% CaCl2 for 10 min at 37°C. Endogenous peroxidases were quenched with 6% H2O2 in PBS for 20 min at room temperature. Affinity-purified anti-Elf5 rabbit IgG was used at 0.25 μg/ml in the blocking buffer supplied with the kit and incubated for 60 min at room temperature. Secondary antibody was a 1:200 dilution of biotinylated goat anti-rabbit IgG (Dako) and incubation was for 40 min at room temperature. Visualisation utilised the DAB substrate kit (Dako). Sections were counterstained with Harris haematoxylin and coverslips mounted with DPX (BDH Laboratory Supplies, Poole, UK). For every tissue, a second serial section on the same slide was used as a negative control, and treated with protein G Sepharose-purified preimmune IgG from the same rabbit.
Results
Design of Elf5 peptide and generation of antibodies
To identify regions within the Elf5 protein that are unique to Elf5 and suitable to use as immunogens, the protein sequences of the DNA binding domains of ets factors related to Elf5 were aligned (Fig. 1). A search of the protein databases showed the peptide CGILEWEDREQG to be unique to Elf5 and completely conserved in human and mouse sequences. This peptide was used to immunise rabbits for the generation of specific anti-Elf5 antibodies. As shown in Fig. 2, the affinity-purified serum IgG specifically recognised the mouse and human Elf5 recombinant proteins on western blot. It did not cross react with other ets proteins, including human Ets-1 and Ets-2 nor with the closely related human Elf3 and mouse Ese-3 (Fig. 2 top). The corresponding affinity-purified preimmune serum did not recognise these proteins (data not shown). Antibody binding was significantly reduced when the serum had been preincubated with human Elf5 recombinant protein, indicating the specificity of the interaction (data not shown). The presence of ets proteins on the western blot was verified by running a duplicate SDS-PAGE and silver-staining the gel (Fig. 2 bottom).
Alignment of the ets domain sequences of ESE proteins, Ets-1 and Ets-2; GenBank accession codes are bracketed: A human Elf5 (AAC79755), B murine Elf5 (AAC79754), C human Elf3 (AAM70481), D mouse Ese-3 (NP_031940), E human Ets-1 (NP_005229) and F human Ets-2 (NP_005230). The numbers along the left side of the sequence indicate the amino acid residue position, taken from the cited GenBank reference. The peptide used to immunise mice is highlighted in pink. Residues conserved in this epitope among the related factors are highlighted in blue
The ELF5-peptide antiserum is specific for Elf5. Recombinant proteins shown, 1 human Elf5, 2 murine Elf5, 3 human Elf3, 4 murine Ese-3, 5 human Ets-1 and 6 human Ets-2, were analysed by western blot with anti-Elf5 peptide antibodies (top) and their presence on the gel was verified by silver-staining (bottom). Protein size markers (M) are indicated. The relevant recombinant protein band in each lane, based on its expected size, is marked by an arrow
Immunolocalisation of Elf5 protein to epithelial cells
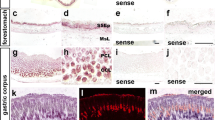
The expression of Elf5 mRNA has been previously detected in organs rich in glandular epithelial cells, such as human and murine kidney, mammary gland and stomach (Oettgen et al. 1999; Zhou et al. 1998). To determine the cell types in which Elf5 protein is expressed, we performed immunohistochemistry on a range of tissues with the anti-Elf5 peptide IgG. Cell types staining positive for Elf5 protein were identified by counterstaining with Harris haematoxylin. In kidney, Elf5 was localised to the renal cortex and was present in the cytoplasm and brush border of the cells of the proximal tubules and in the cells of the convoluted and straight portions of the distal tubules. The cortical glomeruli, interstitium and renal medulla were not found to express Elf5 protein (Fig. 3A). In the submandibular salivary gland the serous and mucinous acini are morphologically distinguishable by light microscopy and haematoxylin and eosin staining. Elf5 staining was observed in the epithelium of the serous acini, while the epithelia of the mucous acini were negative (Fig. 3B). In day 1 lactating mammary gland both ductal and alveolar epithelium stained positive (Fig. 3C). In the stomach (Fig. 3D), Elf5 expression was localised to cells lining the gastric glands in the stomach mucosa. Strong staining was apparent in the larger parietal cells compared with weaker staining in the smaller, more numerous chief cells. Parietal cells secrete HCl, while chief cells secrete pepsinogen from zymogenic granules. All of these mouse tissues stained negative when probed with preimmune IgG (data not shown). Mouse tissues found to be expressing Elf5 mRNA, including bladder, lung, trachea and prostate (Zhou et al. 1998) were also analysed by immunohistochemistry, but the protein was not detected (data not shown). Elf5 mRNA has also been detected in differentiated primary human foreskin keratinocytes (Oettgen et al. 1999), however, we were unable to detect Elf5 protein in mouse skin (data not shown).
Elf5 protein is localised to epithelia of exocrine glands. Immunohistochemistry with anti-Elf5 peptide antibody was performed on sections of murine kidney (A), submandibular gland (B), day 1 lactating mammary gland (C) and human stomach (D). Brown staining indicates positive staining. Features are marked as glomerulus (Gl), proximal tubule (Pt), Distal tubule (Dt), alveolus (Al), duct (Du), mucinous acini (Mu), serous acini (Se), gastric gland (Gg) and gastric pit (Gp). Magnification ×250 in A; ×400 in B–D
Discussion
This study has shown for the first time, using specific anti-Elf5 antibodies and immunohistochemical techniques, that the protein product of the recently cloned ets gene, Elf5, is localised to subsets of epithelial cells in the kidney, mammary gland, submandibular gland and stomach. We found that Elf5 protein is expressed in epithelial cells of the cortical proximal and distal tubules in the kidney, in mammary gland epithelium and in one of the two epithelial lineages in the submandibular salivary gland. Elf5 is also expressed in two types of cell lining the body of the glands in the stomach, showing strong expression in the parietal cells with weaker expression in the chief cells.
Unexpectedly, Elf5 protein was not detectable by immunohistochemistry in other mouse organs in which its mRNA is expressed, including the bladder, lung and prostate (Oettgen et al. 1999; Zhou et al. 1998). There may be several reasons as to why the protein may not be detectable. For instance, our assay may not be sensitive enough to detect low levels of Elf5 or the protein may be unstable in these organs. Elf5 protein may also be undetectable due to translational control. This phenomenon has been observed regarding other ets transcription factors, particularly human Pse, where mRNA can be expressed in both normal and malignant prostatic glandular epithelium and translation takes place most efficiently in normal cells, but not at all in malignant cells. Translational control of the Pse transcript was shown to be due to regions in the 5′UTR and 3′UTR (Nozawa et al. 2000). Interestingly, two transcripts of Elf5, with differing 3′UTR sequences, have been identified in mouse (Zhou et al. 1998). As these may contain the codes for similar regulatory mechanisms, translational control is, therefore, a distinct possibility for the absence of detectable Elf5 protein in some of the tissues examined here.
In conclusion, we have shown for the first time that Elf5 protein is localised to epithelium of exocrine glands including the kidney, mammary gland, stomach and the submandibular salivary gland. The Elf5 peptide antibodies will be a useful tool in elucidating the function of Elf5 in development and disease.
References
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brembeck F, Opitz OG, Libermann TA, Rustgi AK (2000) Dual function of the epithelial specific ets transcription factor, ELF3, in modulating differentiation. Oncogene 19:1941–1949
Chang C-H, Scott GK, Kuo W-L, Xiong X, Suzdaltseva Y, Park JW, Sayre P, Erny K, Collins C, Gray JW, Benz CC (1997) ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene 14:1617–1622
Chen H, Nandi AK, Li X, Bieberich CJ (2002) NKX-3.1 interacts with prostate-derived Ets factor and regulates the activity of the PSA promoter. Cancer Res 62:338–340
Harlow E, Lane D (1988) Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Kas K, Finger E, Grall F, Gu X, Akbarali Y, Boltax J, Weiss A, Oettgen P, Kapeller R, Libermann TA (2000) ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J Biol Chem 275:2986–2998
Kleinbaum LA, Duggan C, Ferreira E, Coffey GP, Buttice G, Burton FH (1999) Human chromosomal localization, tissue/tumor expression, and regulatory function of the ets family gene EHF. Biochem Biophys Res Commun 264:119–126
Neve R, Chang C-H, Scott GK, Wong A, Friis RR, Hynes Ne, Benz CC (1998) The epithelium-specific Ets transcription factor ESX is associated with mammary gland development and involution. FASEB J 12:1541–1550
Ng AY, Waring P, Ristevski S, Wang C, Wilson T, Pritchard M, Hertzog P, Kola I (2002) Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology 122:1455–1466
Nozawa M, Yomogida K, Kanno N, Nonomura N, Miki T, Okuyama A, Nishimune Y, Nozaki M (2000) Prostate-specific transcription factor hPSE is translated only in normal prostate epithelial cells. Cancer Res 60:1348–1352
Oettgen P, Carter KC, Augustus M, Barcinski M, Boltax J, Kunsch C Libermann TA (1997) The novel epithelial-specific Ets transcription factor gene ESX maps to human chromosome 1q32.1. Genomics 445:456–457
Oettgen P, Kas K, Dube A, Gu X, Grall F, Thamrongsak U, Akbarali Y, Finger E, Boltax J, Endress G, Munger K, Kunsch C, Libermann TA (1999) Characterisation of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J Biol Chem 274:29439–29452
Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, et al (2000) PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem 275:1216–1225
Tymms MJ, Ng AY, Thomas RS, Schutte BC, Zhou J, Eyre HJ, Sutherland GR, Seth A, Rosenberg M, Papas T, et al (1997) A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene 15:2449–2462
Wang C-Y, Petryniak B, Ho I-C, Thompson CB, Leiden JM (1992) Evolutionarily conserved Ets family members display distinct DNA binding specificities. J Exp Med 175:1391–1399
Wasylyk C, Kerckaert J-P, Wasylyk B (1992) A novel modulator domain of Ets transcription factors. Genes Dev 6:965–974
Wasylyk B, Hahn SL, Giovane A (1993) The Ets family of transcription factors. Eur J Biochem 211:7–18
Wolvetang EJ, Wilson TJ, Sanij E, Busciglio J, Hatzistavrou T, Seth A, Hertzog PJ, Kola I (2003) ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet 12:247–255
Xu D, Wilson TJ, Chan D, De Luca E, Zhou J, Hertzog PJ, Kola I (2002) Ets1 is required for p53 transcriptional activity in UV-induced apoptosis in embryonic stem cells. EMBO J 21:4081–4093
Yamada N, Tamai Y, Miyamoto H, Nozaki M (2000) Cloning and expression of the mouse Pse gene encoding a novel Ets family member. Gene 241:267–274
Zhou J, Ng AY, Tymms MJ, Jermiin LS, Seth AK, Thomas RS, Kola I (1998) A novel transcription factor, ELF5, belongs to the ELF subfamily of ETS genes and maps to human chromosome 11p13–15, a region subject to LOH and rearrangement in human carcinoma cell lines. Oncogene 17:2719–2732
Acknowledgements
The authors would like to thank Stuart Rodda at Mimotopes for his helpful advice on choosing the Elf5 peptide.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lapinskas, E.J., Palmer, J., Ricardo, S. et al. A major site of expression of the ets transcription factor Elf5 is epithelia of exocrine glands. Histochem Cell Biol 122, 521–526 (2004). https://doi.org/10.1007/s00418-004-0713-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-004-0713-x