Abstract
Background and objectives
Disability and cognitive impairment are known to be related to brain atrophy in multiple sclerosis (MS), but 3D-T1 imaging required for brain volumetrics is often unavailable in clinical protocols, unlike 3D-FLAIR. Here our aim was to investigate whether brain volumes derived from 3D-FLAIR images result in similar associations with disability and cognition in MS as do those derived from 3D-T1 images.
Methods
3T-MRI scans of 329 MS patients and 76 healthy controls were included in this cross-sectional study. Brain volumes were derived using FreeSurfer on 3D-T1 and compared with brain volumes derived with SynthSeg and SAMSEG on 3D-FLAIR. Relative agreement was evaluated by calculating the intraclass correlation coefficient (ICC) of the 3D-T1 and 3D-FLAIR volumes. Consistency of relations with disability and average cognition was assessed using linear regression, while correcting for age and sex. The findings were corroborated in an independent validation cohort of 125 MS patients.
Results
The ICC between volume measured with FreeSurfer and those measured on 3D-FLAIR for brain, ventricle, cortex, total deep gray matter and thalamus was above 0.74 for SAMSEG and above 0.91 for SynthSeg. Worse disability and lower average cognition were similarly associated with brain (adj. R2 = 0.24–0.27, p < 0.01; adj. R2 = 0.26–0.29, p < 0.001) ventricle (adj. R2 = 0.27–0.28, p < 0.001; adj. R2 = 0.19–0.20, p < 0.001) and deep gray matter volumes (adj. R2 = 0.24–0.28, p < 0.001; adj. R2 = 0.27–0.28, p < 0.001) determined with all methods, except for cortical volumes derived from 3D-FLAIR.
Discussion
In this cross-sectional study, brain volumes derived from 3D-FLAIR and 3D-T1 show similar relationships to disability and cognitive dysfunction in MS, highlighting the potential of these techniques in clinical datasets.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Accelerated brain tissue loss occurs from the earliest stages of multiple sclerosis (MS) and is associated with disability and cognitive impairment [1, 2]. Brain markers for neurodegeneration include brain volume measurements on magnetic resonance imaging (MRI). These markers are increasingly used in clinical trials, also as primary outcome measures [3,4,5]. As such trials have been successful and measures of atrophy have been shown to be predictive of long-term disease progression, there is now a need to implement measures for brain atrophy in clinical practice [6].
An important hurdle in this aim is the availability of appropriate MRI sequences in clinical scanning protocols. In general the clinical MRI protocols in MS include high-resolution 3D-FLAIR weighted sequences and post-contrast 3D-T1 sequences, because repeated scanning of lesions is mainly considered to be necessary for the identification and monitoring of inflammatory disease activity [7]. A high-resolution 3D-T1 image without contrast, traditionally considered the best sequence to identify neurodegeneration, is often not present in a clinical protocols due to time constraints.
To translate atrophy measurements to clinical datasets, there are increasing efforts to develop methods to measure brain volumes on clinical sequences. For example ventricular and thalamic volume have been measured on 2D-FLAIR scans, and total brain, grey matter (GM) and white matter (WM) volumes have been calculated on 3D-FLAIR [8,9,10]. However, not all of these methods are open source and they do not provide segmentation of cortical and deep GM structures separately.
Two new open source segmentation methods are Sequence Adaptive Multimodal SEGmentation (SAMSEG) [11] and SynthSeg [12]. Recent work has shown that both SAMSEG and SynthSeg are fast, reliable and reproducible on T1-weighted images [13, 14]. Since they are contrast-adaptive, both methods are promising candidates to evaluate brain atrophy on clinically acquired FLAIR-weighted images, but they still need validation in large MS datasets. Therefore, the objective of this study was to investigate the cross-sectional agreement between 3D-T1 and 3D-FLAIR brain volumetrics in MS using SAMSEG and SynthSeg segmentation methods. Furthermore, we investigated whether the methods on 3D-FLAIR were able to reach similar associations between brain volumes and clinical outcomes in MS as conventional 3D-T1. Lastly, results were externally validated in an independent dataset from a different center.
Methods
Participants
A total of 405 participants were retrospectively included from the Amsterdam MS cohort [15, 16] (The Netherlands) (Table 1). Subjects were selected based on the availability of a 3D-T1 weighted and 3D-FLAIR MRI scan with a voxel size smaller than 1.3 mm3. Included subjects comprised 329 MS patients with an established diagnosis of relapsing–remitting MS (RRMS, n = 242) or progressive MS (PMS, n = 87) and 76 healthy controls (HC). Approval was obtained from the local institutional ethics review board from both centers and written informed consent was provided by all participants.
Clinical assessment
Clinical evaluation consisted of disability assessment by the Expanded Disability Status Scale (EDSS) and a the expanded Brief Repeatable Battery of Neuropsychological tests for cognitive assessment [17]. This BRB-N consists of the selective reminding test (SRT), spatial recall test (SPART), symbol digit modalities test (SDMT), word list generation test (WLG), Stroop color test, memory comparison test (MCT) and test concept shifting test (CST) [16]. Individual Z-scores were calculated for each test based on the means and standard deviations of the healthy controls, with correction for age, sex and education [18]. Z-scores from all tests were averaged to obtain the average cognition score.
MRI acquisition
MRI data were acquired on a 3 Tesla GE Signa HDxt scanner (General Electric, Milwaukee, WI, USA) with an 8-channel head coil. The protocol included a 3D T1-weighted fast spoiled gradient echo (FSPGR) sequence (TR 7.8 ms, TE 3 ms, TI 450 ms, flip angle 12°, 1.0 mm sagittal slices, 0.94 × 0.94 mm2 in-plane resolution) and a 3D FLAIR sequence (TR 8000 ms, TE 125 ms, TI 2350 ms, 1.2 mm sagittal slices, 0.98 × 0.98 mm2 in-plane resolution).
MRI image analysis
This study compared five different brain segmentation approaches. FreeSurfer on the lesion-filled 3D-T1 was used as reference segmentation. The volume output of FreeSurfer was compared to volume outputs of SAMSEG on 3D-FLAIR (SAMSEGFLAIR) and SynthSeg on 3D-FLAIR (SynthSegFLAIR). In order to directly compare 3D-FLAIR vs. 3D-T1 within methods, both methods were also applied to 3D-T1 (SAMSEGT1, SynthSegT1). Volumes of the brain, ventricle, cortex, and total deep gray matter (DGM, including summed bilateral volumes of the thalamus, caudate, putamen, pallidum, nucleus accumbens, hippocampus, and amygdala) were derived from the output of each segmentation method. These volumes were selected because of their clinical relevance in MS and their wide use as outcome measures in MS studies [19].
FreeSurfer
The recon-all pipeline of FreeSurfer 7.1.1 [20] (http://surfer.nmr.mgh.harvard.edu/) was used to automatically perform whole-brain segmentation on 3D-T1 weighted images. This processing includes surface-based parcellation of the cortex [21] and segmentation of the subcortical white matter and DGM [22]. Since the presence of MS lesions affects the accuracy of FreeSurfer’s brain segmentation results, white matter lesions (WMLs) were filled on 3D-T1 prior to running FreeSurfer [23]. In short, WML were automatically segmented with the lesion prediction algorithm (LPA, SPM12) on 3D-FLAIR and filled on 3D-T1 with the SLF toolbox [24, 25].
SAMSEG
SAMSEG is a Bayesian modelling algorithm within the FreeSurfer package (Samseg—Free Surfer Wiki (harvard.edu)), which allows segmentation of neuro-anatomical structures on any MRI contrast without the need for preprocessing [11]. 3D-FLAIRs were segmented with the MS-specific pipeline of SAMSEG (released in FreeSurfer 7.2), which is a dedicated extension of SAMSEG that simultaneously segments brain structures and WMLs (settings: 1 as lesion mask pattern and the default lesion threshold of 0.3). 3D-T1s were segmented with settings 0 as lesion mask pattern and the same lesion threshold of 0.3.
SynthSeg
SynthSeg is a convolutional neural network (CNN) approach trained to segment brain structures on any MRI contrast and resolution, available as part of the FreeSurfer 7.3.2 package (SynthSeg—Free Surfer Wiki (harvard.edu) [12]. Similar to SAMSEG, SynthSeg does not require any preprocessing. While SynthSeg does not segment WMLs, the method has been trained to be robust to the presence of lesions. SynthSeg 2.0 was used to segment both native 3D-FLAIRs and 3D-T1s.
Head size normalization
Normalization of brain volumes by head size is an important step when studying disease-driven neurodegeneration in a cross-sectional study design [26]. The default head size normalization method of FreeSurfer is the so-called “estimated total intracranial volume (eTIV)”, which has been found to be biased by total brain volume and requires manual quality control [27]. Therefore, FreeSurfer-based reference volumes were normalized by the segmentation-based intracranial volume (sbTIV) from the SAMSEG processing stream. sbTIV is proposed as a more robust alternative by FreeSurfer and is less sensitive to brain atrophy [28]. The volumes of all SAMSEG and SynthSeg-derived segmentations were normalized by dividing by the sbTIV of the corresponding pipeline.
Statistical analysis
Agreement between FreeSurfer reference volumes on 3D-T1 and SAMSEG/SynthSeg derived raw volumes on 3D-FLAIR and 3D-T1 was evaluated by calculating intraclass correlation coefficients (ICCs) for consistency (single measures) with a 95% confidence interval (CI) in R statistical software (version 4.0.3; R Foundation for Statistical Computing, Vienna, Austria), to study potential systematic biases between methods. ICCs were calculated separately for HCs and MS. Agreement between head size estimates on 3D-FLAIR and 3D-T1 was analyzed with linear regression analysis with sbTIV on 3D-T1 as reference.
To evaluate to which extent FLAIR-based segmentation can detect differences between patients and controls as well as between different MS subtypes, normalized volumes were compared with analysis of covariance (ANCOVA, Pingouin 0.5.2 [29]) between HC vs. MS and RRMS vs. PMS, with age and sex as covariates. Effect sizes (partial η2) from group comparisons were calculated for each segmentation method. Partial η2 = 0.01 indicates a small effect, η2 = 0.06 indicates a medium effect and η2 = 0.14 a large effect [30]. In addition, linear regression analysis was performed to assess the relation between normalized volumes and EDSS and average cognition as dependent variables in separate models for each segmentation method, corrected for age and sex. All reported beta values were standardized and R2 values were adjusted for the number of variables in each model. P values were false discovery rate (FDR) corrected with an alpha of 0.05 and values of p < 0.05 were considered statistically significant.
Validation cohort
In order to verify the robustness of results across different scanners and centers, analyses were repeated for an independent dataset of 125 MS patients. Approval was obtained from the local institutional ethics review board and written informed consent was provided by all participants. Subjects were tested with an extensive battery of neuropsychological tests, which included SRT, SPART, SDMT, Paced Auditory Serial Addition Task (PASAT), WLG and Stroop color test. Z-scores for each test were calculated based on Italian normative data resulting in scores corrected for age, sex and education [31]. Similar to the Amsterdam cognitive data, average cognition was calculated by averaging Z-scores from all performed tests.
MRI data were acquired on a 3 Tesla Philips Achieva scanner (Philips Medical Systems, Best, The Netherlands) with an 8-channel head coil. The protocol included a 3D T1-weighted turbo field echo (TFE) sequence (TR 8.1 ms, TE 3.7 ms, flip angle 8°, 1.0 mm sagittal slices, 1.0 × 1.0 mm2 in-plane resolution and a 3D FLAIR sequence (TR 4800 ms, TE 291 ms, TI 1650 ms, 1 mm sagittal slices, 0.94 × 0.94 mm2 in-plane resolution).
Data availability
Anonymized data can be shared upon reasonable request from a qualified investigator.
Results
Table 1 shows the clinical characteristics of the Amsterdam and Verona cohorts. The 329 MS patients of the Amsterdam cohort had a mean age of 48.0 ± 11.0 years, mean symptom duration of 14.5 ± 8.4 years and an EDSS of 3.0 [2.5–4.5]). The 125 MS patients of the validation cohort from Verona had a mean age of 38.8 ± 9.7 years, mean symptom duration of 7.8 ± 6.5 years and median EDSS of 2.0 [1.0–3.0].
Volumetric agreement with FreeSurfer
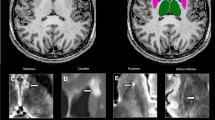
The first step of the analysis was a visual quality check of the different segmentation methods. Figure 1 shows an example of the segmentation outputs of each method. The FreeSurferT1 segmentation showed a large segmentation error for 8 subjects and were excluded from further analyses. SAMSEG and SynthSeg did not show any large segmentation errors. The ICCs for consistency between the non-normalized FreesurferT1 measurements and volumes determined with SAMSEG and SynthSeg on 3D-T1 and 3D-FLAIR are shown in Table 2. All ICC values for brain, ventricle, cortical and DGM volumes were above 0.87 in HC and above 0.90 in MS. For thalamic volumes, the agreement was comparable to the other ICCs for SynthSegFLAIR (HC: ICC = 0.91, MS: ICC = 0.91), but slightly lower for SAMSEGFLAIR (HC: ICC = 0.82, MS: 0.74).
Example of brain segmentation on 3D-T1 and 3D-FLAIR weighted images of an MS subject. FreeSurfer on lesion-filled 3D-T1 was considered the reference method, while SAMSEG and SynthSeg were used to segment unpreprocessed 3D-T1 and 3D-FLAIR weighted images. SAMSEG MS-specific pipeline was used and lesions are displayed in orange
Head size normalization
The agreement between head size normalization estimates were analyzed by performing linear regression analyses. SAMSEGT1 sbTIV was chosen as the reference method based on previous work (see methods) [28]. The R2 between SAMSEG T1 sbTIV and SAMSEG FLAIR sbTIV was 0.95 (β = 0.99, se = 0.011), 0.95 for SynthSegFLAIR sbTIV (β = 0.99, se = 0.011) and 0.96 for SynthSegT1 sbTIV (β = 0.98, se = 0.011) (see Fig. 2). The R2 between FreeSurferT1eTIV and SAMSEGT1 sbTIV was the lowest from all comparisons (R2 = 0.87, β = 0.95, se = 0.018).
Comparison of head size normalization by between SAMSEG segmentation-based total intracranial volume on T1 (SAMSEG T1 sbTIV) versus estimated total intracranial volume on T1 (FreeSurferT1 eTIV), SAMSEG-based sbTIV on FLAIR (SAMSEGFLAIR sbTIV), SynthSeg T1-based sbTIV (SynthSegT1 sbTIV) and on FLAIR (SynthSegFLAIR sbTIV). Comparisons were performed with linear regression analysis and the equation of the regression lines are shown in the plots
Effect sizes MS versus HC
Effect sizes for the differences in normalized MRI volumes for MS vs. HC and RRMS vs. PMS are shown in Fig. 3. Effect sizes between MS and HC were similar for volumes of the brain, ventricles, DGM and thalamus for both FLAIR-based and T1-based methods, with partial η2 ranging from 0.07 (ventricles) to 0.19 (thalamus), all p < 0.001. For cortical volumes, FreeSurferT1 showed a small effect (η2 = 0.03, p = 0.001), which was similarly found by SynthSegFLAIR, (η2 = 0.05, p < 0.001), while SAMSEGFLAIR showed a medium effect (η2 = 0.12, p < 0.001).
Effect sizes RRMS versus PMS
Between RRMS and PMS, FreeSurferT1 showed small effect sizes in brain (η2 = 0.02, p = 0.09), ventricle (η2 = 0.03, p = 0.005) and cortical volumes (η2 = 0.02, p = 0.065). These small effects were similarly found with SynthSegFLAIR (brain: η2 = 0.01, p = 0.109; ventricle: η2 = 0.04, p = 0.003; cortex: η2 = 0.01, p = 0.124), but were absent for SAMSEGFLAIR for brain and cortical volumes, while ventricles were similar (brain: η2 = 0.00, p = 0.331; ventricle: η2 = 0.04, p = 0.002; cortex: η2 = 0.00, p = 0.505).
Relation between volumes and disability
Standardized regression coefficients of the relationship between EDSS and each individual normalized volume are presented in Fig. 4. There were significant associations between EDSS and brain, ventricle, cortical, DGM an thalamic volumes from all different segmentation methods (p < 0.05). Largest variations in association strength between T1 and FLAIR-based volumes were found for the cortex, where FreeSurferT1 and SAMSEGT1 cortical volumes showed the highest association with EDSS (FreeSurferT1:adj. R2 = 0.26, std. β = -0.44, p < 0.001; SAMSEGT1:adj. R2 = 0.26, std. β = -0.48, p < 0.001), and lower association for SynthSegT1 (adj. R2 = 0.24, std. β = -0.33, p = 0.005), SynthSegFLAIR (adj. R2 = 0.24, std. β = -0.30, p = 0.005) and SAMSEGFLAIR (adj. R2 = 0.23, std. β = -0.24, p = 0.020).
Relationship between volumes and average cognition
The association between average cognition and volumes derived from 3D-FLAIR and 3D-T1 are presented in Fig. 5. All methods found an association between average cognition and brain (adj. R2 = 0.24–0.29, std. β = 0.44–0.50, p < 0.001), ventricle (adj. R2 = 0.19–0.21, std. β = -0.33–0.35, p < 0.001), cortex (adj. R2 = 0.11–0.23, std. β = 0.25–0.47, p < 0.001), DGM (adj. R2 = 0.27–0.29, std. β = 0.42–0.46, p < 0.001) and thalamic volumes (adj. R2 = 0.19–0.29, std. β = 0.31–0.45, p < 0.001). Again, the largest variations between methods were observed for the cortex.
Independent validation cohort
Analyses were repeated on an independent validation cohort of 125 participants with MS from Verona (Italy), demographics are shown in Table 1. All ICC values for brain, ventricle, cortical, DGM and thalamic volumes were above 0.86. For the head size normalization strategies, results were highly similar to the Amsterdam results (Supplementary Fig. 1). The R2 between SAMSEG T1 sbTIV and SAMSEG FLAIR sbTIV was 0.95 (β = 0.95, se = 0.020), 0.95 for SynthSegFLAIR sbTIV (β = 0.93, se = 0.019) and 0.95 for SynthSegT1 sbTIV (β = 0.95, se = 0.018). The R2 between FreeSurferT1 eTIV and SAMSEG T1 sbTIV was 0.61 (β = 0.61, se = 0.044), which was lower than the R2 observed in the Amsterdam data (R2 = 0.87, β = 0.95, se = 0.018).
The association between EDSS, average cognition and volumes derived from 3D-FLAIR and 3D-T1 are presented in Supplementary Fig. 2 and 3. For EDSS and average cognition, all methods had similar associations between these outcome measures and brain, DGM and thalamus volumes (p < 0.05) as in the Amsterdam MS cohort. Also similar was the large variation in association strength for the cortical volumes between the different methods and clinical outcomes. While the reference method FreeSurferT1 did not find an association between cortical volume and average cognition (adj. R2 = 0.21, std. β = 0.044, p = 0.671), SAMSEGFLAIR was the only method that found a relation (adj. R2 = 0.27, std. β = 0.25, p = 0.010). The correlation between lesion volumes and normalized tissue volumes for both cohorts are shown in Supplementary Fig. 4.
Discussion
This study investigated the agreement of brain volume measurements on 3D-FLAIR with those on 3D-T1 in a large cross-sectional dataset with MS patients and healthy controls. Our results demonstrated high consistency in the total brain, ventricular and total DGM volumes measured on 3D-FLAIR compared to those measured on standard 3D-T1. However, for cortical and thalamic volumes, the agreement between 3D-FLAIR and 3D-T1 was dependent on the method used, where especially the cortex showed strongest variations for relations to clinical outcome measures. We replicated these findings in an independent validation cohort. Thus, more advanced regional quantifications on 3D-FLAIR require specific choices and further methodological innovation.
A good to excellent consistency was found especially for brain and ventricular volumes [32]. This finding is highly relevant for the adoption of 3D-FLAIR for volume measurements in clinical practice, since whole brain and ventricular volumes are both powerful measures to define, monitor and predict MS severity [2, 33]. For thalamic volumes the agreement was lower using the FLAIR-based methods, especially with SAMSEG. The overall consistency between T1 and FLAIR-based volumes obtained in our study is in agreement with another study using a multi-atlas-based segmentation approach (Geodesic Information Flow, GIF) [10]. However, in that study, only global GM volumes were compared and their method, GIF, was not compared against other standard methods in the field, such as FreeSurfer in this study.
Effect sizes for detecting volume differences in MS compared to controls were stable across methods, which could be expected given the good to excellent relative agreement. Highest effect sizes were found for the thalamus and total DGM, which is consistent with other studies showing the highest atrophy rate in DGM compared to other brain areas [34, 35]. Although we found comparable effect sizes for DGM volumes, another study found systematic differences between DGM segmentation methods (FSL-FIRST, FreeSurfer, GIF and volBrain) compared with manual reference measurements [36]. Moreover, the same study reported reduced accuracy of DGM segmentation in MS versus controls, which could have affected the found effect sizes in the present study since we only used automated segmentation methods. For corticalvolumes, effect sizes between MS and HC were the lowest of all assessed brain structures and showed the largest differences between methods. Cortical segmentation is already a notoriously difficult task on high-resolution 3D-T1 weighted images, with an average variability of 2.5–3% [37] and cortical measurements varying between software [38]. Especially since FLAIR-weighted images are generally not optimized for gray/white matter contrast, a more extensive evaluation of cortical measurements on currently available FLAIR scans is needed.
The most similar correlations for FLAIR-based methods compared to T1-based methods were found for brain and regional volumes with EDSS and average cognition. These correlations were especially highly consistent for the total brain, ventricle, DGM and thalamic volumes. Again, the cortical volumes displayed the largest differences across methods. For example, SAMSEG on 3D-FLAIR falsely detected an association between cortical volumes and average cognition in the validation cohort, while the other T1- and FLAIR-based methods did not show any association. In that regard, SynthSeg cortical segmentation on 3D-FLAIR seems more reliable compared with SAMSEG, although the associations with disability and cognition were less strong compared to FreeSurfer. Since cortical atrophy is an important outcome measure for cognition and has been shown to be clinically predictive for cognitive decline [18], further validation of 3D-FLAIR derived cortical volumes is warranted before using FLAIR derived cortical volumes in relation to cognitive outcomes.
Although 3D-FLAIR sequences are currently recommended for MS diagnosis and monitoring [7], clinical legacy datasets mostly contain 2D-FLAIR scans. Retrospective analysis of these large clinical databases would enable retrospective studies with high sample sizes to gain more insight in MS. SynthSeg has originally been developed with the aim to provide accurate segmentation on low resolution scans of any contrast type, so future work should investigate the segmentation accuracy on 2D-FLAIR compared with 3D-FLAIR. SAMSEG may also be a promising candidate to segment 2D-FLAIR scans, but was designed and validated mostly on high resolution images in MS [11]. The advantage of SAMSEG for application to clinical legacy data is that it currently has a longitudinal pipeline, while SynthSeg is a cross-sectional method.
This study is not without limitations. First, although we replicated our findings in an independent validation cohort from a different center, all data were acquired on 3.0 T MRI scanners. Since clinical MRI scanners often have lower field strength, findings should be replicated on lower field strengths and between scanners as well [39]. Second, the outcomes of this study rely on the surface-based FreeSurfer stream as the reference segmentation. Although FreeSurfer is a well-established research method, manual reference segmentations still remain the golden standard, which was not feasible to create for the large data set used in this study. Third, only cross-sectional correlations of FLAIR images with clinical outcome measures were assessed, but the longitudinal relations of these segmentation methods on FLAIR is unknown. This is especially relevant for facilitating clinical implementation of these techniques since brain atrophy rates are useful measures for assessing disease evolution and treatment response [19]. In addition, in real-world clinical settings, patients are scanned on different scanners and protocols are constantly updated over time. Therefore, the effect of different scanners and protocols on longitudinal measurements should be a topic of future study as well.
Conclusion
Brain volumes segmented on 3D-FLAIR with SynthSeg and SAMSEG show a good to excellent agreement with FreeSurfer-derived 3D-T1 segmentation in MS, especially for total brain and ventricular volumes. Lower volume in MS vs. HC on 3D-FLAIR was relevant for disability and cognitive dysfunction, but effect sizes depended on the segmentation method that was used. While agreement of total DGM, total brain and ventricular segmentation was relatively good, cortical segmentation remains especially difficult, which could be the focus of further improvement of FLAIR-based segmentation methods.
References
Benedict RH et al (2006) Neocortical atrophy, third ventricular width, and cognitive dysfunction in multiple sclerosis. Arch Neurol 63(9):1301–1306
Zivadinov R et al (2016) A serial 10-year follow-up study of brain atrophy and disability progression in RRMS patients. Mult Scler J 22(13):1709–1718
Sormani MP, Arnold DL, De Stefano N (2014) Treatment effect on brain atrophy correlates with treatment effect on disability in multiple sclerosis. Ann Neurol 75(1):43–49
Eshaghi A et al (2018) Progression of regional grey matter atrophy in multiple sclerosis. Brain 141(6):1665–1677
Naismith RT et al (2021) Effects of ibudilast on MRI measures in the phase 2 SPRINT-MS study. Neurology 96(4):e491–e500
Sastre-Garriga J, Pareto D, Rovira À (2017) Brain atrophy in multiple sclerosis: clinical relevance and technical aspects. Neuroimaging Clin 27(2):289–300
Wattjes MP et al (2021) 2021 MAGNIMS–CMSC–NAIMS consensus recommendations on the use of MRI in patients with multiple sclerosis. Lancet Neurol 20(8):653–670
Dwyer MG et al (2017) Neurological software tool for reliable atrophy measurement (NeuroSTREAM) of the lateral ventricles on clinical-quality T2-FLAIR MRI scans in multiple sclerosis. NeuroImage Clin 15:769–779
Dwyer M et al (2021) DeepGRAI (Deep Gray Rating via Artificial Intelligence): fast, feasible, and clinically relevant thalamic atrophy measurement on clinical quality T2-FLAIR MRI in multiple sclerosis. NeuroImage Clin 30:102652
Goodkin O et al (2021) FLAIR-only joint volumetric analysis of brain lesions and atrophy in clinically isolated syndrome (CIS) suggestive of multiple sclerosis. NeuroImage Clin 29:102542
Cerri S et al (2021) A contrast-adaptive method for simultaneous whole-brain and lesion segmentation in multiple sclerosis. Neuroimage 225:117471
Billot B et al (2023) SynthSeg: segmentation of brain MRI scans of any contrast and resolution without retraining. Med Image Anal 86:102789
Sederevičius D et al (2021) Reliability and sensitivity of two whole-brain segmentation approaches included in FreeSurfer–ASEG and SAMSEG. Neuroimage 237:118113
van Nederpelt DR et al (2023) Reliability of brain atrophy measurements in multiple sclerosis using MRI: an assessment of six freely available software packages for cross-sectional analyses. Neuroradiology (Accepted)
Steenwijk MD et al (2014) What explains gray matter atrophy in long-standing multiple sclerosis? Radiology 272(3):832–842
Schoonheim MM et al (2015) Thalamus structure and function determine severity of cognitive impairment in multiple sclerosis. Neurology 84(8):776–783
Rao SM (1990) A manual for the brief repeatable battery of neuropsychological tests in multiple sclerosis. Medical College of Wisconsin, Milwaukee, p 1696
Eijlers AJ et al (2018) Predicting cognitive decline in multiple sclerosis: a 5-year follow-up study. Brain 141(9):2605–2618
Sastre-Garriga J et al (2020) MAGNIMS consensus recommendations on the use of brain and spinal cord atrophy measures in clinical practice. Nat Rev Neurol 16(3):171–182
FreeSurfer FB (2012) FreeSurfer. Neuroimage 62(2):774–781
Fischl B, Sereno MI, Dale AM (1999) Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system. Neuroimage 9(2):195–207
Fischl B et al (2002) Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33(3):341–355
Govindarajan KA et al (2015) Effect of in-painting on cortical thickness measurements in multiple sclerosis: a large cohort study. Hum Brain Mapp 36(10):3749–3760
Schmidt P et al (2012) An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 59(4):3774–3783
Valverde S, Oliver A, Lladó X (2014) A white matter lesion-filling approach to improve brain tissue volume measurements. NeuroImage Clin 6:86–92
Voevodskaya O et al (2014) The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer’s disease. Front Aging Neurosci 6:264
Klasson N et al (2018) Estimated intracranial volume from FreeSurfer is biased by total brain volume. Eur Radiol Exp 2:1–6
Nerland S et al (2022) A comparison of intracranial volume estimation methods and their cross-sectional and longitudinal associations with age. Hum Brain Mapp 43(15):4620–4639
Vallat R (2018) Pingouin: statistics in Python. J Open Source Softw 3(31):1026
Fritz CO, Morris PE, Richler JJ (2012) Effect size estimates: current use, calculations, and interpretation. J Exp Psychol Gen 141(1):2
Amato M et al (2006) The Rao’s Brief Repeatable Battery and Stroop Test: normative values with age, education and gender corrections in an Italian population. Mult Scler J 12(6):787–793
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15(2):155–163
Uher T et al (2021) Evolution of brain volume loss rates in early stages of multiple sclerosis. Neurol Neuroimmunol Neuroinflammation 8(3):e979
Azevedo CJ et al (2018) Thalamic atrophy in multiple sclerosis: a magnetic resonance imaging marker of neurodegeneration throughout disease. Ann Neurol 83(2):223–234
Eshaghi A et al (2018) Deep gray matter volume loss drives disability worsening in multiple sclerosis. Ann Neurol 83(2):210–222
de Sitter A et al (2020) Reduced accuracy of MRI deep grey matter segmentation in multiple sclerosis: an evaluation of four automated methods against manual reference segmentations in a multi-center cohort. J Neurol 267(12):3541–3554
Tustison NJ et al (2014) Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage 99:166–179
Popescu V et al (2016) Grey matter atrophy in multiple sclerosis: clinical interpretation depends on choice of analysis method. PLoS ONE 11(1):e0143942
Battaglini M et al (2019) Lifespan normative data on rates of brain volume changes. Neurobiol Aging 81:30–37
Funding
This project is supported by research Grants from ZonMW (VIDI Grant 09150172010056) and the Dutch MS Research Foundation (08-650, 13-820 and 14-358).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
S. Noteboom is supported by research grants from Atara Biotherapeutics, Merck and Biogen. D. R. van Nederpelt has nothing to disclose. A. Bajrami has nothing to disclose. B. Moraal has nothing to disclose. M. W. A. Caan is shareholder of Nico.lab International Ltd. F. Barkhof serves on the steering committee or iDMC member for Biogen, Merck, Roche, EISAI and Prothena. Consultant for Roche, Biogen, Merck, IXICO, Jansen, Combinostics. Research agreements with Merck, Biogen, GE Healthcare, Roche. Co-founder and shareholder of Queen Square Analytics LTD. Supported by the NIHR biomedical research center at UCLH. M. Calabrese was supported by the GR-2013-02-355322 grant from Italian Ministry of Health as well as the progressive MS alliance and reports grants and personal fees from Biogen Idec, Merck Serono, Novartis, and Roche. H. Vrenken has received research grants from Pfizer, Merck Serono, Novartis, and Teva, speaker honoraria from Novartis, and consulting fees from Merck Serono; all funds were paid directly to his institution. E. M. M. Strijbis has nothing to disclose. M.D. Steenwijk is supported by research grants from Atara Biotherapeutics, Merck and Biogen. M.M. Schoonheim serves on the editorial board of Neurology and Frontiers in Neurology, receives research support from the Dutch MS Research Foundation, Eurostars-EUREKA, ARSEP, Amsterdam Neuroscience, MAGNIMS and ZonMW (VIDI Grant 09150172010056) and has served as a consultant for or received research support from Atara Biotherapeutics, Biogen, Celgene/Bristol Meyers Squibb, Genzyme, MedDay and Merck.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Noteboom, S., van Nederpelt, D.R., Bajrami, A. et al. Feasibility of detecting atrophy relevant for disability and cognition in multiple sclerosis using 3D-FLAIR. J Neurol 270, 5201–5210 (2023). https://doi.org/10.1007/s00415-023-11870-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-023-11870-4