Abstract
Myotonic dystrophy type 2 (DM2) is a common adult onset muscular dystrophy caused by a dominantly transmitted (CCTG) n expansion in intron 1 of the CNBP gene. In DM2 there is no obvious evidence for an intergenerational increase of expansion size, and no congenital cases have been confirmed. We describe the clinical and histopathological features, and provide the genetic and molecular explanation for juvenile onset of myotonia in a 14-year-old female with DM2 and her affected mother presenting with a more severe phenotype despite a later onset of symptoms. Histological and immunohistochemical findings correlated with disease severity or age at onset in both patients. Southern blot on both muscle and blood samples revealed only a small increase in the CCTG repeat number through maternal transmission. Fluorescence in situ hybridization, in combination with MBNL1 immunofluorescence on muscle sections, showed the presence of mutant mRNA and MBNL1 in nuclear foci; the fluorescence intensity and its area appeared to be similar in the two patients. Splicing analysis of the INSR, CLCN1 and MBNL1 genes in muscle tissue demonstrates that the level of aberrant splicing isoforms was lower in the daughter than in the mother. However, in the CLCN1 gene, a heterozygous mutation c.501C>G p.F167L was present in the daughter’s DNA and found to be maternally inherited. Biomolecular findings did not explain the unusual young onset in the daughter. The co-segregation of DM2 with a recessive CLCN1 mutation provided the explanation for the unusual clinical findings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myotonic dystrophies (DMs) are the most common adult onset muscular dystrophy, affecting mainly the skeletal muscle, heart, and the central nervous system [1]. DMs have an autosomal dominant inheritance pattern and are caused by the expansion of similar microsatellites in two functionally unrelated genes. Myotonic dystrophy type 1 (DM1; OMIM no. 160900) is caused by an expansion of a CTG repeat within the 3′ UTR of the DMPK gene located on chromosome 19 [2–4], and myotonic dystrophy type 2 (DM2; OMIM no. 602668) is caused by a CCTG repeat expansion in intron 1 of the CNBP gene located on chromosome 3 [5, 6]. Individuals with DM1 disease have expansions ranging from 50 to >2.000 repeats. The expansion in DM2 ranges in size from 75 to 11.000 CCTG repeats, but the minimum size of a pathogenic expansion is not known [6]. The pathogenic mechanism common to both disorders involves a novel RNA gain of function of the repeat-containing mRNAs transcribed from the mutated DMPK and CNBP alleles. These expanded transcripts form foci that are retained in cell nuclei and alter the functions of RNA binding proteins involved in regulating alternative splicing and mRNA translation [7]. At least two RNA binding proteins have been identified to bind the anomalous CUG/CCUG repeat expansions: muscleblind-like 1 (MBNL1) and CUGBP1. These proteins are antagonist regulators of splicing events, with MBNL1 promoting a switch to adult isoforms and CUGBP1 inducing retention of embryonic isoforms of genes that are misregulated in DMs [8, 9].
DM1 and DM2 share several clinical features; however, there are some differences in the clinical presentation. These differences include the muscle groups prominently affected (distal in DM1 and proximal in DM2) and the presence of a severe congenital form with mental retardation in DM1 but not in DM2 where a neonatal/childhood form has not been confirmed. Moreover, myotonia is marked in DM1, while it is mild and inconsistent in DM2, even by electromyography [10]. The muscle biopsies also show both similar histological features such as central nucleation, fiber atrophy, and nuclear clump fibers, and different findings. In DM2 both central nucleation and fiber atrophy occur preferentially in type 2 fibers, suggesting that DM2 is predominantly a disease of type 2 fibers [11–13].
In DM1, the onset and severity correlate with repeat length, and repeat expansions of >1.500 often result in a severe congenital form of the disease [1, 14]. Intergenerational instability of the CTG expansion provides the molecular basis for the anticipation phenomenon, i.e., a progressively earlier and more severe manifestation of the disease and increase in CTG expansion size in successive generations of DM1 families [15]. In contrast to DM1, in DM2 no correlation between repeat size and disease severity and no evidence for intergenerational expansions have been found [16]. Recent studies have reported DM2 families with heterozygous recessive CLCN1 mutations [17–19]. CLCN1 maps to chromosome 7q35 and when mutated causes myotonia congenita (recessive Becker disease OMIM no. 255700; dominant Thomsen disease OMIM no. 160800). Co-segregation of DM2 mutation and recessive CLCN1 mutations have been suggested to influence the DM2 phenotype [18]. Indeed, Suominen et al. [18] observed that EMG myotonia occurred in all DM2 co-segregating recessive CLCN1 mutations examined, thus concluding that DM2 patients in whom the recessive CLCN1 mutation is present are more likely to be diagnosed than those without this mutation. In DM1 the coexistence of recessive CLCN1 mutations was not more frequent than in controls [18].
In our study we describe the clinical and histopathological features, and provide the genetic and molecular explanation for juvenile onset of myotonia in a 14-year-old female with DM2 and her affected mother presenting with a more severe phenotype despite a later onset of symptoms.
Methods
Patients
Two patients, the proband and her daughter, were clinically evaluated, and reports on other family members were obtained through the patients, but no other family members were investigated. Reportedly, symptoms of muscle weakness and/or muscle stiffness occurred in the proband’s mother and grandmother. They complained of difficulties in rising from a sitting position and in releasing their grip. They died at the ages 64 and 59 years, respectively, because of a non-specified muscular disease.
Muscle pathology
A biceps brachii muscle biopsy was taken from patient 1 at the age of 15 years and from patient 2, her mother, at the age of 45 years. Blood samples and muscle biopsies were used for this study after receiving informed consent from the patients.
Muscle tissue was fresh-frozen in isopentane cooled in liquid nitrogen. Histopathological analysis was performed on serial sections (8 μm) processed for routine histological or histochemical stainings. A standard myofibrillar ATPase staining protocol was used after preincubation at pH 4.3, 4.6, and 10.4 [20].
Immunohistochemistry
Serial transverse muscle cryostat sections 6 μm thick were cut for immunohistochemical staining (IHC). Sections were air-dried and rehydrated in phosphate buffer pH 7.4 (PBS). Non-specific binding sites were blocked with normal goat serum (NGS; DAKO) at a dilution 1:20 in PBS containing 2% bovine serum albumin (BSA; Sigma-Aldrich) for 20 min at room temperature. Mouse monoclonal primary antibodies against two different myosin heavy chain (MHC) isotypes were used at the following dilutions: MHCfast, 1:400 in PBS + 2% BSA (Sigma-Aldrich); MHCslow, 1:400 in PBS + 2% BSA (Sigma-Aldrich). Each antibody was applied for 1 h at room temperature. After washing in PBS 3 times for 5 min, sections were incubated with goat anti-mouse biotinylated secondary antibody diluted 1:300 in PBS + 2% BSA for 1 h at room temperature. Nuclei were counterstained with Mayer’s hematoxylin.
Quantitative evaluation of fiber diameter was made as described previously by Vihola et al. [11] with Scion Image (Scion Corporation, Frederick, MD) on images taken with a light microscope (160×, original magnification). The size of muscle fibers was assessed by measuring the “smallest fiber diameter.” All data were elaborated using Microcal Origin (Microcal Software Inc., Northampton, MA, USA). The metahistograms were normalized to normal mean diameter for men and women. Atrophy and hypertrophy factors were also calculated [20].
Immunohistochemical staining against CLC-1 protein was also done using two different antibodies pooled together, a commercial ClC-1 antibody (CLC11-A, Alpha diagnostic international, San Antonio, TX, USA) and a ClC-1 antibody generated against the 15 C-terminal amino acids (a kind gift from K. Metsikkö, University of Oulu, Finland) both at a dilution 1:50. The double immunohistochemical staining was performed on the BenchMark (Ventana Medical Systems Inc., Tucson, AZ, USA) immuno-stainer. The immunohistochemical stainings were performed using the official protocol of the BenchMark immuno-stainer and visualized with a peroxidase based detection kit (UltraView Universal DAB detection kit, Ventana Medical Systems Inc., Tucson, AZ, USA), and the signal was then amplified (Ventana amplification kit, Ventana Medical Systems Inc., Tucson, AZ, USA).
Fluorescence in situ hybridization (FISH) and immunofluorescence
FISH procedure, using RNA (CAGG)5 Texas red labeled probes (IDT, Coralville, IA, USA), was carried out on muscle sections as previously reported by Cardani et al. [21]. Following the 1XSSC post-hybridization wash without DAPI, the immunofluorescence protocol was performed starting from non-specific binding sites blocking with NGS (DAKO) at a dilution of 1:20 in PBS + 2% BSA (Sigma-Aldrich) for 20 min at room temperature. A polyclonal rabbit anti-MBNL1 (1:1,000 in PBS + 2% BSA; gift from Prof. C.A. Thornton University of Rochester, New York, USA) was applied overnight at 4°C. After washing in PBS three times for 5 min, sections were incubated with Alexa488-labeled goat anti-rabbit secondary antibody (Molecular Probes, Invitrogen) diluted 5 μg/ml in PBS + 2% BSA for 1 h at room temperature. Nuclei were stained with DAPI, and slides were then mounted with ProLong (Invitrogen). Sections were examined on a confocal microscope (Leica TCS SP2 AOBS).
For quantitative measurement of fluorescence intensity and the area of ribonuclear inclusions and MBNL1 nuclear foci, sections were analyzed on a confocal laser scanning microscopy at 630× magnification under identical illumination, exposure, and instrument settings [22].
Genetic analyses
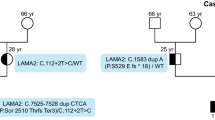
For each subject involved in this study, the DM2 mutations were detected and measured in both peripheral blood and muscle tissues. Genomic DNA was extracted from both tissues using a salting out procedure. Detection of the DM2 mutations was obtained with a long PCR-based method, as previously reported [23]. Characterization of the DM2 mutation was carried out with a Southern blot procedure modified according to the protocol described in Nakamori et al. [24]. Briefly, genomic DNA was digested with HaeIII, and AluI, fragments were resolved on 0.5% agarose gel electrophoeresis and then transferred overnight onto a nylon membrane by alkaline transfer. Blots were then hybridized with a DIG-labeled (CCTG)5 LNA probe. Blots were analyzed by using Storm 860 ImageQuant TL Image Analysis software v7.0 (Amersham Biosciences, Buckinghamshire, UK). Expanded fragments were sized by measuring the bands of major intensity, which presumably correspond to the more representative alleles.
The 23 exons of the CLCN1 gene were amplified in patient 1 by PCR using primers previously described [25]. PCR products were purified with a QIAquick column PCR purification kit (QIAGEN) and analyzed by direct forward and reverse sequencing, using the DNA sequencing Kit (Perkin Elmer Applied Biosystems) on an ABI PRISM 310 DNA automatic sequencer. Sequences were analyzed with Sequencher software (Gene Codes Corporation, Ann Arbor, MI) and compared to the CLCN1 reference sequence NT_079596. The CLCN1 gene was also analyzed by cDNA sequencing in patient 1. For cDNA analysis RNA was extracted from muscle biopsy (Trizol method), and cDNA was generated using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The CLCN1 gene transcript was amplified by polymerase chain reaction using five overlapping primer pairs. The CLCN1 exon 4 was sequenced from genomic DNA of patient 1. Identification of the c.501C<G mutation in patient 2 was attained by direct sequencing of the CLCN1 exon 4 in DNA extracted from both blood and muscle tissues.
Study of INSR, MBNL1, and CLCN1 gene alternative splicing
The RT-PCR splicing assays for the INSR, MBNL1, and CLCN1 genes were performed according to reported protocols [26, 27] using the following pairs of primers (sequences from 5′ to 3′ ends): Fw-GCTGCCCAATACCAGGTCAAC, Rev-TGGTGGGAGAAATGCTGTATGC (MBNL1 gene splicing), Fw-CATCTCTCCCCAGGCTGT, Rev-GCATCCTTGTTCCACACT (CLCN1 gene splicing), Fw-CCAAAGACAGACTCTCAGAT, and Rev-AACATCGCCAAGGGACCTGC (INSR gene splicing). Total PCR products, obtained within the linear range of amplification, were electrophoresed on 3,5 agarose gel for separation. Quantitative analysis of the amplified products was performed using SybrGreenII-stained gels (Perkin-Elmer Life Science, MA, USA) scanned on a fluorimager 595 (Amersham Biosciences, Buckinghamshire, UK). The intensity of each band and the fraction of abnormally spliced (AS) isoforms (AS-isoforms/total) were quantified by densitometry using ImageQuant software. Normalization of the RT-PCR reactions was based on the expression level of the glucose phosphate isomerase housekeeping gene (GPI), and each amplification was carried out in triplicate using independent cDNA samples.
QRT-PCR analysis of CLCN1 mRNA expression
Total RNA was extracted from muscle samples (P1, P2, and two control subjects) using the RNeasy mini kit (Qiagen Co., Valencia, CA, USA). Three micrograms of total RNA was reverse transcribed according to the cDNA protocol of the High Capacity cDNA Archive kit (Applied Biosystem, Foster City, CA, USA). The Hs00163961_m1 Assay-on-demand™ gene expression product, labeled with FAM, was used to quantify CLCN1 transcripts. The β2-miscroglobulin gene (B2M: GenBank accession no. NM_004048) labeled with VIC dye was chosen as the housekeeping, internal control gene. We performed each PCR reaction in triplicate using the Taqman Universal PCR Master Mix and the ABI PRISM 7000 Sequence Detection System. A comparative threshold cycle (Ct) was used to determine CLCN1 expression compared to the calibrator (median value of control subjects). Hence, steady-state mRNA levels expressed a n-fold difference relative to the calibrator. For each sample, the gene Ct value was normalized using the formula ΔCt = CtCLCN1_CtB2M. To determine relative expression levels, the following formula was used: ΔΔCt = ΔCt sample−ΔCt calibrator. The value used to plot relative gene expression was calculated using the expression 2−ΔΔCT.
Results
Patients
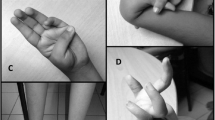
Patient 1. The proband’s 15-year-old daughter was admitted to our department a few months later because she had complained of grip myotonia since the age of 14. On admission she also complained of difficulties in starting leg movements, especially when she had to start climbing stairs and rising from the squatting position. The symptoms improved with repetitive movement. Neurological examination revealed muscles of normal tone and strength. Grip myotonia was evident, and there was a prominent warm-up phenomenon. Lid lag was present. Deep tendon reflexes were normal. Laboratory studies demonstrated normal electrolyte, urea, creatinine, and lactate dehydrogenase levels. Serum creatine and lipid profiles were normal. Routine laboratory studies were normal, including serum creatine levels. EMG showed myotonic discharges in all muscles examined, but no myopathic changes. EKG, Holter electrocardiographic recordings, and echocardiograms were normal.
Patient 2. The proband, a 45-year-old woman, was admitted to our department because of proximal lower limb weakness that had progressed since the age of 30. Onset had been at age 20 with grip myotonia. Family history was positive for lower limb muscle weakness and grip myotonia in her mother and grandmother. Cataracts had been removed at age 40. Neurological examination revealed mild muscle atrophy in the upper limbs but normal muscle strength. Muscles were of normal strength except for the neck flexors (grade 4 MRC) and hip flexors (grade 4 MRC). Deep tendon reflexes were uniformly brisk. Grip myotonia was present, and warm-up was prominent. Routine laboratory studies were normal except for serum creatine levels (240 U/l). EMG showed myotonic discharges in the right abductor pollicis brevis. EKG, Holter electrocardiographs, and echocardiograms were normal.
Histopathological findings
In patient 1 (daughter), routine histological and histochemical stainings of muscle sections did not show the characteristic histopathological feature of DM2. No atrophic fibers, nuclear clumps, or increased centrally nucleated myofibers were observed in muscle sections. Only a slight fiber size variation and predominance of type 2 fibers were present (Fig. 1a, c, e, g). Myopathic changes were instead observed in patient 2 (mother). Muscle sections with increased fiber size variation, internalized nuclei, and many small fibers with nuclear clumps were present (Fig. 1b, d, f, h). A marked atrophy and a preferential central nucleation of type 2 myofibers is also evident (Fig 1d, f). MHCf or MHCs immunostaining allowed us to detect and measure fibers with a diameter smaller than 20 μm and to observe that the majority of the nuclear clumps were type 2 fibers since a thin rim of fast myosin immunostaining was present around the nuclei (Fig. 1h).
a–h Histopathological analysis of muscle biopsy obtained from patient 1 (a, c, e, g) and patient 2 (b, d, f, h). H&E staining (a, b) demonstrates a variation in fiber size more evident in the patient 2 muscle section (mother; b) where central nuclei (white arrow), atrophic fibers (black arrow), and numerous pycnotic nuclear clumps are also present (white arrowheads). These histopathological features are evidenced by ATPase pH 10.4 (c, d) and pH 4.3 (e, f). Fiber size variability is present in patient 1 muscle sections (daughter; c, e) both at type 1 and type 2 fiber levels, whereas in patient 2 atrophic fibers are of type 2 (black arrows; d); central nuclei are predominantly in type 2 fibers (white arrow; f). Pycnotic nuclear clumps are still well evident in patient 2 (white arrowheads; d, f). Fast myosin immunostaining (g, h) shows that atrophic fibers and nuclear clumps in the patient 2 muscle section (black arrows; h) are fast myosin positive. i–l Metahistograms obtained from the analysis of muscle fiber diameters in patient 1 (i) and patient 2 (j). The results are based on sections immunostained for fast or slow myosin. Marked type 2 fiber atrophy is evident in patient 2 only. Tables show the mean diameter of fast or slow fibers and the relative atrophy (A) or hypertrophy (H) factor in patient 1 (k) and patient 2 (l)
Mean diameters of both fast and slow fibers appeared to be in the normal range (normal control range 30–70 μm for women) in both patient 1 and 2 (Fig. 1i–l). The metahistograms based on data obtained from the analysis of muscle fiber diameters on sections immunostained for fast or slow myosin revealed no preferential type 2 fiber atrophy in patient 1 (Fig. 1i, k). On the contrary, in patient 2 the histogram of type 2 fibers had a bimodal distribution due to the presence of both normal-sized fibers and marked atrophic fibers with numerous type 2 nuclear clump fibers (Fig. 1j, l). Indeed, in this patient an increase of the relative atrophic factor of type 2 fibers has been calculated. Hypertrophy of both type 1 and type 2 fibers is also evident (Fig. 1l).
CLC-1 immunohistochemistry showed a reduction of the protein in both patient biopsies when compared to the normal control (Fig. 2).
FISH analysis and MBLN1 immunoflurescence
The ribonuclear inclusions formed by pathologic retention of DM2 repeat expansions were detected by fluorescent in situ hybridization (FISH) in myonuclei in both patient 1 and patient 2 muscle sections (Fig. 3a, d). FISH in combination with MBNL1 immunofluorescence shows that MBNL1 is sequestered in myonuclei as protein foci that co-localize with ribonuclear inclusions (Fig. 3b, c, e, f). The fluorescence intensity (FI) and area of both ribonuclear inclusions and MBNL1 foci have been evaluated in double-stained sections imaged under the same exposure and threshold settings. The mean of FI and area of both types of nuclear foci appear not to be statistically different between the two patients (Fig. 3g, h).
Ribonuclear inclusions and MBNL1 foci in patient 1 (a–c) and patient 2 (d–f). FISH in combination with MBNL1 immunofluorescence reveals that ribonuclear inclusions (red; a, d) and MBNL1 foci (green; b, e) colocalize in nuclei (blue, DAPI) of muscle fibers (c, f). Bars indicate the mean ± SD of area (g) and fluorescence intensity (h) of ribonuclear inclusions and MBNL1 nuclear foci obtained at confocal laser scanning microscopy in patient 1 and 2
Genetic analysis
Genetic characterization of the DM2 mutation was obtained by a combination of long PCR and Southern blot analyses on DNA extracted from both peripheral blood and muscle. The detection of the CCTG expansions in these tissues from patients 1 and 2 was obtained with a long PCR method developed in our laboratory [23]. This analysis revealed an elevated grade of somatic mosaicism and genetic instability in both patients with an increase of the average (CCTG) n number in the muscle tissues of patient 1 of about 300 repetitions compared to patient 2 (not shown). However, long PCR-based techniques can detect only alleles up to 15 kb, depending on the PCR conditions, limiting the detection and sizing of larger DM2 mutations. The average expanded allele sizes were then determined by Southern blot analysis with a digoxigenin-labeled (CCTG)5 LNA probe, measuring the bands of major intensity or at the center of the smear for very diffuse bands, which presumably correspond to the more representative expanded alleles [24]. The molecular characterization of the blood leukocyte DNA allowed us to detect multiple, equally intense bands of about 4.700, 4.000, and 3.000 CCTG repetitions (average of 3.900) in patient 2 and expansions of about 6.400, 4.700, and 1.800 (average 4.300) in patient 1, with a marked prevalence of the lower expanded allele. In the muscle tissue the DM2 mutations appear as very heterogeneous smears, allowing the definition of only an interval of repetitions that was 6.000–3.800 CCTG in patient 2 (average 4.900) and 6.800–4.000 in patient 1 (average 5.400). The molecular sizing of the DM2 mutations, obtained by Southern blot, confirms the long-PCR results, indicating only a small increase in the DM2 mutation size through maternal germline transmission (Fig. 4a).
a Southern blot analysis using a DIG-labeled (CCTG)5 LNA probe of blood and muscle DNA from patient 1 (P1, daughter), patient 2 (P2, mother), and from a healthy control (CN). Arrows indicate sizes (blood) and ranges (muscle) of the ZNF9 expanded alleles. b Panel showing the RT-PCR splicing assay of the INSR, MBNL1, and CLCN1 genes in patients 1 (P1, daughter), 2 (P2, mother), and in a control sample (CTR). Alternatively spliced exons analyzed for each gene are shown in the diagrams on the right. The glucose phosphate isomerase (GPI) gene was used for housekeeping for normalization of cDNA samples
We then considered the possibility that the grip myotonia observed in the young DM2 patient could be related to a second genetic mutation in the CLCN1 gene, genetically linked to myotonia congenita in both the autosomal dominant and recessive forms [28]. The CLCN1 gene is comprised of 23 exons, and disease-causing mutations, including deletions, insertions, frameshift, nonsense, and missense, are distributed all over the gene. Direct sequencing of the coding region and intron/exon boundaries of this chloride channel ion gene in patient 1 revealed a heterozygous mutation c.501C>G p.F167L. This mutation has been previously reported to cause recessive myotonia congenita. Analysis of the CLCN1 exon 4 in the mother’s DNA extracted from both blood and muscle tissues confirmed that this mutation is maternally inherited.
Splicing analysis and quantification of the CLCN1 mRNA
Alternative isoforms of various genes, including insulin receptor (INSR-A), MBNL1 (MBNL1ex7), and CLCN1 (CLCN1ex7a), prevail in affected adult DM1 and DM2 muscle compared to control tissues as a consequence of the in trans effect of the (CCTG) n expansion [8]. Recent evidence also indicates a predominant role of the MBNL1 loss of function due to sequestration and missplicing for the splicing abnormality in DM pathogenesis [26, 29]. In the case of the insulin receptor (INSR), the predominant splice product expressed in DM1 muscle is the exon 11 skipped (non-muscle) isoform, which may contribute to the insulin resistance in DM1 muscle fibers [30, 31]. Misregulated splicing of the muscle-specific chloride ion channel (CLCN1) transcripts may contribute to the myotonic membrane hyperexcitability that characterizes DM muscle cells [32]. We therefore considered these genes as molecular hallmarks of the DM spliceopathy and analyzed their splicing profile in the muscle tissue from patients 1 and 2 and from a control subject. RT-PCR analysis of spliced transcripts was carried out with primers generating fragments of different lengths, according to the inclusion/exclusion of alternatively spliced exons. The panel in Fig. 4b shows the results of this study. As expected, DM2 patients express higher levels (from 35 to 52%) of INSR-A, MBNL1ex7, and CLCN1ex7a isoforms compared to the control (values comprised between 8 and 14%). In muscle from patient 1 the INSR-A/TOT ratio was 41 ± 8%, MBNLex7/TOT ratio was 35 ± 5%, and 37 ± 8% of CLCN1-expressed isoforms contained exon 7a (CLCN1ex7a). In the mother’s muscle tissue the expression profile was slightly more severe and was characterized by INSR-A, MBNLex7, and CLCN1ex7a ratios of 52 ± 5%, 44 ± 7%, and 50 ± 8%, respectively. QRT-PCR analysis to quantify the expression levels of the CLCN1 transcripts showed a considerable downregulation of this gene in the DM2 patients compared to controls. Levels of CLCN1 mRNA were lowered to 45 ± 3% and 38 ± 2% in patient 1 and patient 2 samples, respectively, relative to controls (n = 2, data not shown). These results are in agreement with the previously observed reduction of the CLC-1 protein levels in both DM2 patient biopsies (Fig. 2).
Discussion
In this study we have characterized a 15-year-old DM2 patient and her mother at the clinical, histopathological, genetic, and molecular level in order to further investigate the genotype-phenotype correlation and the unusually young onset in this DM2 family.
Although the age at onset was earlier in the daughter than in the mother, the daughter’s clinical, histopathological, and biomolecular findings did not show greater severity than those observed in her mother. Patient 1 (daughter) presented handgrip myotonia at the age of 14 years. Her muscle biopsy showed no DM2 histopathological features, i.e., no internalized nuclei, type 2 atrophic fibers, or nuclear clump fibers were found in muscle sections. On the contrary, her mother presented clinical and muscle histopathological features typical of DM2. Her myotonia symptoms started around the age of 20 years, whereas the proximal muscle weakness started after age 30. The clinical and histopathological findings showed a more severe phenotype in the mother than in her daughter. This could be explained mainly by the age difference: the daughter (15 years old) has no weakness and a mild histopathological pattern, and when she reaches the age of 45, she could display, like her mother, significant weakness and a significantly altered histopathological pattern. Therefore, in conclusion, the histological and immunohistochemical findings correlated with the disease severity and age at onset in both patients. Southern blot analysis revealed that the CCTG repetition length was larger in skeletal muscle than in blood, especially in the daughter’s tissues, where the DM2 mutation expanded from about 2.000 (the most represented allele in blood) up to 6.000 (CCTG)s (upper alleles in muscle). Comparison of the CCTG repeat number between the daughter’s and mother’s blood and muscle tissues showed only small variation of the DM2 mutation (less than 1.000 repetitions). We can then conclude that the earlier onset observed in the daughter was not linked to a significant increase in the number of CCTG repeats through maternal germline transmission. However, a higher grade of somatic instability seemed to occur in the daughter’s tissues, but the overall cause and biological significance of this observation have yet to be determined. The area and fluorescence intensity of both ribonuclear inclusions and MBNL1 foci appear to be similar in the two patients, suggesting that a similar amount of MBNL1 protein is sequestered in ribonuclear foci.
As expected, INSR, ClCN1, and MBNL1 splicing patterns were altered in both patients with the level of aberrant splicing isoforms slightly lower in the daughter than in the mother. Co-segregation of DM2 mutation and recessive CLCN1 mutations have been shown to occur and modify the DM2 phenotype [18]. The frequency of heterozygous recessive co-segregating CLCN1 mutations in DM2 patients from Finland and Germany is higher than in the controls population [18]. However, this frequency was calculated based not on the total DM2 patient population, but on patients with a molecular diagnosis of DM2, possibly indicating a selection bias in molecular diagnostic referral. It is therefore possible that a large number of DM2 patients without the CLCN1 mutation may remain undiagnosed because of a milder phenotype and that the frequency of recessive CLCN1 mutations would be the same as in the general population. Generally, affected carriers showed more severe muscle stiffness and more severe clinical EMG than those having exclusively the CNBP expansion; hence the CLCN1 mutation may contribute to exaggerating the DM2 phenotype. The co-segregating F167L mutation, found in this family was a C–G transition in CLCN1 exon 4 (position 501) causing an amino acid change in the D2 domain of the protein. This mutation has been described in association with the autosomal recessive form of myotonia congenita and so without a mutation on the other CLCN1 allele would not be expected to cause a disease phenotype. However, functional studies in heterologous expression systems have proven that the effect of this mutation is a slight shift in the Cl potential [33]. Such a background would have been one plausible explanation for the early onset of myotonia in both the mother and in the daughter because DM2 patients already show loss of the chloride channel protein caused by the abnormal splicing of the CLCN1 pre-mRNA, which leads to nonsense-mediated decoys of transcripts containing premature termination codons [32, 34]. Accordingly, the expression levels of the CLCN1 gene are reduced in DM2 muscles, causing a decreased chloride conductance and electrical instability. The analysis of CLCN1 mRNA and protein expression levels in our two DM2 patients demonstrated either CLCN1 missplicing or downregulation. Given that abnormal splicing will affect both the normal and the mutant CLCN1 alleles, the consequent loss of functional CLC-1 resulting from the additive effects of CLCN1 missplicing and mutation will be even more marked than in patients with either the DM2 or CLCN1 mutation alone.
Although both patients were proven to be heterozygous for the c.501C>G mutation in the CLCN1 gene by genomic sequencing of the exons and their boundaries, knowing the frequent occurrence of splicing mutations in the gene, the possibility of co-segregating recessive mutations has still not been fully excluded.
The clinical, histopathological, and biomolecular characterization of the juvenile onset DM2 family presented in this study is helpful to clarify the similarities and distinctions between DM1 and DM2 and therefore better understand the pathogenetic mechanisms of these diseases. Moreover, this study demonstrates that when clinical features are uncommon, additional genes and/or modifying factors need to be explored to account for the phenotype. In this case, the co-segregation of DM2 with a recessive CLCN1 mutation provided the explanation for the unusual clinical findings.
References
Harper PS (2001) Myotonic dystrophy. In: Karpati G, Hilton-Jones D, Griggs RC (eds) Disorders of voluntary muscle. University Press, Cambridge, pp 541–559
Brook JD, McCurrach ME, Harley HG et al (1999) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 69:799–808
Fu YH, Pizzuti A, Fenwick RG Jr et al (1992) An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255:1256–1258
Mahadevan M, Tsilfidis C, Sabourin L et al (1992) Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science 255:1253–1255
Ranum LPW, Rasmussen P, Benzow K et al (1999) Genetic mapping of a second myotonic dystrophy locus. Nat Genet 19:196–198
Liquori CL, Ricker K, Moseley ML et al (2001) Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293:816–817
Kuyumcu-Martinez NM, Cooper TA (2006) Misregulation of alternative splicing causes pathogenesis in myotonic dystrophy. Prog Mol Subcell Biol 44:133–159
Osborne J, Thornton CA (2006) RNA-dominant diseases. Hum Mol Genet 15:R162–R169
Meola G, Cardani R (2009) RNA binding proteins in myotonic dystrophies. In: Denman RB (ed) RNA binding proteins in development and disease. Research Signpost, Kerala, pp 153–166
Meola G, Moxley RT (2004) Myotonic dystrophy type 2 and related myotonic disorders. J Neurol 251:1173–1182
Vihola A, Bassez G, Meola G et al (2003) Histopathological differences of myotonic dystrophy type 1 (DM1) and PROMM/DM2. Neurology 60:1854–1857
Bassez G, Chapoy E, Bastuji-Garin S et al (2008) Type 2 myotonic dystrophy can be predicted by the combination of type 2 muscle fiber central nucleation and scattered atrophy. J Neuropathol Exp Neurol 67:319–325
Pisani V, Panico MB, Terracciano C et al (2008) Preferential central nucleation of type 2 myofibers is an invariable feature of myotonic dystrophy type 2. Muscle Nerve 38:1405–1411
Tsilfidis C, MacKenzie AE, Mettler G, Barcelo J, Korneluk RG (1992) Correlation between CTG trinucleotide repeat length and frequency of severe congenital myotonic dystrophy. Nat Genet 1:192–195
Harper PS, Harley HG, Reardon W et al (1992) Review article: anticipation in myotonic dystrophy: new light on an old problem. Am J Genet 51:10–16
Schoser BGH, Kress W, Walter MC, Halliger-Keller B, ller HL, Ricker K (2004) Homozygosity for CCTG mutation in myotonic dystrophy type 2. Brain 127:1868–1877
Lamont PJ, Jacob RL, Mastaglia FL et al (2004) An expansion in the ZNF9 gene causes PROMM in a previously described family with an incidental CLCN1 mutation. J Neurol Neurosurg Psychiatry 75:343
Suominen T, Schoser B, Raheem O et al (2008) High frequency of co-segregating CLCN1 mutations among myotonic dystrophy type 2 patients from Finland and Germany. J Neurol 255:1731–1736
Sun C, Van Ghelue M, Tranebjaerg L et al (2011) Myotonia congenita and myotonic dystrophy in the same family: coexistence of a CLCN1 mutation and expansion in the CNBP (ZNF9) gene. Clin Genet 80:574–580
Dubowitz V (1985) Muscle biopsy. In: Dubowitz V (ed) A practical approach. Bailliere Tindall, London
Cardani R, Mancinelli E, Sansone V et al (2004) Biomolecular identification of (CCTG)n mutation in myotonic dystrophy type 2 (DM2) by FISH on muscle biopsy. Eur J Histochem 48:437–442
Boi S, Fascio U (1998) A method of quantitative measurement of fluorescence intensity by confocal laser scanning microscopy. J Comput Assist Microsc 10:163–166
Bonifazi E, Vallo L, Giardina E et al (2004) A long PCR-based molecular protocol for detecting normal and expanded ZNF9 alleles in myotonic dystrophy type 2. Diagn Mol Pathol 13:164–166
Nakamori M, Sobczak K, Moxley RT 3rd et al (2009) Scaled-down genetic analysis of myotonic dystrophy type 1 and type 2. Neuromuscul Disord 19:759–762
Lehmann-Horn F, Mailänder V, Heine R et al (1995) Myotonia levior is a chloride channel disorder. Hum Mol Genet 4:1397–1402
Botta A, Rinaldi F, Catalli C et al (2008) The CTG repeat expansion size correlates with the splicing defects observed in muscles from myotonic dystrophy type 1 patients. J Med Genet 45:639–646
Botta A, Bonifazi E, Vallo L et al (2006) Italian guidelines for molecular analysis in myotonic dystrophies. Acta Myol 25:23–33
Pusch M (2002) Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum Mutat 19:423–434
Kanadia RN, Shin J, Yuan Y et al (2006) Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci 103:11748–11753
Savkur RS, Philips AV, Cooper TA (2001) Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet 29:40–47
Savkur RS, Philips AV, Cooper TA et al (2004) Insulin receptor splicing alteration in myotonic dystrophy type 2. Am J Hum Genet 74:1309–1313
Mankodi A, Takahashi MP, Jiang H et al (2002) Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell 10:35–44
Zhang J, Bendahhou S, Sanguinetti MC et al (2000) Functional consequences of chloride channel gene (CLCN1) mutations causing myotonia congenita. Neurology 54:937–942
Charlet BN, Savkur RS, Singh G et al (2002) Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell 10:45–53
Acknowledgment
This work was supported by Grant AFM-MYOTON to Giovanni Meola.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cardani, R., Giagnacovo, M., Botta, A. et al. Co-segregation of DM2 with a recessive CLCN1 mutation in juvenile onset of myotonic dystrophy type 2. J Neurol 259, 2090–2099 (2012). https://doi.org/10.1007/s00415-012-6462-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-012-6462-1