Abstract
Familial primary erythromelalgia is a rare autosomal dominant disease characterized by redness and painful episodes of the feet and hands, which is often triggered by heat or exercise. In this report, a Taiwanese family with the characteristic features of erythromelalgia is described. Genetic linkage studies established that the disease locus maps to human chromosome 2. Sequence analysis indicated that the disease segregates with a novel mutation in the alpha subunit of the voltage-gated sodium channel (SCN9A or Nav1.7). The change observed is predicted to cause the substitution of a highly conserved isoluecine 136 for a valine within the first segment of the transmembrane domain (D1S1). Using immuno-histochemistry to stain a skin biopsy specimen from the affected region, we demonstrate that there is a significant reduction in the number of small fibers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Familial primary erythromelalgia (OMIM: 133020) is a rare autosomal dominant disorder mapping to human chromosome 2q24 [5, 12]. It is characterized by intermittent redness associated with burning pain in the extremities, particularly the hands and feet. Episodes are trigged by warming stimuli or exercise and can be relieved by cooling or raising the affected limbs. Other treatments, such as drug therapy, have met with varying degrees of success [2, 8].
In addition to the overt clinical features of the disease, individuals display an elevation in local skin temperature of the affected region associated with a significant increase in blood flow without significant changes in oxygenation of the tissue [3]. Autonomic reflex responses (e.g., the ability to produce sweat) are often abnormal in individuals with the disease [9]. The pathophysiological basis for the disease is poorly understood as the affected area appears to have both vascular and neural involvement [3].
Recent studies have identified mutations in the alpha subunit of the voltage-gated sodium channel type IX gene (SCN9A or Nav1.7) associated with the disease that have an electrophysiological impact on channel function [4, 6, 12]. This discovery should lead to greater understanding of the disease process and the development of more effective treatments strategies.
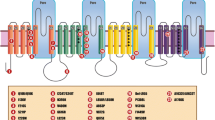
In this report we describe a three-generation Taiwanese family with primary erythromelalgia and characterize the clinical and pathological features. We demonstrate that the disease is inherited in an autosomal dominant manner with the disease locus mapping to chromosome 2. Furthermore, we establish that the disease is associated with a predicted I136V mutation in the SCN9A affecting the first transmembrane domain of the protein (D1S1).
Subjects and methods
Subjects and controls
The proband (IV:4, Figure 1a), a 21 year old man, presented with paroxysmal redness of both feet and hands associated with pain and a burning sensation upon heat stimulation. He described the pain as a tingling sensation similar to that of an electric-shock. Whilst the feet were affected from the age of 11 years, the hands became affected with milder symptoms when he was about 19 years of age. Hot weather, exercise, increased body temperature and wearing shoes would trigger attacks or make them worse. Cooling the affected area by immersion in cold water tended to relieve the pain. The frequency of attacks increased from periodic episodes to 2 or 3 a day with some lasting the whole day. Apart from the aforementioned condition, no other clinical symptoms or signs were detected upon examinations.
Family Pedigree and Genetic Linkage data. a) Pedigree showing the autosomal dominant pattern of inheritance of the disease gene through the family and its segregation with marker loci mapping to chromosome 2q24. The index case is identified with an arrow. Filled symbols represent affected individuals (square = male, circle = female, diamond = sex unknown, slash = deceased). Genetic loci are arranged according to their physical position on the chromosome and the alleles segregating in the family are identified according to their size (base pairs, bp). The haplotype associated with the disease gene is identified with a solid bar. b) HpyCH4III restriction length polymorphism (RFLP) analysis for the family identifying the I136V mutation in SCN9A gene. The wild-type allele is identified by fragment sizes of 264 and 15 bp (not seen), the mutant allele by fragments of 204, 60 and 15 bp (not seen). The segregation of the mutant allele is concordant with the disease gene in this family. c) CLUSTLAW analysis of the peptide sequence in the region of the mutation indicated that Isoleucine at amino acid 136 is conversed within species for different members of the sodium channel family
Members of the proband’s immediate family presented with similar symptoms as did other members of the expanded family through a three/four generation pedigree (Figure 1a) suggesting an autosomal dominant mode of inheritance. Following established ethical codes of practice approved by the local Ethics Committee and involving informed consent, blood samples for DNA extraction were taken from five affected and four unaffected family members and subjected to genetic analysis. Control samples were obtained from a random panel of DNA samples collected for this purpose.
Genetic and mutational analysis
DNA was extracted from blood using standard extraction techniques. Linkage analysis was confined to chromosome 2q24.2-2q24.3 as defined by microsatellite loci D2S237, D2S382, D2S2363, D2S2330, D2S1395 and D2S2345. Fluorescent-labeled primer pairs for each locus were used for PCR amplification of the sample DNA employing the condition recommended by the manufacturer (AmpliTaq Gold® DNA Polymerase, Applied Biosystems, Foster City, CA, USA). Resolution of the amplification product was performed on a Beckman Coulter CEQ8000™ automated DNA sequencer (Beckman Coulter Inc., Fullerton, CA, USA) and analysed using software supplied by Beckman Coulter.
SCN9A gene sequence (GenBank: NM_002977) analysis employed exon primers and amplification condition described by Yang and others (2004) [12]. The amplification product was purified, sequenced using BigDye® sequencing chemistry (Applied Biosystems) and analysed on an automated ABI3100™ DNA sequencer (Applied Biosystems). Additional genetic analysis of the SCN9A gene involved the use of a HypCH4III restriction site polymorphism. HypCH4III was used according to the manufacturer’s recommendation (New England Biolabs, Ipswich, MA, USA) to digest PCR product, which was resolved by agarose gel electrophoresis, stained with ethidium bromide and visualized by UV trans-illumination.
Immuno-histopathology
Following established ethical procedures and protocols approved by the Ethics Committee of the National Taiwan University Hospital, a 3 mm diameter skin punch biopsy specimen was taken from the affected region and processed for immuno-histopathology using standard protocols. Nerve fibres were identified using rabbit antibody to protein gene product 9.5 (PGP9.5 UltraClone, Isle of White, UK) and visualized using the Vector® SG peroxidase kit with avidin-biotin complex and biotinylated goat anti-rabbit immunoglobulin (Vector, Burlingame, CA, USA) counter stained with eosin.
The number of nerve fibres/mm epidermal length in each section was counted at 40x magnification on a BX40 Olympus microscope. Epidermal length was measured using Image-Pro Plus software (Media Cybernetics, Silver Spring, MD, USA).
Results
Genetic analysis
As the clinical features and the family history fulfilled the diagnostic criteria for familial primary erythromelalgia, we decided to ascertain whether the disease locus mapped to human chromosome 2q24.2–24.3. Six markers (D2S2370, D2S382, D2S2363, D2S2330, D2S1395 and D2S2345) spanning a 9.3 Mb region around the candidate gene SCN9A were selected and family members genotyped for the segregation of alleles at these loci. The analysis showed that affected family members shared a common haplotype on chromosome 2q24.2-24.3 that was not present in unaffected members (Figure 1a). These findings are not incompatible with the linkage to 2q24.2-24.3 suggesting that a mutation in SCN9A might be the cause of the disease in this family.
Direct sequencing of the SCN9A gene in the proband identified a novel A→G transversion at nucleotide 406 of the coding sequence (reference sequence GeneBank NM_002977) encoded by exon 3. This is predicted to cause the substitution of an isoluecine for a valine at amino acid 136. Isoluecine at 136 is conserved across mammalian species for this gene and among other members of this sodium channel family within species (Figure 1c).
The “A” to “G” transversion at nucleotide 406 creates a new restriction site for the restriction endonuclease HpyCH4 III. Thus, we were able to use this change to determine whether the mutation segregated with the disease haplotype in the other members of the family. Sample DNA from each family member was amplified by PCR with primers to exon 3. The resultant amplification product was digested with HypCH4III and analysed on an agarose gel (Figure 1b). The normal allele generates two fragments of 264 and 15bp (not seen in Figure 1b) whereas the mutant allele generates three fragments of 204, 60 and 15bp (again the latter is beyond the resolution of the gel in Figure 1b). As figure 1b shows, the mutation in SCN9A segregates with the disease gene. Furthermore, this change was not observed in 100 normal Taiwanese control samples suggesting that it is not a normal polymorphism within the population.
Immuno-histopathology
Using antibody (PGP9.5) that identify the small nerve fibers, we observed that the patient’s epidermis showed a marked reduction in the number of small nerve fibres with some denervation of the sweat glands (Figure 2a) compare with the control sample (Figure 2b). Epidermal nerve density (END) in the patient was estimated to be 0.44 ± 0.07 fibres/mm (mean ± s.d.) compared with 11.16 ± 3.7, 5.8, 4.2 fibres/mm (mean ± s.d., the fifth and first percentile values) for normal individuals less than 60 years of age (t(10) = 2.897, p < 0.05). The Congo red staining of the skin was negative and no inflammatory process was identified in the skin pathology.
Immuno-histochemical staining of the skin for the presence of protein gene product (PGP) 9.5 identifying small nerve fibres. a) Biopsy sample from the affected region of the patient. Inset, an enlarged portion of the small nerve fibre (arrow). b) A similar region from a control sample. The number of PGP9.5 positive small nerve fibres is significantly reduced in the sample taken from the patient compared with the control sample. Scale bar = 50 μM
Discussion
In this report, we describe a three-generation Taiwanese family with symptoms characteristic of autosomal dominant primary erythromelalgia. We established that the disease gene segregates with a specific haplotype described by markers that flank the SCN9A gene, which is known to be responsible in other cases of primary erythromelalgia [5, 6]. Sequences and restriction length polymorphism (RFLP) analysis of SCN9A revealed that the disease segregated with a novel A→G transversion at nucleotide 406 of the coding sequence. This changed, encoded by exon 3, is predicted to cause the replacement of a highly conserved isoluecine at amino acid 136 for a valine located within the first segment of the transmembrane domain (D1S1). As this change was not detected in 100 population control samples, we concluded that it is likely to be the pathological cause of the disease in our family.
Why mutations in the SCN9A gene cause the symptoms characteristic of primary erythromelalgia is unclear. However, in vitro expression studies of other SCN9A mutations (F1449V, I848T and L858H) demonstrated that they change the action potential of the channel leading to a potential gain of function [1, 4, 7, 11]. Whether the mutation observed in our family has a similar electro-physiological effect remains to be established. None-the-less, our histo-pathological studies on skin from the affected region and those of others [10], demonstrates that there is a significant reduction in the number of small nerve fibres associated with autonomic function such as sweating, shivering and vasodilatation. Thus, it is reasonable to speculate that the cause of small fibre degeneration could be the result of milieu change around the terminal axons or an excessive discharge caused by the mutant sodium channel. It also seems responsible to speculate that the use of sodium channel blockers might be a reasonable adjunct in the treatment of this disease.
References
Cummins TR, Dib-Hajj SD, Waxman SG (2004) Electrophysiological properties of mutant Nav1.7 sodium channels in a painful inherited neuropathy. J Neurosci 24:8232-8236
Davis MD, Sandroni P (2002) Lidocaine patch for pain of erythromelalgia. Arch Dermatol 138:17–19
Davis MD, Sandroni P, Rooke TW, Low PA (2003) Erythromelalgia: vasculopathy, neuropathy, or both? A prospective study of vascular and neurophysiologic studies in erythromelalgia. Arch Dermatol 139:1337–1343
Dib-Hajj SD, Rush AM, Cummins TR, Hisama FM, Novella S, Tyrrell L, Marshall L, Waxman SG (2005) Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain 128:1847–1854
Drenth JP, Finley WH, Breedveld GJ, Testers L, Michiels JJ, Guillet G, Taieb A, Kirby RL, Heutink P (2001) The primary erythermalgia-susceptibility gene is located on chromosome 2q31–32. Am J Hum Genet 68:1277–1282
Drenth JP, te Morsche RH, Guillet G, Taieb A, Kirby RL, Jansen JB (2005) SCN9A mutations define primary erythermalgia as a neuropathic disorder of voltage gated sodium channels. J Invest Dermatol 124:1333–1338
Han C, Rush AM, Dib-Hajj SD, Li S, Xu Z, Wang Y, Tyrrell L, Wang X, Yang Y, Waxman SG (2006) Sporadic onset of erythermalgia: A gain-of-function mutation in Na(v)1.7. Ann Neurol 59:553–558
Kuhnert SM, Phillips WJ, Davis MD (1999) Lidocaine and mexiletine therapy for erythromelalgia. Arch Dermatol 135:1447–1449
Mork C, Kalgaard OM, Kvernebo K (2002) Impaired neurogenic control of skin perfusion in erythromelalgia. J Invest Dermatol 118:699–703
Uno H, Parker F (1983) Autonomic innervation of the skin in primary erythermalgia. Arch Dermatol 119:65–71
Waxman SG, Dib-Hajj SD (2005) Erythromelalgia: A hereditary pain syndrome enters the molecular era. Ann Neurol 57:785–788
Yang Y, Wang Y, Li S, Xu Z, Li H, Ma L, Fan J, Bu D, Liu B, Fan Z, Wu G, Jin J, Ding B, Zhu X, Shen Y (2004) Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet 41:171–174
Acknowledgements
We thank Dr. James Hill for his comments during the preparation of the manuscript. These experiments were supported by a grant (No. NTUH 92A03-5) from the National Taiwan University Hospital. DAS is grateful for finical support of the MRC UK.
Author information
Authors and Affiliations
Corresponding author
Additional information
Received in revised form: 31 March 2006
Rights and permissions
About this article
Cite this article
Lee, MJ., Yu, HS., Hsieh, ST. et al. Characterization of a familial case with primary erythromelalgia from Taiwan. J Neurol 254, 210–214 (2007). https://doi.org/10.1007/s00415-006-0328-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-006-0328-3