Abstract
Identification of body fluids found at crime scenes provides important information that can support a link between sample donors and actual criminal acts. Previous studies have reported that DNA methylation analysis at several tissue-specific differentially methylated regions (tDMRs) enables successful identification of semen, and the detection of certain bacterial DNA can allow for identification of saliva and vaginal fluid. In the present study, a method for detecting bacterial DNA was integrated into a previously reported multiplex methylation-sensitive restriction enzyme-polymerase chain reaction. The developed multiplex PCR was modified by the addition of a new semen-specific marker and by including amplicons for the 16S ribosomal RNA gene of saliva- and vaginal fluid-specific bacteria to improve the efficacy to detect a specific type of body fluid. Using the developed multiplex system, semen was distinguishable by unmethylation at the USP49, DACT1, and PFN3 tDMRs and by hypermethylation at L81528, and saliva could be identified by detection of saliva-specific bacteria, Veillonella atypica and/or Streptococcus salivarius. Additionally, vaginal fluid and menstrual blood were differentiated from other body fluids by hypomethylation at the PFN3 tDMR and the presence of vaginal fluid-specific bacteria, Lactobacillus crispatus and/or Lactobacillus gasseri. Because the developed multiplex system uses the same biological source of DNA for individual identification profiling and simultaneously analyses various types of body fluid in one PCR reaction, this method will facilitate more efficient body fluid identification in forensic casework.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Body fluids found at a crime scene are one of the most important pieces of forensic evidence that can provide valuable information to identify suspects or victims and also can provide important information supporting a link between sample donors and actual criminal acts [1].
Various methods to identify the type of tissues or body fluids have been continually developed in forensic science. Catalytic, enzymatic and immunologic tests have generally been used [2], and advances in forensic genetics have led to the development of methods that detect tissue-specific messenger RNA or micro RNA expression [3–5]. Recently, DNA-based body fluid identification has begun to receive attention [6, 7]. Because DNA has mainly been used as a biological source for personal identification profiling, DNA-based body fluid identification seems promising. Two DNA-based body fluid identification methods have been reported: a DNA methylation-based method [6–11] and a bacterial DNA-based method [12–17].
DNA methylation affects gene expression regulation without DNA sequence changes. Different cell types have different DNA methylation patterns [18]. Specifically, chromosome segments called tissue-specific differentially methylated regions (tDMRs) show different DNA methylation profiles according to cell or tissue type [19–21]. Body fluid identification by analysing the DNA methylation status of these regions has been reported as a new method in forensic science [6, 7]. An et al. [8] described a body fluid identification method using methylation-sensitive restriction enzyme-polymerase chain reaction (MSRE-PCR) and methylation SNaPshot, which can identify semen and can distinguish vaginal fluid and menstrual blood from blood and saliva. In addition, the Nucleix DSI-Semen kit (Nucleix, Tel Aviv, Israel) is a recently developed commercial kit that makes it possible to distinguish semen from other body fluids with a small amount of DNA using MSRE-PCR [9, 10]. These methods can be used to analyse aged samples by applying a small amplicon strategy and are compatible with commercially available human identity testing kits and instrumentation [8–10]. However, DNA methylation-based analysis is in its early stage of development and thus needs to be improved such that it can utilize more body fluid-specific DMRs for detailed and reliable body fluid identification.
Another DNA-based method to identify the type of body fluids uses the DNA of bacteria that are present in a specific body fluid by amplifying bacterial species-specific regions of the 16S ribosomal RNA (rRNA) gene or the 16S-23S rRNA intergenic spacer region [12–17]. In particular, amplification of hypervariable regions in the 16S rRNA gene can facilitate detection of tissue-specific bacteria [22, 23]. Forensic body fluid identification by detection of bacterial DNA has focused primarily on saliva and vaginal fluid. Saliva can be identified by the presence of Streptococcus salivarius and/or Streptococcus mutans [12–14], while vaginal fluid can be distinguished from other body fluids based on detection of Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners and/or Lactobacillus jensenii [15–17]. In addition, Gardnerella vaginalis and Atopobium vaginae have been suggested as useful for vaginal fluid identification because they are associated with bacterial vaginosis, the most frequent vaginal infection in fertile women [15, 23]. However, this method is limited in that it can be used to distinguish among only a few types of body fluids containing bacteria.
The goal of the present study was to integrate DNA methylation-based analysis and bacterial DNA-based analysis to improve the efficacy of existing DNA-based body fluid identification methods. We expect integration of the two methods in a single PCR reaction will allow more efficient discrimination of body fluids since the two methods complement each other.
Materials and methods
Samples
Blood, saliva and semen from 20 males and vaginal fluid and menstrual blood from 14 female volunteers were collected using procedures approved by the Institutional Review Board of Severance Hospital, Yonsei University in Seoul, Korea. The 34 donors provided written informed consent after the goals and procedures of the study were explained. During sample collection, it was determined that 2 of the 20 male volunteers had undergone a vasectomy. Blood was collected in a syringe, and 200 μL aliquots were stored frozen. Saliva and freshly ejaculated semen were collected in a microcentrifuge tube and plastic cups, respectively, and 200 μL aliquots of each were stored frozen. Vaginal fluid and menstrual blood were collected using sterile cotton swabs and allowed to dry at room temperature. Dried swabs were stored frozen until use.
DNA was extracted from each aliquot of blood, saliva and semen using a QIAamp® DNA Mini Kit (QIAGEN, Hilden, Germany) and from each swab of vaginal fluid and menstrual blood using a QIAamp® DNA Investigator Kit (QIAGEN) following the manufacturer's instructions. Extracted DNA was quantified using a Quantifiler® Duo DNA Quantification Kit (Applied Biosystems, Foster City, CA, USA).
Selection of markers and primer design for the development of multiplex PCR
For DNA methylation-based body fluid identification, two tDMRs for the USP49 and DACT1 genes were selected as a semen-specific unmethylation markers and the PFN3 tDMR was selected as a vaginal fluid and menstrual blood-specific hypomethylation marker [7, 8]. A previously reported semen-specific methylation marker, L81528 [6], was also added for more accurate identification of semen. To determine the methylation status of these four tDMRs, primers flanking the Hha I recognition sites (GCGC) of the four tDMRs were designed using the Primer 3 program (http://frodo.wi.mit.edu/primer3/). The amplicon sizes were smaller than 150 bp and one forward or reverse primer was labelled with the fluorescent dye FAM (Table 1).
For body fluid identification based on the detection of bacterial DNA, Streptococcus salivarius [12–14] and Veillonella atypica [24] were selected as saliva-specific bacteria, and L. crispatus and L. gasseri [15–17] were selected as vaginal fluid-specific bacteria. Primers for the amplification of body fluid-specific bacterial DNA were designed to target species-specific regions of the 16S rRNA gene using the Primer 3 program and the probe match program of the Ribosomal Database Project (http://rdp.cme.msu.edu/). The amplicon sizes were 98–127 bp, and one forward or reverse primer was labelled with the fluorescent dye TAMRA.
Primers for the amplification of amelogenin, D3S1358, an amplification control, and a digestion control were also designed using the Primer 3 program and were labelled with FAM or HEX fluorescent dye. The artificial DNA templates of the amplification and digestion controls for PCR success and restriction enzyme digestion, respectively, were obtained by PCR amplification of the 481-bp portion of the pCR®2.1-TOPO® vector (Invitrogen, Carlsbad, CA, USA) [8].
Restriction enzyme treatment and multiplex PCR
Before conducting multiplex PCR, 1 ng of genomic DNA was digested with Hha I in a 10-μL reaction containing 10 U of Hha I (New England Biolabs, Ipswich, MA, USA), 1 μL of Gold ST*R 10× Buffer (Promega, Madison, WI, USA) and 1 μL of artificial DNA template (equivalent to 1 ng of genomic DNA) at 37 °C for 30 min, and then the enzyme was heat-inactivated by incubation at 65 °C for 20 min. Multiplex PCR was carried out in a 20-μL reaction volume containing 10 μL of enzyme-digested DNA, 2.0 U of AmpliTaq Gold® DNA Polymerase (Applied Biosystems), 1 μL of Gold ST*R 10× Buffer, 5 % dimethyl sulphoxide (Sigma-Aldrich Inc., St. Louis, MO, USA) and 0.12–1.0 μM of each primer (Table 1). PCR cycling was conducted on a PTC-200 DNA engine (MJ Research, Waltham, MA, USA) under the following conditions: 95 °C for 11 min; 28 cycles of 94 °C for 20 s, 59 °C for 60 s, and 72 °C for 30 s; and a final extension at 60 °C for 60 min. One microlitre of amplification product in a mix containing 20 μL HiDi formamide (Applied Biosystems) and 0.2 μL LIZ-500 size standard (Applied Biosystems) was denatured by incubation at 95 °C for 5 min immediately followed by 3 min on ice, and run on an ABI 310 Genetic Analyzer (Applied Biosystems) according to the manufacturer's instructions. All resulting electropherograms were analysed using GeneMapper® ID software v.3.2 (Applied Biosystems). The threshold for a positive signal was set to 100 relative fluorescent units.
Sensitivity test and analyses of aged samples, mixtures and artificial casework samples
Sensitivity testing was carried out using a series of diluted DNA (1 ng, 500 pg, 250 pg, 125 pg, 62 pg, 31 pg and 15 pg) derived from saliva, semen and vaginal fluid samples from two volunteers per body fluid. One microlitre of each concentration was tested in triplicate to evaluate the minimum quantity required to obtain the predictable body fluid profile.
DNA obtained from body fluid samples that had been exposed to ambient temperature for 75 days in the previous report [8] were used as an aged sample test of our detection method. These samples were analysed under the same conditions except for the use of 29 amplification cycles.
In addition, vaginal swabs spotted with 5 or 10 μL of semen and with 5 or 10 μL of saliva, and spots of saliva and semen mixed at ratios of 1:1 and 1:2 (i.e. 10 μL saliva and 10 μL semen, and 10 μL saliva and 20 μL semen) on sterile cotton tissues were left to dry overnight for a mixture test.
For a casework validation, some typical artificial casework samples were collected from anonymous contributors. Three post-coital vaginal samples and a post-coital penile sample were obtained with a sterile cotton swab or clean tissue paper, and a skin swab sample was obtained from a kissing site.
DNA from all aged samples, mixtures and artificial casework samples were extracted using a QIAamp® DNA Investigator Kit (QIAGEN) following the manufacturer's instructions. In some cases which have mixed semen and vaginal secretions, differential DNA extraction protocol provided in the QIAamp® DNA Investigator Kit (QIAGEN) was also used.
Results and discussion
Multiplex PCR system development
To facilitate body fluid identification using DNA, a multiplex PCR system was developed by integrating the method that detects DNA methylation status at several tDMRs and the method that checks for the presence of body fluid-specific bacteria.
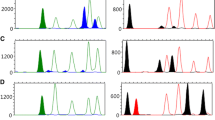
The developed multiplex PCR system comprised 12 amplicons containing four tDMRs and four bacterial DNA targets, and each amplicon was labelled with FAM, TAMRA or HEX. The FAM fluorescence signal represented amelogenin typing results and DNA methylation status at the four tDMRs (USP49, DACT1, L81528 and PFN3) to identify semen and to differentiate vaginal fluid and menstrual blood from blood and saliva. The TAMRA fluorescence signal represented the presence of L. crispatus or L. gasseri in vaginal fluid and the presence of Streptococcus salivarius or V. atypica in saliva, and the HEX fluorescence signal represented the D3S1358 typing results and amplification of controls providing confirmation of PCR success and complete restriction enzyme digestion (Fig. 1).
This multiplex PCR system, which is based on MSRE-PCR, has some improvements in comparison with previously reported methods because it allows simultaneous detection of bacterial DNA with human genomic DNA. In order to integrate the bacterial DNA detection method with DNA methylation analysis using MSRE-PCR, differences in the methylation sites of human genomic DNA and bacterial DNA were considered. Human DNA methylation generally occurs at the C5 position of cytosines that are followed by a guanine while bacterial DNA methylation occurs at the N6 position of adenines in the sequence 5′-GATC-3′ and at the C5 position of the cytosines in the sequences CCAGG and CCTGG [25, 26]. Therefore, the MSRE Hha I recognition site, GCGC, is always unmethylated in bacterial DNA. In the developed multiplex system, however, bacterial DNA segments not containing the Hha I restriction site were targeted and thereby the amplification of bacterial DNA was not influenced by MSRE treatment.
In comparison with the MSRE-PCR reported by An et al. [8], this multiplex PCR system facilitates definite identification of semen including sperm cells. Even with semen samples showing a low methylation signal at the DACT1 and USP49 tDMRs, semen identification could be carried out by amplification of the semen-specific methylation marker, L81528. In addition, this newly developed multiplex PCR system is different from previously reported bacterial DNA-based methods [12–17] because of the use of V. atypica instead of Streptococcus mutans as saliva-specific bacteria. In fact, Streptococcus mutans DNA is generally obtained using a Gram-positive bacterial DNA extraction method involving dithiothreitol, and the detection of Streptococcus mutans DNA is almost impossible using DNA extraction methods that are commonly used in forensic laboratories (data not shown). Therefore, V. atypica was substituted for Streptococcus mutans for the identification of saliva because its DNA can be easily extracted with commonly used genomic DNA extraction methods [24].
Body fluid identification with the developed multiplex PCR
The developed multiplex PCR was successfully used to differentiate between blood, saliva, semen and vaginal fluid–menstrual blood obtained from 20 males and 14 females. Through DNA methylation analysis, this multiplex PCR system could classify samples as blood–saliva, semen or vaginal fluid–menstrual blood. Bacterial DNA analysis was used to further distinguish saliva and vaginal fluid–menstrual blood (Fig. 2). DNA obtained from blood and saliva produced amplicons for three tDMRs, USP49, DACT1 and PFN3, and then saliva was further differentiated by amplification of saliva-specific bacterial DNA from Streptococcus salivarius and/or V. atypica. DNA extracted from vaginal fluid and menstrual blood also produced amplicons for the USP49, DACT1 and PFN3 tDMRs, but with much lower yield at the PFN3 tDMR. Moreover, bacterial DNA amplification of L. crispatus and/or L. gasseri confirmed the presence of vaginal fluid or menstrual blood. None of the semen samples showed an amplicon for bacterial DNA, but unlike the other body fluids, the representative results of the DNA methylation profile of semen appeared in three patterns (Fig. S1). Ten of the eighteen samples from non-vasectomized males produced an amplicon only at the L81528 tDMR. The other eight semen samples gave very low yields but detectable amplicons at the three semen-specific unmethylation markers, USP49, DACT1 and PFN3 tDMRs in addition to the L81528 amplicon. Nonetheless, semen identification was not problematic because of the high amplification yield of L81528 in all 18 semen samples with spermatozoa. The simultaneous analysis of methylation markers and unmethylation marker facilitated accurate semen identification even with the samples that were difficult to analyse using the MSRE-PCR method developed previously by An et al. [8]. On the other hand, the semen samples from two vasectomized males showed DNA methylation profiles similar to those of vaginal fluids or menstrual blood with no amplicon for bacterial DNA. Taken together, these findings indicate that this multiplex PCR system was very efficient in the differentiation of blood, saliva, semen and vaginal fluid–menstrual blood.
Further examination revealed that checking for the presence of bacterial DNA complements DNA methylation analysis for body fluid identification. While most of the saliva samples had a very high yield for the PFN3 tDMR, two of the samples produced low amplicon yields for the PFN3 tDMR (<70 % of the yield without enzyme treatment). However, detecting saliva-specific bacteria enabled these samples to be accurately identified as saliva. Saliva-specific bacteria were detected in all but two saliva samples; Streptococcus salivarius was discovered in 18 saliva samples and V. atypica was identified in 12 saliva samples (Table 2). Therefore, integration of DNA methylation analysis and bacterial DNA detection facilitated exact identification of saliva. However, the vaginal fluid-specific bacterial species, L. crispatus, was detected in the saliva of one donor in addition to saliva-specific bacteria. However, the sample showed a much higher amplicon yield for Streptococcus salivarius and V. atypica than for L. crispatus, and thereby was predicted to be saliva rather than vaginal fluid or menstrual blood. Giampaoli et al. [17] reported that Lactobacillus species could be detected in postprandial oral cavity fluid and in yogurt, suggesting that the donor ate prior to donating the sample. Vaginal fluid-specific bacteria facilitated discrimination of vaginal fluid and menstrual blood from other body fluids by amplifying sequences from one or both of the two vaginal fluid-specific bacteria. L. crispatus was detected in eight vaginal fluid and eight menstrual blood samples, and L. gasseri was detected in nine vaginal fluid and eight menstrual blood samples. Vaginal fluid and menstrual blood from the same donor did not always contain the same bacteria, but the predominance of one Lactobacillus species almost always coincided between samples. Since the population of Lactobacillus species can change during the course of the menstrual cycle [27–29], different types of vaginal fluid-specific bacteria may be found in vaginal fluid or menstrual blood from the same donor. There was one vaginal fluid and three menstrual blood samples with no detectable bacterial DNA. A bacterial DNA amplicon was also not detected in the menstrual blood sample from the donor who had no detectable bacterial DNA in her vaginal fluid. We speculate that the reason for this may be either the presence of other predominant species of bacteria such as L. jensenii and L. iners or the intake of antibiotics could cause this result. Our results showing more menstrual blood with no detection of target bacteria compared with vaginal fluid was similar to the result of a previous report [30].
Overall, the results indicate that this multiplex PCR system enables discrimination of more types of body fluid in one multiplex PCR reaction than hitherto reported body fluid identification methods using DNA. Still, there is yet much to be solved for the routine use of the developed methods in common forensic laboratories. In particular, with regard to blood and semen identification, addition or subtraction of markers might improve the analysis. Because blood and semen do not have any detectable bacteria, their identification quite relies on the DNA methylation pattern at tDMRs. However, in the developed multiplex PCR system, blood has no specific tDMR, and semen is identified by the presence of a single tDMR signal with very low or no methylation at the rest of three tDMRs. This may be mitigated by adding a second semen-specific marker because relying on a single positive signal or on the negative signals only can lead to false negatives or false positives. Moreover, bacterial DNA analysis also showed false negatives and a false positive in some samples. Therefore, identification and selection of more appropriate markers will be necessary for the future body fluid identification using DNA.
Sensitivity test
Sensitivity was tested to verify the detection limit of the developed multiplex PCR method. Among the five types of body fluid, saliva, semen and vaginal fluid were subjected to the test because of their different DNA methylation and body fluid-specific bacteria profiles (Fig. S2). Saliva and semen were successfully identified with 500 pg of DNA or more, and vaginal fluids could be identified with 250 pg of DNA or more without any drop-in or drop-out. Allele drop-in or drop-out was observed at some tDMRs with 125 pg of DNA, but appropriate body fluid-specific bacteria were detected in most saliva and vaginal fluid samples. Although the type and amount of bacteria in body fluids would vary from person to person, bacterial DNA analysis seems to be more sensitive than DNA methylation analysis. Therefore, the developed multiplex PCR system, which detects DNA methylation and bacterial presence simultaneously, not only allows accurate body fluid identification but also surpasses the previous MSRE-PCR in terms of sensitivity.
Aged sample test
To test the efficacy of the developed multiplex system with aged samples, DNA extracted from samples exposed to the environment for 75 days were analysed (Fig. S3). Similar to a previous publication [8], DNA methylation analysis was possible for all aged samples except saliva. However, the presence of saliva was indicated by detection of Streptococcus salivarius DNA. The same result had also been observed for low amounts of templates in sensitivity test (Fig. S2b), which suggested the possibility that bacterial DNA analysis can be more sensitive than DNA methylation analysis. Therefore, inferring the type of body fluids first based on the bacterial DNA profile should facilitate more rapid identification of body fluids, particularly with saliva. In this case, DNA methylation profile will, nevertheless, be very useful to confirm the identification results and to check for the presence of other body fluid types. On the other hand, an aged menstrual blood sample did not show Lactobacillus amplicons, consistent with the very low signal for bacterial DNA in the same fresh sample. Thus, this multiplex PCR system could be used to successfully analyse most aged body fluid samples with improved accuracy, suggesting the potential for its application to forensic casework.
Mixture sample test
Body fluids are often found at a crime scene as mixtures. Therefore, mixed samples including saliva–semen, saliva–vaginal fluid and semen–vaginal fluid were tested using the newly developed multiplex PCR method (Fig. S4). Saliva–semen mixtures in 1:1 and 1:2 ratios could be clearly identified by amplifying L81528 and Streptococcus salivarius DNA. Saliva mixed with vaginal fluid swabs produced both of the Lactobacillus species and Streptococcus salivarius amplicons, but the yield of the Streptococcus salivarius amplicon was very low. Similarly, semen mixed with vaginal fluid swabs showed a profile consistent with vaginal fluid when DNA was extracted using general DNA extraction methods (data not shown). However, differential DNA extraction, which can be used in sexual assault cases, enabled identification of semen and vaginal fluid from the resultant pellet and supernatant, respectively. Therefore, the multiplex PCR method developed in this study is useful in mixture analysis as well as analysis of single source samples.
Artificial casework sample test
Since the vaginal swab spotted with semen showed a profile consistent with vaginal fluids in a mixture sample test using general DNA extraction methods, three post-coital vaginal samples and a penile sample were collected for an artificial sexual assault casework sample test. When DNA was extracted using general DNA extraction methods, two of three post-coital vaginal samples and a penile sample showed a mixed sample profile by producing semen-specific L81528 amplicon in addition to the vaginal fluid-specific bacterial signal and a low peak for PFN3 tDMR (Fig. 3). Amelogenin and D3S1358 also displayed a tendency of mixed sample with low AMELY amplicon signal and three or more peaks at the STR locus (Fig. S5). However, another post-coital vaginal sample obtained with tissue paper showed a profile that is consistent with vaginal fluids. In this case, differential DNA extraction enabled identification of semen and vaginal fluid from the resultant pellet and supernatant, respectively. In artificial casework samples, the ratio of male to female DNA varied from case to case. Only when male DNA was less than 10 % of total DNA in DNA quantification with a Quantifiler® Duo DNA Quantification Kit (Applied Biosystems), the developed multiplex PCR system showed a profile consistent with vaginal fluid.
A skin swab from kissing site successfully confirmed the presence of saliva by producing saliva-specific bacterial DNA amplicon and methylation profile. The skin swab obtained from clean skin surface did not produce any DNA profile because extracted DNA was scarce. Although additional validation will be needed, the newly developed multiplex PCR system is considered to facilitate actual casework analysis.
Conclusion
Analyses of tDMRs and detection of body fluid-specific bacteria have been proposed as a promising new method for body fluid identification using DNA. In this study, we presented a highly reliable and sensitive multiplex PCR system that allows simultaneous analysis of tDMRs and bacterial DNA by integrating the two methods. The multiplex PCR method developed in this study can be used to discriminate between four types of body fluid: blood, saliva, semen and vaginal fluid–menstrual blood. Five hundred picogram of DNA from saliva and semen and 250 pg of DNA from vaginal fluid were enough to identify the type of body fluid, while body fluid-specific bacteria were detected in much lower concentrations of DNA (125 pg). In addition, the multiplex PCR allowed successful body fluid identification with artificial casework samples as well as aged or mixed samples.
The multiplex PCR developed in the present study was designed to use a standard capillary electrophoresis platform; thus, special training is not required for these protocols. Moreover, the multiplex system uses the same biological source of DNA for personal identification profiling and simultaneously analyses various body fluids in one PCR reaction. Although it remains to be investigated whether this approach is more sensitive and more practical than RNA- or peptide-based assays, the newly developed multiplex method ensures forensic applicability as well as high specificity, reliability and sensitivity, thereby facilitating more efficient body fluid identification in forensic casework. A few false negatives and a false positive were observed, but this problem could be solved by identification and addition of more appropriate markers.
References
Virkler K, Lednev IK (2009) Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci Int 188:1–17
An JH, Shin KJ, Yang WI, Lee HY (2012) Body fluid identification in forensics. BMB Rep 45:545–553
Zubakov D, Hanekamp E, Kokshoorn M, van IJcken W, Kayser M (2008) Stable RNA markers for identification of blood and saliva stains revealed from whole genome expression analysis of time-wise degraded samples. Int J Legal Med 122:135–142
Hanson EK, Lubenow H, Ballantyne J (2009) Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal Biochem 387:303–314
Fleming RI, Harbison S (2010) The development of a mRNA multiplex RT-PCR assay for the definitive identification of body fluids. Forensic Sci Int Genet 4:244–256
Frumkin D, Wasserstrom A, Budowle B, Davidson A (2011) DNA methylation-based forensic tissue identification. Forensic Sci Int Genet 5:517–524
Lee HY, Park MJ, Choi A, An JH, Yang WI, Shin KJ (2012) Potential forensic application of DNA methylation profiling to body fluid identification. Int J Legal Med 126:55–62
An JH, Choi A, Shin KJ, Yang WI, Lee HY (2013) DNA methylation-specific multiplex assays for body fluid identification. Int J Legal Med 127:35–43
Wasserstrom A, Frumkin D, Davidson A, Shpitzen M, Herman Y, Gafny R (2013) Demonstration of DSI-semen—a novel DNA methylation-based forensic semen identification assay. Forensic Sci Int Genet 7:136–142
Larue BL, King JL, Budowle B (2013) A validation study of the Nucleix DSI-Semen kit—a methylation-based assay for semen identification. Int J Legal Med 127:299–308
Madi T, Balamurugan K, Bombardi R, Duncan G, McCord B (2012) The determination of tissue-specific DNA methylation patterns in forensic biofluids using bisulfite modification and pyrosequencing. Electrophoresis 33:1736–1745
Nakanishi H, Kido A, Ohmori T, Takada A, Hara M, Adachi N et al (2009) A novel method for the identification of saliva by detecting oral streptococci using PCR. Forensic Sci Int 183:20–23
Nakanishi H, Ohmori T, Hara M, Takada A, Shojo H, Adachi N et al (2011) A simple identification method of saliva by detecting Streptococcus salivarius using loop-mediated isothermal amplification. J Forensic Sci 56(Suppl 1):S158–S161
Hsu L, Power D, Upritchard J, Burton J, Friedlander R, Horswell J et al (2012) Amplification of oral streptococcal DNA from human incisors and bite marks. Curr Microbiol 65:207–211
Akutsu T, Motani H, Watanabe K, Iwase H, Sakurada K (2012) Detection of bacterial 16S ribosomal RNA genes for forensic identification of vaginal fluid. Legal Med (Tokyo) 14:160–162
Benschop CC, Quaak FC, Boon ME, Sijen T, Kuiper I (2012) Vaginal microbial flora analysis by next generation sequencing and microarrays; can microbes indicate vaginal origin in a forensic context? Int J Legal Med 126:303–310
Giampaoli S, Berti A, Valeriani F, Gianfranceschi G, Piccolella A, Buggiotti L et al (2012) Molecular identification of vaginal fluid by microbial signature. Forensic Sci Int Genet 6:559–564
Ohgane J, Yagi S, Shiota K (2008) Epigenetics: the DNA methylation profile of tissue-dependent and differentially methylated regions in cells. Placenta 29:S29–S35
Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H et al (2005) Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A 102:3336–3341
Kitamura E, Igarashi J, Morohashi A, Hida N, Oinuma T, Nemoto N et al (2007) Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics 89:326–337
Illingworth R, Kerr A, Desousa D, Jørgensen H, Ellis P, Stalker J et al (2008) A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6:e22
Chakravorty S, Helb D, Burday M, Connell N, Alland D (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69:330–339
De Backer E, Verhelst R, Verstraelen H, Alqumber MA, Burton JP, Tagg JR et al (2007) Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol 7:115
Kang JG, Kim SH, Ahn TY (2006) Bacterial diversity in the human saliva from different ages. J Microbiol 44:572–576
Marinus MG, Morris NR (1973) Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol 114:1143–1150
May MS, Hattman S (1975) Analysis of bacteriophage deoxyribonucleic acid sequences methylated by host- and R-factor-controlled enzymes. J Bacteriol 123:768–770
Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UM, Zhong X et al (2012) Temporal dynamics of the human vaginal microbiota. Sci Transl Med 4:132ra52
Witkin SS, Linhares IM, Giraldo P (2007) Bacterial flora of the female genital tract: function and immune regulation. Best Pract Res Clin Obstet Gynaecol 21:347–354
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL et al (2011) Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108:4680–4687
Fleming RI, Harbison S (2010) The use of bacteria for the identification of vaginal secretions. Forensic Sci Int Genet 4:311–315
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0005208).
Conflicts of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, A., Shin, KJ., Yang, W.I. et al. Body fluid identification by integrated analysis of DNA methylation and body fluid-specific microbial DNA. Int J Legal Med 128, 33–41 (2014). https://doi.org/10.1007/s00414-013-0918-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-013-0918-4