Abstract
Necrophagous blow fly larvae can provide accurate estimates of the minimum postmortem interval in death investigations. During larval development, predictable morphological changes occur and measurements of weight, length, and width are compared to species-specific growth curves for reliable age estimates. However, aging blow fly pupae is more challenging because morphological and anatomical changes are not visible with the naked eye. Thus, delicate preparation of the pupae or rearing to the adult stage seems unavoidable. Conversely, metamorphosis evokes a remodelling of the larval shape to adult structures, and gene expression analysis potentially serves as a molecular tool to mirror the ageing process of a pupa. The present study focuses on the differential expression of two newly described, arbitrarily named genes (15_2, 2014192) and two previously identified genes (actin, arylphorin receptor) during Calliphora vicina (Diptera: Calliphoridae) metamorphosis. Quantification through real-time PCR revealed significant up- and downregulation of these transcripts found to be temperature dependent and age specific, hence, a new possibility to age forensically important blow fly pupae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Insects play a major role in forensic investigations as they can be used to estimate the minimum postmortem interval (PMImin) of a corpse [1]. Blow flies (Diptera: Calliphoridae) are known to be among the first colonizers of a corpse and their offspring can give a good estimate of the time a body has been exposed to insects [2–4]. Larvae undergo predictable changes during development. Besides anatomical changes of the posterior spiracles, for instance, they increase in length, width, and weight. The age of the specimens can be calculated based on measurements of these morphological characters and comparison to species-specific growth curves [5–9]. Following the feeding stage, larvae of certain species disperse from the food source in order to pupate. The pupal stage can last up to 50 % of the whole juvenile development and low temperatures elongate this period up to several weeks for specific species [e.g., 10]. During this stage, age estimation is increasingly difficult because length, for instance, does not significantly change during metamorphosis [11]. Typically, pupae are reared to adulthood for subsequent age estimation in death investigations. This method, however, is not feasible for preserved pupae or specimens which die during the rearing process. Dissection of the puparium to reveal key developmental processes during metamorphosis is possible but it is time consuming and difficult as it depends on experience. Though color of the puparium has been suggested as a tool for aging, the color change is no longer predictable after the first few hours of puparium formation [12, 13]. Methods for reliable age estimations of forensically relevant pupae are necessary, especially when pupae represent the oldest specimens present at a crime scene or even the only source of entomological evidence available.
The analysis of differentially expressed genes (DEGs) during blow fly metamorphosis can lead to an understanding of the process of formation of the adult fly and may also represent a new approach to predict pupal age in death investigations. It has been demonstrated that metamorphosis is induced and controlled by the hormone ecdysone which activates a hierarchy of response genes differentially expressed throughout pupal development [e.g., 14, 15] and that cell growth, cell death, and cell differentiation are responsible for changes of the larval body structures [e.g., 16–18]. So far expression profiles of developmentally regulated genes have been used by Tarone et al. [19] and Tarone and Foran [20] to age forensically important Lucilia sericata (Diptera: Calliphoridae) specimens throughout development. However, identified DEGs have not succeeded to accurately age blow fly pupae [20–23].
The present research focuses on the temporal gene expression of four genes during the metamorphosis of Calliphora vicina (Diptera: Calliphoridae), an abundant and widely distributed forensically relevant blow fly [1]. Transcripts were either obtained by differential display PCR based on ACP™ technology [24, 25] or from previous studies. The assumption tested here is that the correlation of gene expression with specific points in time during pupal development expands the possibilities of PMImin estimation based on conventional entomological approaches.
Material and methods
Specimen collection and fly rearing
Two colonies were established from wild-caught C. vicina individuals collected in 2007 and 2010 in Frankfurt am Main, Germany. Species determination was conducted visually using the identification key by Rognes [26].
Adult flies were held in cages (74 × 70 × 60 cm) under nonconstant temperature conditions (average temperature of approximately 19°C) and a photoperiod of 16:8 (L/D). Fresh water and sugar were provided ad libitum. Beef blood was provided once a week as a protein meal. Fresh beef liver served as an oviposition medium and was offered once a week. After 24 h, the liver containing eggs and larvae (between 123 and 1,121 individuals) was transferred to a 300-mL plastic cup and placed in a rearing chamber with no light at constant temperatures of 15, 20, and 25°C, respectively. Larvae were continuously supplied with ground meat (half beef, half pork) ad libitum. To avoid intraspecific competition and overcrowding, larvae were split onto several plastic cups. Upon completion of the feeding stage, larvae crawled out of the 300-mL cup and into pupation medium (sawdust) within a 5-L plastic container. Maggots remained in the post-feeding stage for several days, depending on rearing temperature.
When approximately 100 post-feeding maggots had pupated (within a time range of 17 h maximum) they were separated from the remaining individuals and their age was set to “zero”. Five pupae were collected at 24 h intervals starting from age zero until more than five adult flies were observed. Experiments were repeated at the same temperature two to three times within 1 to 10 months using either one of the colonies. A total of 510 pupae were collected.
Detection and isolation of differentially expressed genes
Whole pupae were homogenized in 1 mL TRIzol® Reagent (Invitrogen) and stored at −80°C until RNA extraction. Total RNA was extracted following manufacturer’s instructions and eluted in 100 μl RNA Storage Solution (Ambion). Residual DNA was digested using the TURBO DNA-free™ kit (Ambion) and the RNA concentration was determined spectrophotometrically (NanoDrop ND-1000).
Complementary DNA (cDNA) synthesis and subsequent GeneFishing™ PCR was conducted through ACP™ technology [24, 25]. First-strand cDNA was synthesized by reverse transcription in a total reaction volume of 20 μl following manufacturer’s instructions [24, 25, 27] containing 3 μg total RNA as template. First-strand cDNA was diluted by adding 80 μl DNAse-free water and either served as template for quantitative real-time PCR approaches or GeneFishing™ PCR. For the latter, 20 ACP primers (included in GeneFishing™ DEG 101 & 102 Premix Kits) were used in single PCR reactions to amplify differentially expressed genes. Here, 5 μl of first-strand cDNA products served as templates for second-strand cDNA synthesis. Thermal cycler conditions and PCR set-up for first- and second-strand syntheses are described by Kim et al. [24]. PCR amplifications were performed in a T3000 thermal cycler (Biometra, Göttingen, DE).
All gene fishing experiments were performed with pupae reared at 20°C. Intensity of the bands yielded in an agarose gel (between 0.5 and 2.5 %) stained with ethidium bromide and visualized under UV light, displayed the potentially up- and downregulation of specific transcripts. These genes of interest (GOIs) and also potential constitutively expressed genes were isolated by molecular cloning of PCR products into a vector with the Invitrogen, TOPO TA Cloning Kit (Life Technologies GmbH, Darmstadt, DE) following the recommended steps. Plasmids were purified with the Invitrogen S.N.A.P.™ MiniPrep Kit. Purified plasmid DNA was sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Weiterstadt, DE) following manufacturer’s instructions. The protocol included a total reaction volume of 20 μl consisting of 3 μl Big Dye, 2.5 μl 5× sequencing buffer, 1 μl primer, and 1 μl PCR product.
Sequencing products were purified using gel-filtration columns (Qiagen, DyeEx 2.0 Spin Kit, Hilden, DE) and run on an ABI3130 Genetic Analyzer equipped with a 36-cm Capillary Array/POP-4 (Applied Biosystems).
The two genes encoding for actin and the arylphorin receptor were included in this study as they have been shown to yield different transcription levels during metamorphosis [21, 28].
Quantitative real-time PCR
For quantification, a real-time PCR was conducted on a StepOnePlus™ Real-Time PCR System (Applied Biosystems). Custom TaqMan® Gene Expression Assays were established by ABI technology and primers and probes were designed based on the nucleotide sequences of our transcripts of interest (Table 1). Quantitative RT-PCR reaction consisted of 1 μl Custom TaqMan® Gene Expression Assays (20× concentration mix including unlabeled forward and reverse primer and an 6FAM™ dye-labeled MGB probe), and 10 μl TaqMan® Fast Universal PCR Mastermix (2×). Nine microliters of 1:10 diluted cDNAs were used as templates. All samples were measured in triplicates. The PCR program was the following: 20 s at 95°C followed by 40 cycles of 95°C for 1 s and 60°C for 20 s. Throughout the study, qRT-PCR cycle thresholds were set to 0.5 for analysis so that quantities of mRNA levels are comparable within and between experiments.
Processing of qRT-PCR data
To quantify relative changes in gene expression, we followed the approach given in Livak and Schmittgen [29] by using PCR efficiency (E) and cycle threshold values (CT) to calculate the change in gene expression at a certain point in time relative to a reference time (“age zero”, see above). This so-called fold induction or fold change (FC) was calculated for all four genes of interest and two constitutively expressed, housekeeping genes (HKGs) in 24-h intervals after onset of pupariation. To account for variable PCR efficiencies, FC computations were adjusted according to Pfaffl [30] by dropping the assumption that PCR efficiency remains constant in all samples (formulas (1–3), below). We applied the modification given by Vandesompele et al. [31] by taking the geometric mean of the FCs of both HKGs to obtain the normalized FC of our GOIs (formulas (4–5), below). Finally, normalized FC values of GOIs were log-transformed to the base of 2, thus, an increase or decrease in GOI mRNA levels are represented by positive or negative values, respectively (formula (6), below).
At each point in time, CT values from each triplicate were averaged. PCR efficiency for each sample was calculated with the software LinRegPCR version 12.11 using default settings [for details see 32]. Efficiencies for every temperature-dependent experiment were averaged.
Statistical analysis
LogFC values of GOIs were subjected to a two-way ANOVA to explore the influence of both pupal age and rearing temperature on relative gene expression. A Shapiro–Wilk test was applied to assure normal distribution of logFC values. P values for multiple testing were adjusted according to the Sidák correction. All statistical computations were conducted in R (R Foundation for Statistical Computing).
Results
Based on the application of a differential display PCR (ACP™ technology, Seegene, described in [24, 25]), we isolated approximately 30 transcripts potentially representing age-specific marker genes. Many transcripts, however, displayed similar expression profiles in qRT-PCR experiments or low variation in expression levels during metamorphosis (data not shown). Thus, the absolute number of GOIs was reduced to four. Two transcripts could not be identified through BLAST after sequence analysis and were arbitrarily named 15_2 and 2014192, respectively. The nucleotide sequences for actin and for the receptor of the storage protein arylphorin (arylphorin receptor), were obtained from GenBank [Accession numbers: AY954916 and X79100, respectively].
Two transcripts were found to be expressed at constant levels throughout pupal development, thus likely to represent housekeeping genes. One HKG was identified through BLAST and showed similarities in its amino acid sequence to Drosophila melanogaster RpL23, a ribosomal gene (query coverage, 93 %; maximum identity, 100 %; E value, 5e-62). The second constitutively expressed gene revealed reduced similarities (BLAST, 83 % maximum identity at a query coverage of 77 %; E value, 7e-32) in its amino acid sequence to D. melanogaster RpS26, another ribosomal protein. For this reason, we chose to continue referring to this gene as 19_2. Although HKGs previously described for another blow fly, L. sericata, [19] were initially included, the software tool geNorm [31] identified the combination of 19_2 and RpL23 as best for normalization of gene expression data in C. vicina pupae. Gene expression data were generated for 506 specimens. Four specimens did not yield a sufficient amount of RNA for cDNA synthesis and were not included.
Overall expression profiles during pupal development
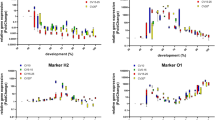
Normalized LogFC values of each informative gene were averaged among the three temperature-dependent experiments and mean values were plotted against development percent (Fig. 1).
Expression of 15_2 increased steadily until it reached its maximum at approximately 22 % of pupal development followed by a decrease to levels below our age zero reference after 60 % completed development. The transcripts 2014192 and actin yielded similar expression patterns and a steady increase in mRNA quantity is observed after 50 % of metamorphosis is completed. The expression level of the arylphorin receptor was high in age zero pupae but mRNA levels decreased rapidly after the initial hours of puparium formation and remain at a constant level during pupal development.
Detailed expression profiles during pupal development
Figure 2 plots gene expression against age (days) to visualize the correlation of temperature, gene expression, and development. FC values were averaged among each temperature. Not surprisingly, the higher the temperature, the faster the pupal development thus resulting in a faster progression of gene expression and changes in mRNA levels. Patterns of gene expression per se are fairly the same for all temperature regimes. Metamorphosis lasted 18 days at 15°C, 11 days at 20°C, and 9 days at 25°C, respectively.
15_2 gene expression levels within the first 2 to 3 days were similar in pupae reared at all three temperatures. The maximum expression is reached at day 2 for pupae reared at 20 and 25°C and at day 3 for pupae reared at 15°C, respectively. A similar phenomenon was observed for 2014192. FC values of the arylphorin receptor are similar throughout metamorphosis and not dependent on temperature as seen in Table 2.
Gene expression is significantly influenced by age in all loci (two-way ANOVA, P < 0.001, see Table 2). Rearing temperature also significantly affected gene expression in all analyzed genes except the arylphorin receptor. The combination of age and temperature influenced FC values significantly in all genes indicating that the interaction term age × temperature is a critical component determining gene expression.
Supplementary material, Fig. 3 displays the normalized fold induction values of all pupae obtained for either one of the temperature-dependent experiments versus age (days). The spread in the data is indicated by the lower and upper whisker of the boxplots. Outliers outside the 25 and 75 % quartile are represented by points. It appears that the overall spread in the gene expression profiles is sufficiently low to allow the inference of tight confidence intervals on age using inverse prediction. This statistic methodology may represent a further step toward precise age prediction based on statistical evaluation of gene expression data.
Discussion
The focus of this investigation was to identify informative marker genes whose expression profiles would be predictive of age during C. vicina metamorphosis. A precise age determination of morphologically similar blow fly pupae would improve PMImin estimations based on entomological evidence. To date, morphological and genetic landmarks have been identified to divide Diptera metamorphosis into distinct developmental units [11–13, 20, 28]. Moreover, the application of temporal gene expression patterns to forensic entomology has been shown to increase the precision in age estimation of L. sericata specimens throughout development [20]. However, identification of appropriate marker genes and application of this methodology to age blow fly pupae in practice is still in its infancy [20–22]. Nonetheless, aging (i.e., the processing of specific developmental steps) is dependent on temporal gene activity and is predominantly regulated by hormones such as the steroid hormone ecdysone, a key hormone in insect development. Burmester and Scheller [33] demonstrated that ecdysone initiates the uptake of the hexamerin storage protein arylphorin in Calliphora third instar larvae through cleavage of the receptor. This supports the high arylphorin receptor peak in late third instar larvae as shown by Ames et al. [21]. After onset of puparium formation, however, hexamerin endocytosis processes are not further required and the amount of arylphorin receptor mRNA ceases rapidly. Thus, arylphorin receptor mRNA levels are promising as molecular marker for age estimation as we are able to distinguish between very young pupae and pupae of higher ages.
At least two genes responsible for the formation of the cytoskeleton have been found to be expressed at a high rate in Drosophila pupae in their last third of development [28]. In accordance with this, we found a steady accumulation of actin mRNA after 50 % of metamorphosis is completed.
The biological functions of 15_2 are unknown so far. Interestingly, expression patterns correlate with the quantification levels of the ecdysone receptor protein within the first half of pupal development (data not shown). The ecdysone receptor is necessary for ecdysone-mediated gene expression and relative expression levels of 15_2 in the first half of metamorphosis may thus be ecdysone responsive. The gene encoding for the ecdysone receptor itself was not included in this work as other genes were more informative in age prediction.
With the exception of 15_2, all graphs show a monotonous rise or fall in gene expression over time (Fig. 2). Thus, it should be possible to deduce pupal age from FC values as both terms are essentially in a one-to-one relationship. It may not be possible to obtain unambiguous age estimates based on the expression pattern of a single gene alone for time periods when expression remains fairly constant (e.g., days 2–5 in 2014192 at 25°C; Fig. 2). As other genes, however, may prove to be highly dynamic during the same period (e.g., 15_2), the combined logFC data can be used to overcome this lack of prediction resolution.
The rate of gene expression patterns of almost all analyzed GOIs were significantly influenced by temperature. Temperature is the most dominant factor in development of poikilothermic animals; the higher the temperature, the faster developmental rates [10]. Consequently, low temperatures result in a slower increase or decrease in gene expression compared to high temperatures. Age as a second variant also significantly influences FC values.
Based on the presented temporal changes in gene expression it is possible to divide metamorphosis in smaller developmental units supported by genetic landmarks. However, besides temperature, the geographical origin of a specific population may have impact on developmental rates and thus result in longer or shorter duration of a certain developmental stage or the complete development as shown for L. sericata [34, 35]. This might explain the duration of the pupal stage observed here compared to other studies [e.g., 10]. Future studies focussing on population diversity will help to better assess variation in developmental rates and gene expression for a certain species.
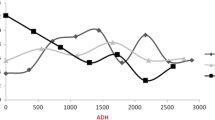
In addition to age and temperature alone, the interaction between temperature and age also proved to have a significant impact on gene expression. Very likely, this reflects the dependency of gene expression on accumulated degree hours (ADH), since ADH is a multiplicative interaction of temperature and age. Thus, the amount of degree hours accumulated at a certain age mirrors the percent level of completion of development. Since pupal development requires differential gene expression, gene expression also mirrors the percent level of completion specific developmental units. Thus, the dependency of gene expression on ADH is in accordance with ADH being a developmental determinant. In a next step, we focus on the application of a combined gene expression to predict pupal age in ADH (Boehme et al., The application of temporal gene expression to postmortem interval estimation: introducing PUPAC - a statistical tool to predict the age of forensically important Calliphora vicina pupae (Diptera: Calliphoridae)). A verification of the benefit of this method is necessary for further, standardized application to death investigations. However, the age-dependent expression of at least four genes in combination with the availability of powerful statistical methods should allow the establishing of standardized protocols to infer statistically corroborated age estimations of forensically important C. vicina pupae.
References
Smith KGV (1986) A manual of forensic entomology. Cornell University Press, London
Villet MH, Amendt J (2011) Advances in entomological methods for death time estimation. In: Turk EE (ed) Forensic Pathology Reviews doi:10.1007/978-1-61779-249-6_11.
Amendt J, Richards CS, Campobasso CP, Zehner R, Hall MJR (2011) Forensic entomology: applications and limitations. Forensic Sci Med Pathol doi:10.1007/s12024-010-9209-2
Forbes SI, Dadour I (2010) The soil environment and forensic entomology. In: Byrd JH and Castner JL (eds) Forensic entomology—The utility of arthropods in legal investigations, 2nd edn. CRC, Boca Raton, p 416
Richards CS, Crous KL, Villet M (2009) Models of development for blowfly sister species Chrysomya chloropyga and Chrysomya putoria. Med Vet Entomol 23:56–61
Grassberger M, Reiter C (2001) Effect of temperature on Lucilia sericata (Diptera: Calliphoridae) development with special reference to the isomegalen- and isomorphen-diagram. Forensic Sci Int 120:32–36
Wells JD, LaMotte LR (1995) Estimating maggot age from weight using inverse prediction. J Forensic Sci 40:585–590
Greenberg B (1991) Flies as forensic indicators. J Med Entomol 28:565–577
Reiter C (1984) Zum Wachstumsverhalten der Maden der blauen Schmeißfliege Calliphora vicina. Zeitschrift für Rechtsmedizin, 91:295–308
Marchenko MI (2001) Medicolegal relevance of cadaver entomofauna for the determination of the time of death. Forensic Sci Int 120:89–109
Zajac BK (2011) Morphologische und histologische Methoden zur Bestimmung des Alters forensisch relevanter Fliegenpuppen. Diplomarbeit. Goethe-Universität Frankfurt am Main
Bainbridge SP, Bownes M (1981) Staging the metamorphosis of Drosophila melangaster. J Embryol exp Morph 66:57–80
Robertson CW (1936) The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. J Morphol 59:351–399.
Beckstead RB, Lam G, Thummel CS (2005) The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol 6:R99
Thummel CS (1996) Flies on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet 12:306–310
Buszczak M, Segraves WA (2000) Insect metamorphosis: out with the old, in with the new. Curr Biol 10:R830–R833
White KP, Rifkin SA, Hurban P, Hogness DS (1999) Microarray analysis of Drosophila development during metamorphosis. Science 286:2179–2184
Bowen ID, Mullarkey K, Morgan SM (1996) Programmed cell death during metamorphosis in the blow-fly Calliphora vomitoria. Microsc Res Tech 34:202–217
Tarone AM, Jennings KC, Foran DR (2007) Aging blow fly eggs using gene expression: a feasibility study. J Forensic Sci 52:1350–1354
Tarone AM, Foran DR (2011) Gene expression during blow fly development: improving the precision of age estimates in forensic entomology. J Forensic Sci 56:S112–S122.
Ames C, Turner B, Daniel B (2006) Estimating the post-mortem interval (II): the use of differential temporal gene expression to determine the age of blowfly pupae. International Congress Series 1288:861–863
Gaudry E, Blais C, Annick M, Dauphin-Villemant C (2006) Study of steroidogenesis in pupae of the forensically important blow fly Protophormia terraenovae (Robineau-Desvoidy) (Diptera: Calliphoridae). Forensic Sci Int 160:27–34
Mösch SA (2005) Molekularbiologische Altersbestimmung an Puppen der forensisch relevanten Schmeißfliege Lucilia sericata. Diplomarbeit, Fachhochschule Aachen.
Kim YJ, Kwak CI, Gu YY, Hwang IT, Chun JY (2004) Annealing control primer system for identification of differentially expressed genes on agarose gels. BioTech 36:424–434
Hwang IT, Kim YJ, Kim SH, Kwak CI, Gu YY, Chun JY (2003) Annealing control primer system for improving specificity of PCR amplification. BioTech 35:2–6
Rognes K (1991) Blowflies (Diptera, Calliphoridae) of Fennoscandia and Denmark. E.J. Brill/Scandinavian Science, Leiden
GeneFishing™ DEG Premix Kit, User Manual, Seegene
Beckmann B (2004) Studien der Genexpressionsänderung während der Entwicklung von Drosophila melanogaster mittels DNA-Microarrays. Dissertation. Ruprecht-Karls-Universität Heidelberg
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta (CT)) method. Methods 25:402–408
Pfaffl MW (2001) A new mathematical model for relative quantificationin real-time RT-PCR. Nucleic Acids Res 29: e45
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034.1-research0034.11
Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Burmester T, Scheller K (1997) Developmentally controlled cleavage of the Calliphora arylphorin receptor and posttranslational action of the steroid hormone 20-hydroxyecdysone. Eur J Biochem 247:695–702
Tarone AM, Picard CJ, Spiegelman C, Foran DR (2011) Population and temperature effects on Lucilia sericata (Diptera: Calliphoridae) body size and minimum developmental time. J Med Entomol 48:1062–1068.
Gallagher MB, Sandhu S, Kimsey R (2010) Variation in developmental time for geographically distinct populations of the common green bottle fly, Lucilia sericata (Meigen). J Forensic Sci 55:438–442
Acknowledgments
This project was financially supported by the Deutsche Forschungsgemeinschaft (project number: ZE 501/2-1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boehme, P., Spahn, P., Amendt, J. et al. Differential gene expression during metamorphosis: a promising approach for age estimation of forensically important Calliphora vicina pupae (Diptera: Calliphoridae). Int J Legal Med 127, 243–249 (2013). https://doi.org/10.1007/s00414-012-0699-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-012-0699-1