Abstract
Insertion–deletion (INDEL) markers are very frequent in the human genome and present several advantages for population and forensic studies, such as low mutation rates, easy interpretation, small amplicons, easy genotyping, and the possibility of using multiplex PCR. The great adaptability of INDELs for amplification of low copy number or degraded DNA allows its using as an interesting platform of genetic identity by DNA in forensic cases. In the present study, we tested the ability of 48 diallelic INDEL markers on genotyping forensic samples collected from different biological samples related to criminal cases. Moreover, we evaluated the lowest DNA concentration with which there was amplification of all markers from each one of three indel-plex panels. When comparing the performances obtained by the indel-plex panels described in this study with results obtained using Identifiler® kit (Applied Biosystems) related to forensic samples, as well as to control samples with different concentrations of DNA, we observed superior efficiency on samples with low copy number or in the presence of inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The advent of multiplex short tandem repeats (STRs) has granted an increase in test sensitivity and speed, allowing the simultaneous amplification of more than 15 STR loci in a single analysis. Besides STRs, two groups of human polymorphisms have shown to be promising in forensic genetics: polymorphisms based on single nucleotide substitution (SNPs) and polymorphisms generated by insertion or deletion of one or more nucleotides (INDELs). In 2002, Weber et al. [1] identified and characterized 2,000 diallelic insertion–deletion polymorphisms (INDELs) in the human genome.
Unlike STRs, both SNPs and INDELs yield very short amplification products that can be identified when DNA is much degraded, as is the case of cadavers which are in a very advanced stage of decomposition or carbonization [1].
Recent advances in forensic genetics have focused on the development of genotyping assays using short amplicons, in order to improve the successful amplification of degraded samples, as illustrated by the development and application of SNP sets and reengineered mini-STRs. Nevertheless, in contrast with standard STR strategies, the current SNP assays usually involve complex genotyping protocols, needing more steps and/or the implementation of new methods and expensive high-throughput technologies [2].
Insertion/deletion polymorphisms can combine desirable characteristics of both SNPs and STRs. This kind of polymorphisms present interesting features as genetic markers: (1) INDELs are spread throughout the human genome (autosomes and sex chromosomes), apparently with the same density (although there are hotspots); (2) the polymorphism derives from a single mutational event; (3) many of them display significant differences in allele frequencies among geographically separated population groups (therefore, they can be used as ancestral informative markers (AIMs); (4) small INDELs can be analyzed in short amplicons, opening perspectives for good multiplexing ability and, on the other hand, improving successful amplification of degraded DNA; (5) the genotyping of small INDELs is relatively easy and inexpensive with a simple dye-labeling electrophoretic approach, but also suitable for automation and analysis with high-throughput technologies [2, 3].
Santos et al. [4] described a 48-INDEL–AIM panel that allows fast and cost-effective genotyping that can be used to distinguish major ethnic populations, specifically Europeans, Africans, and Native Americans. INDELs also identify substructure in mixed populations. They also demonstrated that in mixed populations from different ancestral groups, the marker panel allows estimating, in an accurate and reliable manner, the individual and global interethnic admixture relative to those groups [4].
The main objectives of this work were to evaluate the efficiency of three panels of INDEL markers in amplifying different forensic samples, identify the lowest DNA concentration in which indel-plex panels are still able to amplify all markers, and compare the performances obtained by the evaluated multi-INDELs to the obtained by an STR kit.
Material and methods
Samples
This study was carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association. Twenty-five forensic samples obtained from the Forensic Biology and Forensic Anthropology Laboratories of Scientific Police of Amapá (POLITEC-AP) in the city of Macapá (0°02′20″ N; 51°03′59″ W), State of Amapá, northern Brazil were investigated as follows: (a) three vaginal secretion samples (VS01, VS02, and VS03) with positive results for the presence of sperm or prostate-specific antigen, collected from three sex crime victims; (b) cartilage, bone, and tooth totalized eight samples collected from five cadavers of unknown identity (CD01, CD02, CD03, CD04A, CD04B, CD05A, CD05B, CD05C); (c) one sample of muscle collected from a fetus (M01); (d) tree samples of oral cells collected in a case of criminal paternity of the alleged father, mother, and son (PM01A, PS01A, PAF01A); (e) thee samples of blood collected in a case of criminal paternity of the alleged father, mother, and son (PM01B, PS01B, PAF01B); (f) eight dried blood spots (B01A, B01B, B01C; B02A, B02B, B02C, B03, B04). The ancestry estimates were made using the structure software (http://pritch.bsd.uchicago.edu/software/structure2_1.html), and parental populations samples used were described in Francez et al. 2011 [5].
Using 1 μL of DNA of each concentrations obtained from a serial dilution of the calibration curve DNA of the Quantifiler Y® kit (Applied Biosystems), which has concentrations of 50, 16.7, 5.6, 1.8, 0.6, 0.2, 0.056, and 0.02 ng/μL, we evaluated the lowest concentration of DNA with which there was amplification of all markers in each one of the three indel panels evaluated. Using the commercial Identifiler® kit, the experiment was replicated with the same conditions and serial dilution.
DNA extraction and quantification
Table 1 shows the nature of the biological samples used and the extraction techniques employed in this study. After the extraction, the samples were quantified by real-time PCR, using a BIORAD iQ5 equipment (Hercules, CA, USA) and the Quantifiler DUO MR kit (Applied Biosystems).
PCR amplification
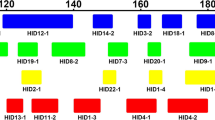
To analyze the genetic profile of the investigated samples, we used 48 diallelic autosomal markers formed by the insertion–deletion of small DNA fragments (3–30 bp) and genotyped using three multiplex PCR procedures (one for each set of African, European, and Native American ancestry markers—identified of indel-plex 01, indel-plex 02, and indel-plex 03, respectively) and a GeneAmp® PCR system 9,700 thermocycler (Applied Biosystems). Marker identification, primers used, and PCR conditions were as described by Santos et al. [4] (see Supplementary Table 1, Online Resource).
All forensic samples examined also underwent amplification of autosomal STR markers (D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16539, D2S1338, D19S433, vWA, TPOX, D1851, D5S818, FGA, and Amel) using Identifiler® kit (Applied Biosystems).
Genotyping
Electrophoresis and typing were performed in an ABI 3130 Avant Automated Sequencer (Applied Biosystems, Foster City, CA, USA). Data acquisition was performed with the ABI PRISM™ 3130–Avant Data Collection v2.0 software (Applied Biosystems), and for profiles analysis, we used the GeneMapper ID v3.2 software (Applied Biosystems). Typing quality and allele designation were assured by simultaneous electrophoretic analysis of a control sample of known size and sequences. Allele designations were made with the ABIGS LIZ 500 reference ladder (Applied Biosystems) as size standard.
Results and discussion
Aiming to evaluate the utility and performance of the three indel-plex panels in practical situations, particularly in the analysis of low copy number or highly degraded DNA (common in forensic samples) and as complementary tool in kinship analysis, we tested these three indel-plex on 25 forensic samples (Table 1) and on a control DNA sample with serial dilutions of eight different concentrations. The results of these two tests can be seen in Tables 2 and 3.
In evaluating the lowest DNA concentration with which all systems from the three indel-plex still amplified, it was observed that over the indel-plex 01, this concentration was 0.6 ng/μL, while for the indel-plex 02 and 03, the lowest concentration was 0.2 ng/μL. Replicated the analyses with a STR kit, as expected, the short-amplicon strategy revealed an enhanced amplification success when compared to standard STRs, since at concentrations of 0.6 ng/μL obtained STR profiles, unlike the three indel-plex panels, were already partials (see Supplementary Figs. 1–28, Online Resource and Table 2).
In relation to forensic samples, after DNA quantification by real-time PCR, we observed concentrations ranging from 0.2 ng/μL in the CD03 sample to 19.5 ng/μL in the B04 sample (Table 3). Of the 25 forensic samples investigated, 22 (see Supplementary Figs. 29–64, Online Resource) showed complete profiles of the three indel-plex evaluated; samples CD03 and B02A showed partial profiles for the indel-plex 01 and complete to indel-plex 02 and 03. Only sample B04 showed partial profile for the three indel-plex investigated (indel-plex 01 = 4 amplified systems; indel-plex 02 = 13 amplified systems, and indel-plex 03 = 11 amplified systems) (Table 3).
For samples CD03 and B02A, low DNA concentration caused partial genotypes evidenced on indel-plex 01 which had the lowest sensitivity among the three multiplex evaluated. Regarding sample B04, the results were due to the presence of inhibitors and excess DNA (19.5 ng/μL) observed by real-time PCR (Table 3).
Amplifying the same forensic samples using STR systems through Identifiler® kit, we found that 17 electropherograms had complete profile, 4 had partial profile, and 4 showed no amplification. These results demonstrate the potential use of indel-plex as a complementary tool to STRs in the analysis of forensic samples.
Paternity investigations showing few STR transmission incompatibilities between alleged father and child are not an uncommon scenario and often become problematic. In some cases, relatively high W values are obtained even when accounting for the incompatibilities explainable as mutational events or silent alleles. With such paternity indexes, a doubt can persist on whether the tested man is a close relative of the true father (e.g., his brother/father/uncle). This problem can be largely overcome by using a complementary high number of markers with low mutation rates, which is the case of both SNP and indel multiplex strategies [6]. Estimates based on analysis of offspring in humans suggest a mutation rate of 10−3 events per locus per generation for STRs [7]. While the mutation rate of SNPs (2.3 × 10−8) and INDELs (2.3 × 10−9) show significantly lower [8].
Indel-plex markers may be useful as additional tools to STRs in paternity cases. Pereira et al. [2], in a recent paternity investigation (typical trio), the testing father presented two incompatibilities at D5S818 and D13S317. Using a battery of 21 STRs and accounting for mutational events, the estimated W was 99.998%. The doubt then arose on whether the testing man was the true father of the child (and two STR mutations had occurred), or a close relative of the true father presenting only two incompatibilities and achieving similarly high W values. The complementary study of 38 slower evolving bi-allelic indels in this case showed no further incompatibilities and allowed raising W to 99.99998%, thus, corroborating the hypothesis of paternity. In Supplementary Figs. 65–70 (Online Resource), it is possible to observe the electropherograms related to indel-plex 02 and 03 in a paternity case examined in the present study.
The fact that bi-allelic systems employed in the three indel panels analyzed in this study have shown, in general, great differences in allele frequencies (δ = 40%) between African, European, and/or Native American populations [4], therefore characterized as AIMs (Ancestry-Informative Marker), allows a potential use of these systems in population and forensic studies in which analysis of the overall and individual interethnic admixture proportions are needed (see Supplementary Table 2, Online Resource).
The genetic ancestry for the three parental ethnic groups, African, European, and Native American, calculated for the forensic samples analyzed were estimated at 0.328 ± 0.0115, 0.29 ± 0.0131, and 0.39 ± 0.0113, respectively. The individual admixture components estimated for this samples varied substantially—between 8% and 81% for African DNA, from 11% to 48% for European DNA, and from 8% to 72% for Native American DNA (Tables 4 and 5).
In another study [5], we estimated the genetic ancestry and the forensic parameter of 130 inhabitants of the same Brazilian Amazon population. The averages calculated were polymorphism information content (PIC) = 33.3%, power of discrimination (PD) = 56.7%, power of exclusion (PE) = 14.2%, observed heterozygozity (Ho) = 42%. The power of discrimination and the power of exclusion for the 48 INDELs studied were 99.9999999999999998035% and 99.9456321%, respectively (see Supplementary Table 3, Online Resource).
Conclusion
In conclusion, the use of this 48 INDEL polymorphisms that are informative of ancestry and that can be genotyped using three multiplex PCRs and gel electrophoresis, with small length variation between alleles constitutes a valuable approach in forensic genetics, gathering advantageous characteristics similar to those of SNPs (e.g., short-amplicon analysis and low mutation rates) while at the same time keeping the chemistry and simple workflow already established for STRs in all forensic labs.
References
Weber JL, David D, Heil J, Fan Y, Zhao C, Marth G (2002) Human diallelic insertion/deletion polymorphisms. Am J Hum Genet 71:854–862
Pereira R, Phillip C, Alves C, Amorim A, Carracedo A, Gusmão L (2009) Insertion/deletion polymorphisms: a multiplex assay and forensic applications. Forensic Sci Int Genetics Suppl 2:513–515
Pereira R, Phillips C, Alves C, Amorim A, Carracedo A, Gusmão L (2009) A new multiplex for human identification using insertion/deletion polymorphisms. Electrophoresis 30:3682–3690
Santos NPC, Ribeiro-Rodrigues EM, Ribeiro-dos-Santos AKC, Pereira R, Gusmão L, Amorim A, Guerreiro J, Zago M, Matte C, Hutz MH, Santos SEB (2010) Assessing individual interethnic admixture and population substructure using a 48–insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat 31:184–190
Francez PAC, Ribeiro-Rodrigues EM, Santos SEB (2011) Allelic frequencies and statistical data obtained from 48 AIM INDEL loci in an admixed population from the Brazilian Amazon. Forensic Sci Int Genet. doi:10.1016/j.fsigen.2011.04.002
Phillips C, Fondevila M, Garcia-Magarinos M, Rodriguez A, Salas A, Carracedo A, Lareu MV (2008) Resolving relationship tests that show ambiguous STR results using autosomal SNPs as supplementary markers. Forensic Sci Int Genet 2:198–204
Weber JL, Wong C (1993) Mutation of human short tandem repeats. Hum Mol Genet 2:1123–1128
Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304
Comey CT, Koons BW, Presley KW, Smerick JB, Sobieralski CA, Stanley DM, Baechtel FS (2004) DNA extraction strategies for amplified fragment length polymorphism analysis. J Forensic Sci 39:1254–1269
Sambrook J, Fritsch EF, Maniatis T (1989) Isolations of DNA from mammalian cells. In: Ford N, Nolan C, Ferguson M (eds) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York, pp 916–919
Sweet D, Lorente M, Valenzuela A, Lorente JA, Alvarez JC (1996) Increasing DNA extraction yield from saliva stains with a modified Chelex method. Forensic Sci Int 83:167–177
Acknowledgments
The authors wish to thank the assistance of the Criminal Experts Ciro Fernandes de Oliveira Penido and José Maria Ferreira Faro in the extraction and quantification of DNA from forensic samples used in this study. There was no financial association that might pose any potential conflict of interest to disclose.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
da Costa Francez, P.A., Rodrigues, E.M.R., de Velasco, A.M. et al. Insertion–deletion polymorphisms—utilization on forensic analysis. Int J Legal Med 126, 491–496 (2012). https://doi.org/10.1007/s00414-011-0588-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-011-0588-z