Abstract
DNA typing of 8 recently described STRs on the Y chromosome was carried out by means of 2 multiplex amplification reactions for 134 unrelated males from Cantabria, a region in northern Spain. Multiplex 1 included loci DYS460 (GATA A7.1), GATA A10, GATA H4 and DYS439; multiplex 2 included DYS461 (GATA A7.2), GATA C4, DYS437 and DYS438. Haplotype diversity was found to be 99.36%, similar to that obtained with the standard 9-STR set ("minimal haplotype") of the European Y-user group (99.35%). The 13-locus haplotype resulting from the combination of the standard minimal haplotype and the 4-locus multiplex 1 showed a 99.89% diversity. Further inclusion of the 4 loci in multiplex 2 resulted in a haplotype diversity of 99.93%. The combination of the "minimal haplotype" and the multiplex 1 in the present study may be an efficient way of increasing the power of discrimination in forensic cases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polymorphisms on the Y chromosome have become a very useful tool in forensic and population genetics. A number of short tandem repeat (STR) polymorphisms have been widely studied in Europe and other areas, and large databases have been made publicly available [1]. Thus, population frequency data of a standard set of STRs combined in a so-called minimal haplotype are now available through the internet database of the Y-user group [1]. Such a haplotype is established by typing 9 loci: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, and DYS385I-II. Other STRs have also been recently suggested as interesting for forensic purposes [2, 3, 4]. However, given the linkage among different loci on the non-recombining part of the Y chromosome, it is unclear if they add useful information to the widely studied minimal haplotype, or result in redundant information.
Therefore, we planned to study allele population distribution and forensic power of a recently described set of eight STRs, and compare them with the minimal haplotype.
Materials and methods
Subjects
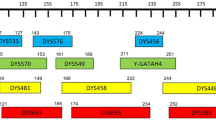
Unrelated male individuals (n=134) living in Cantabria, a region in northern Spain with a population about 500,000, were studied. Genomic DNA was extracted from peripheral blood by a commercial method according to the manufacturer's instructions (Qiagen, Germany) and quantified by light absorbance (Genequant, Pharmacia, Sweden). Aliquots containing 10–30 ng DNA were used to amplify the regions of interest by PCR, with the primers reported by González-Neira et al. [4], except for loci DYS460 (GATA A7.1), DYS461 (GATA A7.2) and GATA H4. These loci were amplified with new primers designed to decrease the size range of the alleles, as recently reported in the collaborative exercise by the GEP-ISFG (Spanish and Portuguese Group of the International Society of Forensic Genetics) [5, 6] (Table 1). Two multiplex reactions were carried out. Multiplex 1 included FAM-labelled primers to amplify GATA A7.1 (0.3 μM), GATA A10 (0.2 μM), GATA H4 (0.5 μM),and DYS439 (0.2 μM). Multiplex 2 contained TET-labelled primers to amplify GATA A7.2 (0.2 μM), GATA C4 (0.5 μM), DYS437 (0.16 μM), and DYS438 (0.5 μM). Both reaction mixes contained 2 mM MgCl2, 200 μM dNTPs and 0.5 U Taq Gold polymerase (Applied Biosystems, USA) in a 25 μl volume. Cycling conditions for both multiplexes were: pre-incubation at 95° for 7 min, followed by 32 cycles of 30 s at 94°, 20 s at 60° and 30 s at 70°, with a final extension step of 45 min at 70°C.
DYS385 locus was analysed with the protocol proposed by Schneider et al. [7]. Other loci in the minimal haplotype were amplified with the primers described by Gusmão et al. and Kayser et al. [8, 9, 10], as previously reported [11].
Amplicon analysis
Aliquots containing 1.2 μl of PCR product were mixed with 24 μl formamide and 1 μl TAMRA as a internal size standard, heated for 5 min at 95°C, quenched in an ice bath for 10 min and injected into an ABI 310 capillary electrophoresis system (Applied Biosystems). Amplicon size was determined with the local Southern method implemented in Genescan software. Allele designation followed the ISFG and other previously published recommendations [12, 13]. Sequenced alleles were used as controls.
Data analysis
Allele and haplotype frequencies were estimated with Arlequin software (Schneider et al, University of Geneva; available at http://anthro.unige.ch/arlequin). Locus and haplotype diversities were calculated as (1-ΣPi2)(n/n-1), where Pi is the allele or haplotype frequency [14]. The haplotype matching probability was estimated as 1−haplotype diversity.
Results and discussion
Allele frequencies of the loci included in multiplex 1 are shown in Table 2; and those of loci included in multiplex 2 in Table 3. If only the 4 loci included in multiplex 1 were considered, 59 different haplotypes were found among the 134 subjects studied, whereas multiplex 2 determined 41 different haplotypes. Thus, loci included in multiplex 1 were more useful for discrimination purposes. The combined analysis of both multiplexes resulted in 98 different haplotypes: 2 haplotypes were found in 6 individuals, 1 haplotype in 5, 6 in 3, 10 in 2, and 79 in a single individual. Haplotype diversity was 0.973 for multiplex 1, 0.891 for multiplex 2, and 0.992 for the combined 8-locus haplotype.
Allele frequency distributions in this population group were similar to the distribution in other Iberian populations [6, 12], but different from the Oriental ones [15].
A population subset (n=104) was also genotyped for the standard minimal haplotype, in order to compare haplotype diversity of the different allele combinations (population data for some of those loci have been previously published [11]). The results obtained are shown in Tables 4 and 5. A similar haplotype diversity was obtained with the combined two newer multiplexes reported here, and with the standard minimal haplotype. Adding allele information derived from multiplex 1 to the standard minimal haplotype increased discrimination ability, but further addition of multiplex 2 resulted in only a small gain (Table 5).
The lack of recombination and the subsequent linkage among different loci on the Y chromosome means that marginal utility (i.e., the gain of discrimination power attained by including new loci) decreases as more STRs are included into the analysis. The ideal number of loci to study has not been firmly established, and is probably different depending on the purposes of the study and, in forensic cases, on the availability of other genetic data. Certainly, there is some degree of compromise between the power of discrimination attained and the cost and effort of allele typing.
In the present study, the power of discrimination was similar to that found in a recent European study by Bosch et al. who reported a minimal haplotype diversity of 0.9896, which increased to 0.9988 after including data from 10 additional loci [16]. Somewhat larger diversity values were found in Asian populations. Tsai et al. reported a 0.9999 diversity for the minimal haplotype plus the DYS388 locus in Taiwan [17]. Shin et al. found that a combined haplotype including the minimal loci plus DXYS156 and DYS388 displayed a 0.9995 diversity in Korea [18].
Our results suggest that the 9-locus standard minimal haplotype, as defined by the Y-user group [1] and the 8-locus haplotype determined by using multiplexes 1 and 2 reported here have a similar power of discrimination. Both are likely to be discriminative enough in many routine cases. Combining the minimal haplotype with the multiplex 1 (DYS460, GATA A10, GATA H4, DYS 439) into a 13-locus Y-STR set further improves discrimination power, up to a figure similar to that recently reported for the combined analysis of 19 loci [16]. However, little is gained by the further inclusion of loci in multiplex 2.
References
Roewer L, Krawczak M, Willuweit S et al. (2001) Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci Int 118:106–113
Ayub Q, Mohyuddin A, Qamar R, Mazhar K, Zerjal T, Mehdi SQ, Tyler-Smith C (2000) Identification and characterisation of novel Y-chromosomal microsatellites from sequence database information. Nucleic Acids Res 28:e8
White PS, Tatum OL, Deaven LL, Longmire JL (1999) New, male-specific microsatellite markers from the human Y chromosome. Genomics 57:433–437
González-Neira A, Elmoznino M, Lareu MV, Sánchez-Diz P, Gusmão L, Prinz M, Carracedo A (2001) Sequence structure of 12 novel Y chromosome microsatellites and PCR amplification strategies. Forensic Sci Int 122:19–26
Sánchez-Diz P, Gusmão L, Beleza S et al. (2003) Results of the GEP-ISFG collaborative study on two Y-STR tetraplexes: GEPY I (DYS461, GATA C4, DYS437 and DYS438) and GEPY II (DYS460, GATA A10, GATA H4, DYS439). Forensic Sci Int, in press
Beleza S, Alves C, González-Neira A, Lareu M, Amorim A, Carracedo A, Gusmão L (2003) Extending STR markers in Y chromosome haplotypes. Int J Legal Med 117:27–33
Schneider PM, Meuser S, Waiyawuth W, Seo Y, Rittner C (1998) Tandem repeat structure of the duplicated Y-chromosomal STR locus DYS385 and frequency studies in the German and three Asian populations. Forensic Sci Int 97:61–70
Gusmão L, Gonzalez-Neira A, Pestoni C, Brion M, Lareu MV, Carracedo A (1999) Robustness of the Y STRs DYS19, DYS389 I and II, DYS390 and DYS393: optimization of a PCR pentaplex. Forensic Sci Int 106:163–172
Gusmão L, Gonzalez-Neira A, Sanchez-Diz P, Lareu MV, Amorim A, Carracedo A (2000) Alternative primers for DYS391 typing: advantages of their application to forensic genetics. Forensic Sci Int 112:49–57
Kayser M, Caglia A, Corach D et al. (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110:125–133
Zarrabeitia MT, Riancho JA, Sánchez-Diz P, Sánchez-Velasco P (2001) 7-Locus Y chromosome haplotype profiling in a northern Spain population. Forensic Sci Int 123:78–80
Gusmão L, Alves C, Beleza S, Amorim A (2002) Forensic evaluation and population data on the new Y-STRs DYS434, DYS437, DYS438, DYS439 and GATA A10. Int J Legal Med 116:139–147
Gusmão L, Gonzalez-Neira A, Lareu MV, Costa S, Amorim A, Carracedo A (2002) Chimpanzee homologous of human Y specific STRs. A comparative study and a proposal for nomenclature. Forensic Sci Int 126:129–136
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Hou YP, Zhang J, Li YB, Wu J, Zhang SZ, Prinz M (2001) Allele sequences of six new Y-STR loci and haplotypes in the Chinese Han population. Forensic Sci Int 118:147–152
Bosch E, Lee AC, Calafell F, Arroyo E, Henneman P, Knijff P de, Jobling MA (2002) High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forensic Sci Int 125:42–51
Tsai LC, Yuen TY, Hsieh HM, Lin M, Tzeng CH, Huang NE, Linacre A, Lee JCI (2002) Haplotype frequencies of nine Y-chromosome STR loci in the Taiwanese Han population. Int J Legal Med 116:179–183
Shin DJ, Jin HJ, Kwak KD et al. (2001) Y-chromosome multiplexes and their potential for DNA profiling in Koreans. Int J Legal Med 115:109–117
Acknowledgement
Supported in part by a grant from Fundación Marqués de Valdecilla.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zarrabeitia, M.T., Riancho, J.A., Gusmão, L. et al. Spanish population data and forensic usefulness of a novel Y-STR set (DYS437, DYS438, DYS439, DYS460, DYS461, GATA A10, GATA C4, GATA H4). Int J Legal Med 117, 306–311 (2003). https://doi.org/10.1007/s00414-003-0393-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-003-0393-4