Abstract
Validation studies were carried out using the commercially available PCR multiplex system genRESMPX-2. In addition to amelogenin, this system comprises the complete set of eight STR systems which are components of the German DNA database established in 1998 by the Federal Criminal Office of Germany (BKA). The minimum amount of template DNA which gave a complete DNA pattern ranged between 100 pg and 200 pg. Mixed samples could clearly be assigned from ratios between 1:5 (ACTBP2) and 1:20 (VWA, FGA). Experimental investigations with different forensic materials, environmental studies, reproducibility and precision data as well as practical casework analysis revealed that the genRESMPX-2 kit can be regarded as a sensitive, reliable and robust multiplex system even in the case of samples containing limited amounts or degraded DNA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the past a variety of different PCR multiplex systems were established for forensic purposes, which allow the simultaneous amplification of different STR (short tandem repeat) loci and which are a powerful and rapid method for individual identification (Cotton et al. 2000; Lederer et al. 2000; Frank et al. 2001). Such systems allow the reduction of time, cost and material and, therefore, permit the maximization of the forensic expressiveness even in cases of limited evidential material (Kimpton et al. 1993; Urquhart et al. 1995). The number of loci which can be amplified in a single reaction has increased up to 15 autosomal STR systems plus the sex-specific amelogenin system (Oldroyd et al. 1995; Cotton et al. 2000; Finis 2001; Moretti et al. 2001). Most of the systems have been developed on the basis of fluorescence detection (Lins et al. 1996; Moretti et al. 2001) and are constructed to fulfil the demands for usage in the Combined DNA Indexing System (CODIS; Budowle et al. 1998) representing the standard for forensic applications in the USA (Budowle and Sprecher 2001).
Following the TWGDAM (Technical working group on DNA analysis methods) guidelines (Budowle 1995), validation studies were carried out with the genRESMPX-2 kit. This kit comprising the systems VWA, ACTBP2, TH01, D21S11, D8S1179, D3S1358, FGA, D18S51 and amelogenin, includes all loci of the German DNA database established in 1998 by the Federal Criminal Office of Germany (BKA). Detailed information will be given concerning sensitivity, specificity, reproducibility, robustness and forensic applicability.
Materials and methods
Materials
Validation studies were carried out with blood and saliva samples from paternity cases, micro-bloodstains, tissue samples, samples of external trials of the German DNA profiling group (GEDNAP) and casework samples.
DNA extraction and quantitation
DNA was extracted from blood samples using either Chelex 100 (Walsh et al. 1991) or the salting-out method according to Miller et al. (1988). Extraction of saliva samples, blood and saliva stains was performed using Chelex 100 with additional proteinase K digestion. With the exception of blood and saliva stains, DNA was extracted from casework and tissue samples using the All-tissue DNA kit (GEN-IAL, Troisdorf, Germany). DNA quantitation was performed using the ACES 2.0 Human DNA Quantitation system (GIBCO BRL, Life Technologies, Gaithersburg, MD).
Amplification
Depending on the different extraction methods the following amounts of template DNA were used:
-
1.
1–5 µl out of 200 µl extract solution for the Chelex 100 method
-
2.
1–5 µl out of 50 µl for the GEN-IAL extraction method
-
3.
1–2 ng of spectrophotometrically quantified DNA.
PCR was performed as recommended by the manufacturer. Amplification was carried out in a TRIO-Thermoblock (Biometra, Göttingen, Germany). Primer sequences are available on request from Serac GmbH, Bad Homburg, Germany.
Electrophoresis
Electrophoresis was carried out on an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Darmstadt, Germany) using a 47 cm×50 µm ID capillary with the polymer POP4 or POP6. Electrophoresis was done using the module GS STR POP6 A or GS STR POP4 A, depending on the polymer used. From each sample 1–5 µl was mixed with 12 µl HiDi-formamide (Applied Biosystems) and 0.5 µl of the internal standard genRES LS500ROX. After denaturation (5 min, 95°C) the samples were cooled on ice and immediately transferred to the autosampler. The data were collected using the ABI Prism 310 collection software and analyzed with the 310 GeneScan software (version 2.1).
Studies on fresh blood and saliva samples
The genotypes of 57 blood and 66 saliva samples were determined using to the above mentioned extraction, amplification and analysis parameters.
Sensitivity and mixture studies
Sensitivity studies were carried out with different amounts of quantified human DNA samples ranging from 5 to 0.05 ng. For mixture studies, 5–0.05 ng of one sample were added to a constant amount (5 ng) of another sample resulting in mixture ratios of 1:1, 2:1, 5:1, 10:1, 20:1, 50:1 and 100:1.
External trials of the German DNA Profiling Group (GEDNAP)
The samples of the external trials 20, 21, 22 and 23 of the German DNA Profiling Group (GEDNAP; Rand et al. 2002) were analysed. Each trial consisted of dried blood samples on cotton from three different individuals and four stains of various origins. The DNA was extracted using the Chelex 100 method.
Studies on tissue samples
Fresh tissue samples from autopsies (n=3, muscle and liver), which were frozen at −20°C prior to analysis were used. Extraction of approximately 100 mg of each sample was performed using the GEN-IAL extraction kit according to the standard protocol. In a first approach 1 µl of the extract was analysed and in a second approach the DNA extract was diluted 1:2, 1:10, 1:50 and 1:100. Blood samples of the respective corpses were used for comparisons of the amplification patterns.
Environmental studies using different extraction methods
Human DNA from 8 blood stains on cotton, stored in dark and dry place at room temperature and 37°C (drying chamber) for 6 years, were extracted using Chelex100 and the GEN-IAL extraction kit, 2 µl of each extract was used for analysis.
Reproducibility studies
Intra-day reproducibility was tested by 10 re-injections of 1 sample using a new capillary (0 injections) and a capillary with 250 injections. The inter-day reproducibility was tested by injection of one freshly prepared DNA sample over a period of time using the same capillary, but with an increasing number of injections (0–300 injections). In addition, 1 ng of 1 DNA sample was amplified 10 times using identical amplification and electrophoresis conditions. The precision was calculated according to Frazier et al. (1996).
Non-human studies
DNA samples from the following non-human animals were analysed: primates including gorilla (1 male, 1 female), chimpanzee (1 male, 1 female) and orangutan (1 male, 1 female) and non-primates including tiger (n=3), dog, horse (n=2), cow, pig, condor (n=4) and wild boar. The samples of the primates were kindly provided by the Institute of Legal Medicine, Münster, Germany, and those of the non-primates by BIOPSYTEC AG (Rheinbach, Germany). Amplification of 2–5 ng of primate DNA and 1–50 ng of non-primate DNA was carried out according to the standard PCR protocol.
Stutter intensities and allele balance
Genotypes of 80 human individuals were determined as described above. The stutter intensity was calculated from 42–65 genotypes at each locus. In the case of heterozygous genotypes, alleles were at least two repeat units apart (8 bp). Relative stutter peak heights were calculated by dividing the peak height at the stutter position by the height of the associated allele signal. Heterozygous allele balance of 33–59 heterozygous genotypes was calculated by dividing the height of the second peak by the height of the first allele signal. The total signal height used for calculation was <4500 relative fluorescence units (rfu).
Casework samples
A total of 11 cases with 59 different samples were examined using the genRESMPX-2 kit. These cases included fresh blood samples (n=2), blood samples from corpses (n=3), fresh saliva samples on buccal swabs (n=11), bloodstains on different matrices (n=16), muscle (n=10), humerus (n=1), femur (n=1), saliva stains on tooth brushes (n=3) on cigarettes (n=3) and on bottlenecks (n=3), hair roots (n=3), epithelial cells of the skin (n=1) and remnants of a meal (n=2). The age of the samples ranged from 16 days up to 33 months.
Results and discussion
Blood and saliva samples
A total of 57 blood and 66 saliva samples were investigated in order to obtain experience with amplification patterns depending on the amount of template DNA used. With the exception of ACTBP2, all loci showed homogeneous amplification for both types of samples. In a few cases, additional signals appeared within the allele range of the ACTBP2-locus which did not occur using a singleplex amplification of this locus (data not shown). These non-specific signals could be eliminated by further optimisation of the buffer composition and without influencing the overall performance of the assay. For this reason, the buffer originally used has been replaced by the improved one, which is now an inherent part of the genRESMPX-2 kit.
Amplification of more than 5 ng of template DNA led to non-specific peaks especially at loci with shorter amplicon length. In addition, fragment lengths of alleles could not be accurately assigned as the detection system became saturated. Artefacts such as incomplete 3’-adenosine addition, stutter and poor spectral and electrophoretic separation were observed when conditions were applied (e.g. too much template DNA) which resulted in peak heights greater than 4500 rfu.
Sensitivity and mixture studies
All alleles could be detected and assigned to the allelic ladder down to 100 pg template DNA (Table 1). Therefore, the overall sensitivity of the genRESMPX-2 kit was at least as high as that of the PowerPlex 16 system (>250 pg, Finis 2001), the AmpFlSTR Profiler Plus/AmpFlSTR Cofiler multiplex system (>160 pg, LaFountain et al. 2001), the AmpFlSTR Blue amplification kit (>250 pg, Wallin et al. 1998) and the MPX-1 multiplex kit (>125 pg, Lederer et al. 2000). Nevertheless, preferential amplification and low signal intensities of 100–150 rfu were partly observed applying DNA levels below 200 pg. Although these signals could be clearly distinguished from the background and, therefore, can be assigned as real alleles, these phenomena should be kept in mind especially for the interpretation of forensic samples containing limited amounts of DNA.
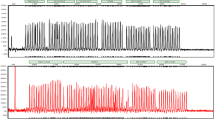
In studies with different mixture ratios the detection limit of the minor component greatly depended on the locus. The ratio ranged from 1:5 (ACTBP2) to 1:20 (VWA and FGA; Fig. 1) and was similar to that of other commercial multiplex systems (PowerPlex16 1:10/AmpFlSTR Blue 1:10–1:30/MPX-1 1:20).
GEDNAP trials
The examination of reference samples and the stain material of several GEDNAP trials showed consistent and typable results (Fig. 2). Saliva stains on three cigarettes, one buccal swab, one chewing gum and one stamp as well as the bloodstains gave full and homogeneous DNA profiles. The two cigarettes (one with 10 µl saliva and one smoked) showed preferential amplification and allele drop-out for the ACTBP2 system. Blood mixtures of two different persons with mixture ratios of 3:1 and 4:1 were detected and full DNA profiles could be obtained.
Analysis of the stains 5 (A bloodstain on cotton), 6 (B bloodstain on cotton consisting of two components, mixture ratio 4:1), 7 (C secretion stain consisting of saliva and sperm, mixture ratio 3:1) and 8 (D saliva stain on a cigarette filter) from GEDNAP 23. For amplification of stains 5 and 6, 2 µl was used and 5 µl was used for stains 7 and 8. Only the loci TH01, D21S11 and D8S1179 are shown
Tissue samples
Studies were carried out on fresh muscle (n=3) and liver (n=3) samples. With the exception of the ACTBP2 locus, all loci showed a homogeneous allele pattern with balanced peak heights over a broad range of template DNA. Insufficient amplification was observed at the ACTBP2 locus resulting in low signal intensities, ladder bands or extra bands. In order to get a clear pattern at this locus, 1:2, 1:50 and 1:100 dilutions of the extracts were prepared and amplified. While none of the dilutions gave satisfactory results with the multiplex system, amplification in a singleplex PCR showed clear amplification products at this locus.
Environmental studies and different extraction methods
Exposure of forensic specimens to different environmental conditions can greatly affect the integrity of the DNA samples derived from this material. In addition, DNA purity may additionally influence the sensitivity of the assay. For this reason stains which were stored under different conditions were analysed using two different extraction methods.
In all cases, storage at 37°C for 6 years followed by Chelex100 extraction resulted in a complete drop-out of all systems. When the same stains were extracted with the All-tissue DNA extraction kit, allelic signals could be observed. Nevertheless, peak heights only ranged between 100 and 700 rfu and, in addition, preferential amplification, allelic drop-out and locus drop-out were observed especially for STR systems with higher amplicon lengths.
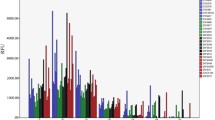
The analysis of bloodstains stored at room temperature for 6 years was successful for DNA extracted with the All-tissue DNA extraction kit. Only one DNA sample extracted with this kit showed a locus drop-out at the ACTBP2-locus. DNA samples extracted with Chelex100 showed one locus drop-out at each of the loci D21S11 and D3S1358, two at D8S1179 and three at ACTBP2, FGA and D18S51 (Fig. 3).
Analysis of a 6-year-old bloodstain stored at room temperature. Only the sex-specific system amelogenin (AMG) and the STR loci VWA and ACTBP2 are shown. Amplification pattern of the control DNA K562 (A). Extraction was carried out using Chelex 100 (B) and the All-tissue DNA kit (C), 2 µl of each extract was amplified using the genRESRMPX-2 kit
Reproducibility and precision
In Tables 2 and 3 reproducibility and precision data for amplification and subsequent analysis are summarised. The mean values of the standard deviation for calculated fragment lengths ranged between 0.02 and 0.03 bp. Similar values have recently been determined by Finis (2001) and LaFountain et al. (2001). The precision of the fragment length calculation was always above 99.98% and independent of the fragment length itself. With the exception of ACTBP2 the average peak height ratio for each system lay between 0.85 and 1.01. This demonstrates that alleles were amplified to a similar extent and without significant preference. Similar observations have recently been reported for other multiplex systems (Frank et al. 2001; LaFountain et al. 2001; Moretti et al. 2001). The relative peak height ratios of the ACTBP2 locus indicate the occurrence of preferential amplification which may, at least in part, be a consequence of heterozygous alleles which differ greatly in size. The minimum and maximum precision value of the relative peak height ratios ranged between 88% and 99% indicating that heterozygous alleles of one locus were amplified in a reliable manner. The relative peak heights between different loci were calculated in order to get information about the homogeneity of simultaneous amplification of alleles of different pairs of loci. With the exception of the systems VWA/ACTBP2 all other peak height ratios varied between 1.6 (TH01/D21S11) and 0.83 (D3S1358/D18S51). For the combination VWA/ACTBP2 the peak height ratios were determined to be 0.46 (allele 1) and 0.38 (allele 2). This demonstrates that although the VWA locus was not over-amplified (3000 rfu) the ACTBP2 alleles were amplified with comparatively less efficiency.
Species specificity
Non-human studies were carried out to evaluate the species specificity of the genRES MPX-2 kit. No reproducible signals were found using DNA amounts of 0.5–2 ng. Nevertheless, using more than 15 ng of primate or non-primate DNA, amplicons within the range of 100 bp–350 bp were found for samples from pig, orang-utan, chimpanzee and gorilla. However, only few of the detectable signals could be assigned to the allelic ladder. In addition, amplicons were confined to individual loci and, therefore, no characteristic profiles could be obtained. Similar observations have recently been described for several other multiplex systems (Sparkes et al. 1996; Wallin et al. 1998; LaFountain et al. 2001; Levedakou et al. 2002). At least for primates, the appearance of these amplification products may be attributed to a partial homology within the primer binding regions of the same loci (Wallin et al. 1998). As a consequence, interpretation of samples which may contain DNA of unknown species should be done carefully. Nevertheless, the risk of wrong typing due to the existence of non-human DNA seems to be unlikely for the following reasons: firstly, DNA amounts of more than 15 ng, with which additional signals exclusively appeared, are not normally used for PCR amplification. Secondly, the respective signals could not be regarded as characteristic human profiles because of severe allelic imbalance and occurrence at isolated loci.
Stutter intensity and allele balance
In Table 4 (left half) the relative intensities of stutter signals are shown for each of the amplified STR loci. The lowest average percentage stutter was observed for the TH01 system (3.6±1.4%). The highest value was found at the ACTBP2 locus (12.0±3.0%). For all loci an average stutter of 9.4% was determined. Nevertheless, in individual cases, relative intensities of up to about 19% (ACTBP2, D21S11 and D3S1358) were detected. The average peak balance is summarised in the right half of Table 4. The values ranged from a low of 50.9% to a high of 138.8%, both observed at the ACTBP2 locus. The average peak height ratio for all loci was 90.2%.
The occurrence of stutter bands and unbalanced signal intensities may complicate the interpretation of mixed specimens containing DNA from more than one person. Stutter peaks, which are 4 bp shorter than the attendant allele peak, may concur with the main signal of any 4 bp shorter allele of a minor component. Consequently, it might not be possible to distinguish the respective allele peak from the stutter peak at the same position. The average relative stutter intensity of all loci tested in this study was 9.4±2.5%. Nevertheless, upper and lower range values greatly varied between the different loci, which may be a consequence of the individual sequence composition of the repeat structures (Lazaruk et al. 2001). To facilitate quantitative interpretation of stutter and minor mixture components, a conservative threshold can be calculated adding the average percentage stutter observed at a locus and the 3-fold standard deviation. For the genRESMPX-2 amplified loci these limits would range from 7.8% for the TH01 locus to 21% for the ACTBP2 system. As a consequence, although the detection of minor components of as low as 5% is theoretically possible, the correct interpretation depends on the analysed locus as well as the number of contributors and alleles in stutter peak positions.
Not only the presence of more than two alleles per locus but also a considerable difference in signal intensity may indicate concomitance of more than one individual. Nevertheless, efficiency of the amplification of two heterozygous alleles within one locus may differ substantially, thereby complicating the interpretation of the results. On average, the observed peak height ratio of the genRESMPX-2 system was 90%, indicating that amplification of the second (longer) allele is slightly worse than the first allele. This effect was especially pronounced at the ACTBP2 locus, where heterozygous alleles may greatly differ in size. This observation can mainly be seen as a consequence of preferential amplification as it has also been shown for other loci spanning a long range of fragment sizes (Walsh et al. 1992, 1996). The minimal observed peak height ratio was 51% at the ACTBP2 locus, whereas values of >58% appeared at all other loci. Except for the ACTBP2 system, similar ratios have recently been found for other multiplex PCR systems (Moretti et al. 2001; LaFountain et al. 2001). Because peak height ratios may vary within individual amplification reactions (Moretti et al. 2001) repeated analysis can be a reasonable way to ensure reliable interpretation of the results particularly when samples containing limited amounts of DNA have to be analysed. In these cases, stochastic effects during amplification have to be taken into account (Budowle et al. 1995), possibly resulting in unbalanced signal height or, even worse, allele drop-out.
Casework samples
A total of 11 cases with 59 different samples were analysed using the genRESMPX-2 kit. The results are summarised in Table 5. The profiles were closely inspected with respect to allele and/or locus drop out, non-allelic peaks and stutter bands. Only the amplification of one saliva stain on a bottleneck completely failed using the genRESMPX-2 kit. Interpretation of the ACTBP2 amplification pattern in part was complicated by the occurrence of non-specific signals. However, these artefacts could be eliminated with a new buffer system.
References
Budowle B (1995) Technical working group on DNA analysis methods. Guidelines for a quality assurance program for DNA analysis. Crime Lab Digest 22:21–43
Budowle B, Sprecher CJ (2001) Concordance study on population database samples using PowerPlexTM 16 kit and AmpFlSTR Profiler Plus kit and AmpFlSTR Cofiler kit. J Forensic Sci 46:637–641
Budowle B, Lindsey JA, DeCou JA, Koons BW, Giusti AM, Comey CT (1995) Validation and population studies of the loci LDLR, GYPA, HBGG, D7S8, and Gc (PM loci), and HLA-DQ alpha using a multiplex amplification and typing procedure. J Forensic Sci 40:45–54
Budowle B, Moretti TR, Niezgoda SJ, Brown BL (1998) CODIS and PCR-based short tandem repeat loci: law enforcement tools. In: Proceedings of the Second European Symposium on Human Identification. Promega Corporation, Madison, WI, pp 73–88
Cotton EA, Allsop RF, Guest JL, Frazier RRE, Koumi P, Callow IP, Seager A, Sparkes RL (2000) Validation of the AmpFlSTR SGM Plus system for use in forensic casework. Forensic Sci Int 112:151–161
Finis CJ (2001) Megaplex analysis from a single amplification: validation of the PowerPlexR16 system. Profiles DNA 4:3–6
Frank WE, Llewellyn BE, Fish PA et al. (2001) Validation of the AmpFlSTRTM Profiler Plus PCR Amplification kit for use in forensic casework. J Forensic Sci 46:642–646
Frazier RRE, Millican ES, Watson SK et al. (1996) Validation of the Applied Biosystems PrismTM 377 automated sequencer for forensic short tandem repeat analysis. Electrophoresis 17:1550–1552
Kimpton C, Gill P, Walton A, Urquhart A, Millican ES, Adams M (1993) Automated DNA profiling employing multiplex amplification of short tandem repeat loci. PCR Methods Appl 3:13–22
LaFountain MJ, Schwartz MB, Svete PA, Walkinshaw MA, Buel E (2001) TWGDAM validation of the AmpFlSTR Profiler Plus and AmpFlSTR COfiler STR multiplex systems using capillary electrophoresis. J Forensic Sci 46:1191–1198
Lazaruk K, Wallin J, Holt C, Nguyen T, Walsh PS (2001) Sequence variation in humans and other primates at six short tandem repeat loci used in forensic identity testing. Forensic Sci Int 119:1–10
Lederer T, Seidl S, Graham B, Betz P (2000) A new pentaplex PCR system for forensic casework analysis. Int J Legal Med 114:87–92
Levedakou EN, Freeman DA, Budzynski MJ et al. (2002) Characterization and validation studies of PowerPlex 2.1, a nine-locus short tandem repeat (STR) multiplex system and penta D monoplex. J Forensic Sci 47:757–772
Lins AM, Sprecher CJ,Puers C, Schumm JW (1996) Multiplex sets for the amplification of polymorphic short tandem repeat loci—silver stain and fluorescence detection. Biotechniques 20:882–889
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Moretti TR, Baumstark AL, Defenbaugh DA, Keys KM, Smerick JB, Budowle B (2001) Validation of short tandem repeats (STRs) for forensic usage: performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. J Forensic Sci 46:647–660
Oldroyd NJ, Urquhart AJ, Kimpton CP, Millican ES, Watson SK, Downes T, Gill PD (1995) A highly discriminating octoplex short tandem repeat polymerase chain reaction system suitable for human individual identification. Electrophoresis 16:334–337
Rand S, Schürenkamp M, Brinkmann B (2002) The GEDNAP (German DNA profiling group) blind trial concept. Int J Legal Med 116:199–206
Sparkes R, Kimpton C, Watson S et al. (1996) The validation of a 7-locus multiplex STR test for use in forensic casework (I). Mixtures, ageing, degradation and species studies. Int J Legal Med 109:186–194
Urquhart A, Oldroyd NJ, Colin PK, Gill P (1995) Highly discriminating heptaplex short tandem repeat PCR system for forensic identification. Biotechniques 18:116–121
Wallin JM, Buoncristiani MR, Lazaruk KD, Fildes N, Holt CL, Walsh PS (1998) TWGDAM validation of the AmpFlSTR Blue PCR amplification kit for forensic casework analysis. J Forensic Sci 43:854–870
Walsh PS, Metzger DA, Higuchi R (1991) ChelexR 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Walsh PS, Erlich HA, Higuchi R (1992) Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl 1:241–250
Walsh PS, Fildes NJ, Reynolds R (1996) Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res 24:2807–2812
Acknowledgements
The authors wish to acknowledge Marianne Steevens and Waltraud Hargesheimer for helpful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Junge and T. Lederer contributed equally to the data presented
Rights and permissions
About this article
Cite this article
Junge, A., Lederer, T., Braunschweiger, G. et al. Validation of the multiplex kit genRESMPX-2 for forensic casework analysis. Int J Legal Med 117, 317–325 (2003). https://doi.org/10.1007/s00414-003-0385-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-003-0385-4