Abstract
In eukaryotic phyla studied so far, the essential centromeric histone H3 variant (CENH3) is loaded to centromeric nucleosomes after S-phase (except for yeast) but before mitotic segregation (except for metazoan). While the C-terminal part of CENH3 seems to be sufficient for mitotic centromere function in plants, meiotic centromeres neither load nor tolerate impaired CENH3 molecules. However, details about CENH3 deposition in meiocytes are unknown (except for Drosophila). Therefore, we quantified fluorescence signals after the immunostaining of CENH3 along meiotic and mitotic nuclear division cycles of rye, a monocotyledonous plant. One peak of fluorescence intensity appeared in the early meiotic prophase of pollen mother cells and a second one during interkinesis, both followed by a decrease of CENH3. Then, the next loading occurred in the male gametophyte before its first mitotic division. These data indicate that CENH3 loading differs between mitotic and meiotic nuclei. Contrary to the situation in mitotic cycles, CENH3 deposition is biphasic during meiosis and apparently linked with a quality check, a removal of impaired CENH3 molecules, and a general loss of CENH3 after each loading phase. These steps ensure an endowment of centromeres with a sufficient amount of correct CENH3 molecules as a prerequisite for centromere maintenance during mitotic cycles of the microgametophyte and the progeny. From a comparison with data available for Drosophila, we hypothesise that the post-divisional mitotic CENH3 loading in metazoans is evolutionarily derived from the post-divisional meiotic loading phase, while the pre-divisional first meiotic loading has been conserved among eukaryotes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Correct segregation of chromosomes during cell division is a prerequisite for stable genome inheritance. In all eukaryotes, active centromeres are responsible for this essential task. Centromeres are characterised by the kinetochore protein complex which interacts with microtubule fibres of the spindle apparatus. Assembly and maintenance of kinetochores at active centromeres, except for the ‘point centromeres’ of budding yeasts (Clarke and Carbon 1985), is not primarily determined by DNA sequence, but is epigenetically specified (Karpen and Allshire 1997). The basic step of kinetochore formation is the incorporation of the centromeric histone H3 variant CENH3 (first described as human CENP-A by Earnshaw and Rothfield (1985)) instead of H3 into nucleosomes at centromere positions.
Contrary to canonical histone H3, CENH3 is not incorporated in a replication-dependent manner (again with the exception of budding yeast). Fungi (Pearson et al. 2004; Takayama et al. 2008), red algae (Maruyama et al. 2007), protozoa (Dubin et al. 2010) and higher plants tested so far (Lermontova et al. 2006; 2007; Nagaki et al. 2005) revealed CENH3 deposition before the separation of sister chromatids during mitotic division. In contrast, human tumour cells (HeLa) (Jansen et al. 2007; Hemmerich et al. 2008), Drosophila embryonic cells (Schuh et al. 2007), Drosophila brain stem and non-stem cells (Dunleavy et al. 2012), chicken DT40 cells (Silva et al. 2012) and Xenopus leavis egg cell extracts (Bernad et al. 2011; Moree et al. 2011) do so after sister chromatid separation (from anaphase to G1). However, for Drosophila, tissue culture cells also loading during G2 (Ahmad & Hennikoff 2001) and metaphase (Mellone et al. 2011) has been reported (see also Lidsky et al. 2013). What happens with the centromeric nucleosomes from replication until the loading of CENH3 is still unclear (discussed in e.g. Lermontova et al. 2011b; Padeganeh et al. 2013a, b). Several options have been proposed: (1) transient formation of hemisomes (tetrameric nucleosomal histone cores containing one CENH3 molecule), (2) octameric histone cores containing one molecule each of H3 and of CENH3 (heterosomes), (3) a halved nucleosome density or (4) canonical nucleosomes which, during CENH3 loading, get substituted by CENH3 nucleosomes or experience an exchange of H3 by CENH3. For human cells Dunleavy et al. (2011) suggested H3.3 deposition at S as a space holder until its substitution by CENH3 in G1. Whatever will be the ultimate answer(s), a stunning problem remains: why metazoans compensate the replication-mediated CENH3 dilution not before but after the main centromere function (i.e. correct segregation of sister chromatids) has been fulfilled?
Other questions concern CENH3 loading during meiosis, when first homologues, and subsequently sister chromatids, are separated without an intermediate replication. For instance, does the deposition of new CENH3 occur before or after the separation of homologs? Are both meiotic divisions linked with a CENH3 deposition event although quantitatively one should be sufficient?
Previous studies have shown that CENH3 is required for meiosis as well. Only in the worm Caenorhabditis elegans, having holocentric chromosomes, meiocytes do not require CENH3, at least not in amounts needed in mitotic cells (Monen et al. 2005). In A. thaliana, the depletion of CENH3 via RNA interference (RNAi) does not disturb segregation during mitosis. Below a certain threshold, however, mitotic division in plants ceases. RNAi plants with a low level of CENH3 are semi-sterile. Meiosis is not blocked but rather shows distorted chromosome segregation (Lermontova et al. 2011a). The same effect is observed when either N-terminally truncated EYFP-tagged CENH3 (Lermontova et al. 2011a) or ‘tail-swapped’ GFP-tagged CENH3 (with the N-terminal part of H3 exchanged against that of CENH3, Ravi et al. 2011) largely or completely substitute the wild-type protein. Thus, the C-terminal part of CENH3 is sufficient for deposition at and correct function of somatic centromeres. In contrast to somatic cells, pollen mother cells in G2 do not reveal N-terminally truncated or modified CENH3 (Lermontova et al. 2011a; Ravi et al. 2011). The N-terminal part alone cannot target meiotic centromeres (I. Lermontova, unpublished results). Taking together, these results indicate that likely CENH3, lacking the correct N-terminus, cannot be loaded to and is actively removed from centromeres in meiocytes. Because immunostaining of meiocytes revealed no meiotic stage in A. thaliana and in barley (Sanei et al. 2011) during which centromeres were free of CENH3, not all CENH3 is exchanged, at least not simultaneously. Rather, a stringent quality check before the first meiotic division ensures the absence of impaired CENH3 by preventing incorporation and by removal of such molecules from centromeric nucleosomes of pre-meiotic nuclei. Here, we report on CENH3 deposition dynamics at centromeres in male plant meiotic and gametophytic cells by the quantification of CENH3 signals during and after meiosis and speculate about the evolution of CENH3 deposition pathways.
Materials and methods
Preparation of nuclei and immunolocalization
To analyse the structure and amount of CENH3 signals during mitosis and meiosis of rye Secale cereale L. (Petkuser Sommerroggen), root tips and anthers were fixed in 4 % paraformaldehyde/3.6 % sucrose and squashed onto slides. G1, S and G2 phase nuclei (Fig. S1) were isolated and flow-sorted according to their DNA content from rye root meristems after formaldehyde fixation using a FACS Aria (BD Biosciences) as described (Pecinka et al. 2004).
To evaluate the CENH3 amount, immunostaining was performed according to Jasencakova et al. (2000). CENH3 was detected with rabbit anti-rice CENH3 antibodies (Nagaki et al. 2004) and goat anti-rabbit rhodamine (1:300; Jackson Immuno Research Laboratories). Nuclei were counterstained with DAPI (1 μg/ml) in Vectashield (Vector Laboratories).
To analyse the post-meiotic loading of EYFP-CENH3(C) to A. thaliana centromeres, transgenic plants expressing N-terminally truncated EYFP-tagged CENH3 (Lermontova et al. 2011a) were used. Flower buds were fixed and immunostained with rabbit anti-GFP antibodies (1:500; Clontech) and goat anti-rabbit rhodamine as described above to detect EYFP-CENH3(C).
Microscopic evaluation, image processing and statistics
To analyse the structures of immunosignals and chromatin at an optical resolution of ∼100 nm (super-resolution), structured illumination microscopy (SIM) was applied using a C-Apo 63x/1.2 W Korr objective of an Elyra microscope system and the software ZEN (Zeiss, Germany). Images were captured separately for each fluorochrome using appropriate excitation and emission filters and merged using the ZEN software.
To determine the amount of CENH3 within somatic and meiotic nuclei, fluorescence intensities were measured using the TINA 2.0 software in maximum intensity projections generated from stacks of optical SIM sections through the specimens by the ZEN software. An intensity threshold was set to computationally subtract the background pixels from the image. The corrected sum of grey values of all signals within the nucleus was used to determine the CENH3 content.
The mean CENH3 amounts, based on fluorescence intensity, and its confidence intervals (95 %) were calculated with the SigmaPlot 2000 software. Non-overlapping confidence intervals were regarded as significant differences between the mean values.
Results
In mitotic cereal cells CENH3 deposition occurs before separation of sister chromatids
Immunofluorescence intensity measurement of endogenous CENH3 of G1, S, G2 and mitotic cells of rye root tip meristems showed that CENH3 signals double their intensity and split into sister kinetochores during late G2 to pro-metaphase (Figs. 1 and 2a). A similar behaviour was observed in Arabidopsis, barley (Lermontova et al. 2006, 2007) and Luzula (Nagaki et al. 2005) indicating a pre-mitotic loading mechanism in higher plants.
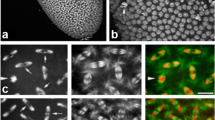
Relative CENH3 amount in somatic and meiotic nuclei of rye measured as fluorescence intensity of immunosignals. To compare directly the amount of CENH3 in mitotic (sporophytic and gametophytic), meiotic and somatic nuclei, the maximum average signal intensity per nucleus in early meiotic prophase I was set 100 %. The numbers inside each bar indicate the numbers of nuclei scored. The bars represent the total signal intensities per nucleus. i.e. the sum of signals per chromosome of a nucleus. In interkinesis, metaphase II and tapetum, the bars show the mean CENH3 amount present per daughter nuclei. For the structure of signals within the nuclei of different developmental stages, see Fig. 2
CENH3 distribution in somatic root tip nuclei, during mitosis (a), during meiosis and in somatic anther nuclei (b) of rye. During somatic G2-metaphase, CENH3 loading is accompanied by kinetochore splitting. During meiosis, the kinetochores of paired homologous centromeres (each containing two sister centromeres) split in the late prophase I, and the sister kinetochores split in the telophase I. In interphase nuclei, centromeres may associate (see pre-replicative tetrad and tapetum nuclei)
In meiocytes loading of CENH3 occurs before and after meiosis I
Pollen mother cells (PMC) of rye were investigated to analyze CENH3 loading during meiosis of higher plants (Figs. 1, 2 and 3). Flow-sorted G1, S and G2 cells of rye root tip meristems yielded fluorescence intensities of about half of that measured for meiotic prophase cells, but about equal to that of individual daughter nuclei during anaphase. PMC in early prophase displayed similar fluorescence intensities as mitotic pro-metaphase nuclei, indicating a pre-meiotic CENH3 loading in PMCs. The immunofluorescence intensity decreases significantly (P < 0.05) towards metaphase I. This decrease could be the result of a purifying quality check for intact CENH3, which starts during leptotene and zygotene, when the signals for N-terminally altered CENH3 gradually disappear and are then absent until gametophytic mitoses start (Ravi et al. 2011). Possibly, the removal of imperfect e.g. N-terminally damaged CENH3 causes the fluorescent intensity decrease from early prophase I till metaphase I. Telophase I daughter nuclei revealed less than half of the fluorescence intensity of prophase I nuclei. Then, at a similar degree of chromatin compactness, a significant (P < 0.05) fluorescence intensity increase of ∼16 % occurs in interkinesis nuclei. This increase after the separation of homologues might represent a ‘reloading’ to compensate for the loss of incomplete or damaged CENH3 during the preceding quality check. This meiotic reloading seems not to accept N-terminally modified CENH3 molecules, because such molecules are not detectable in that stage in A. thaliana (Lermontova et al. 2011a; Ravi et al. 2011). Thus, in rye meiocytes, a pre-divisional loading takes place during pre-meiotic G2, and a second compensatory loading occurs after the first meiotic division during interkinesis. Between metaphase II and the replication of tetrad nuclei, a further decrease of CENH3 amount occurs (similar to that which occurs towards metaphase I) and becomes compensated in late post-replicative tetrad nuclei until the kinetochores split before the onset of the first pollen grain mitosis. This suggests that meiotic and mitotic CENH3 loading are differently regulated processes and that removal and reloading of CENH3 takes place prior to meiosis and prior to gametophytic mitosis. Within transgenic A. thaliana plants expressing N-terminally truncated EYFP-tagged CENH3 (EYFP-CENH3(C)), we found the recombinant protein absent from meiotic cells (Lermontova et al. 2011a) until the early tetrad stage (Fig. 4a). At later tetrad stages, it appeared first in nucleoplasm (Fig. 4b), then associated to heterochromatin around the nucleolus (Fig. 4c) and eventually, before the first pollen mitosis, at distinct chromocenters (Fig. 4d). According to Borg et al. (2009), the tetrad stages in Fig. 4c, d correspond to late S/G2 before the first gametophytic mitosis. The first gametophytic mitosis proceeds like a sporophytic mitosis (Houben et al. 2011; Banaei-Moghaddam et al. 2012), and in A. thaliana again tolerates tail-swapped (Ravi et al. 2011) or truncated CENH3 molecules.
Comparison of CENH3 deposition timing during the somatic and meiotic cell cycles in different eukaryotic phyla. In the mitotic cycle, budding yeast (Pearson et al. 2004) loads CENH3 during S, and fission yeast (Takayama et al. 2008) biphasic during S and G2; plants (Lermontova et al. 2006, 2007; Nagaki et al. 2005) depending on species, from the late G2 to early prophase, red algae (Maruyama et al. 2007) and the protozoic amoeba Dictyostelium (Dubin et al. 2010) before sister chromatid separation, and metazoans, depending on the species, from anaphase to G1 i.e. after the separation of sister chromatids; however, for Drosophila (Mellone et al. 2011) also loading during metaphase has been reported. In meiocytes of rye, loading occurs before prophase I, accompanied by the removal of imperfect or damaged molecules and followed by a decrease of CENH3 towards metaphase I (upward arrow). A reloading of exclusively perfect molecules occurs during interkinesis, followed again by a loss or removal of CENH3 in pre-replicative tetrad nuclei before regular loading prior to the first pollen mitosis. In Drosophila, loading occurs also before prophase I and at the exit of meiosis (broken arrow)
Loading of N-terminally truncated EYFP-CENH3(C) to centromeres of A. thaliana nuclei prior to the gametophytic mitosis. No EYFP-CENH3(C) immunosignals were detected at centromeres of post-meiotic cells in early tetrad stage (a). At later tetrad stages, EYFP-CENH3(C) accumulated within nucleoplasm (b) and then first at heterochromatic regions around the nucleolus (c) and later at distinct chromocenters (d). For comparison with signals in somatic interphase nuclei, see Fig. 2. On (b) and (d) the green colour represents autofluorescence of the exine and cytoplasm of the pollen grains
Somatic nuclei of rye anthers, and in particular individual nuclei of binucleate tapetum cells, showed lower fluorescence intensity than meristematic interphase nuclei. This is in accordance with the observation that old (senescent) differentiated somatic cells of A. thaliana (Lermontova et al. 2011b), and, in particular, tapetum cells which no longer divide (Talbert et al. 2002), display a reduced CENH3 content as reflected by fluorescence intensity.
Discussion
Here, we describe a biphasic CENH3 deposition during meiosis in monocotyledonous plants.
The only other organism with monocentric chromosomes for which CENH3 loading has been quantitatively traced during meiosis is Drosophila melanogaster (Dunleavy et al. 2012; Raychaudhury et al. 2012). For this species, both groups describe a CENH3 loading during male pre-meiotic G2 to prophase I and, similar to what we found in rye, a drop between both meiotic divisions which was explained by the reorganisation of the kinetochore between meiosis I and II. Whether a quality control of CENH3 molecules takes place is unknown. Contradictory results were obtained regarding a loading at the exit of the second meiotic division. Dunleavy et al. (2012) describe a loading after telophase II, while Raychaudhury et al. (2012) did not observe CENH3 loading at the exit of meiosis. In particular, the results of Dunleavy et al. (2012) indicate a similarity of meiotic CENH3 loading between plants and metazoans. While plants show a pre-divisional loading in meiotic as in mitotic cells, animals seem to switch from a pre-divisional loading in meiotic to a predominantly post-divisional one in mitotic cells. Mitoses cease when centromeres do not contain, at least in some chromosomes, a sufficient amount of CENH3, but meiotic division apparently proceeds under conditions of insufficient CENH3 amounts with disastrous consequences for chromosome segregation (Lermontova et al. 2011a; Ravi et al. 2011). The amount of CENH3 may drop below a critical threshold if e.g. the quality check led to a strong decrease of CENH3 molecules by removing imperfect CENH3 molecules. Therefore, two phases of meiotic loading make sense. The first one, before the onset of meiosis I, is similar to mitotic CENH3 loading in plants, but is linked with the quality check. A quality check as in PMC seems to occur also in female meiocytes. Pre-meiotic interphase nuclei of A. thaliana megaspore mother cells release C-terminally linked CENH3-GFP fusion molecules (She et al. 2013). The second later loading phase still does not tolerate modified molecules and apparently compensates for the removal of imperfect CENH3 molecules, which occurs before, during or immediately after pre-meiotic loading. Presumably, a post-divisional loading, corresponding to the second meiotic CENH3 deposition became the ‘typical’ somatic loading mode in animals. Post-divisional loading in animals apparently became acceptable since the CENH3 level after one replication-mediated CENH3 dilution is still sufficient for kinetochore formation and correct chromosome segregation during the next mitotic cycle. However, the altered ‘geometry’ of centromeres (Watanabe 2012) during the separation of homologues and the presumed quality check at the beginning of meiosis might be the reason for retaining a pre-divisional meiotic loading also in animals.
Taking these observations together, we conclude: For eukaryotic phyla except for mitosis in metazoa, pre-divisional CENH3 loading seems to be the rule. An additional quality check for CENH3 completeness is apparently included into the pre-meiotic loading before the first meiotic division. A later, post-divisional reloading (after the first or the second meiotic division) again does not accept imperfect CENH3 molecules. Both these CENH3 deposition phases together, although followed by a decrease of CENH3 content, guarantee a sufficient amount of perfect CENH3 molecules as well as functioning centromeres from generation to generation. The second meiotic deposition makes also sense in the light of the observation made for Drosophila that the pre-existing CENH3 level determines the amount of subsequent CENH3 loading cycles (Raychaudhuri et al. 2012). The post-divisional loading, as required for meiocytes, apparently became predominant for dividing somatic cells in animals. However, the meiotic pre-divisional CENH3 loading is maintained also in metazoans most likely to handle the mono-oriented sister centromeres during the separation of homologues and also for the quality check of CENH3 before gamete formation.
References
Ahmad K, Henikoff S (2001) Centromeres are specialized replication domains in heterochromatin. J Cell Biol 153:101–109
Banaei-Moghaddam AM, Schubert V, Kumke K, Weiss O, Klemme S, Nagaki K, Macas J, Gonzalez-Sanchez M, Heredia V, Gomez-Revilla D, Gonzalez-Garcia M, Vega JM, Puertas MJ, Houben A (2012) Nondisjunction in favor of a chromosome: the mechanism of rye B chromosome drive during pollen mitosis. Plant Cell 24:4124–4134
Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A (2011) Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol 192:569–582
Borg M, Brownfield L, Twell D (2009) Male gametophyte development: a molecular perspective. J Exp Bot 60:1465–1478
Clarke L, Carbon J (1985) The structure and function of yeast centromeres. Ann Rev Genet 19:29–55
Dubin M, Fuchs J, Graf R, Schubert I, Nellen W (2010) Dynamics of a novel centromeric histone variant CenH3 reveals the evolutionary ancestral timing of centromere biogenesis. Nucl Acid Res 38:7526–7537
Dunleavy EM, Almouzni G, Karpen GH (2011) H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2:146–157
Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, Karpen GH (2012) The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol 10:e1001460
Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321
Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S (2008) Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol 180:1101–1114
Houben A, Kumke K, Nagaki K, Hause G (2011) CENH3 distribution and differential chromatin modifications during pollen development in rye (Secale cereale L.). Chromosome Res 19:471–480
Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176:795–805
Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I (2000) Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12:2087–2100
Karpen GH, Allshire RC (1997) The case for epigenetic effects on centromere identity and function. Trends Genet 13:489–496
Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I (2006) Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18:2443–2451
Lermontova I, Fuchs J, Schubert V, Schubert I (2007) Loading time of the centromeric histone H3 variant differs between plants and animals. Chromosoma 116:507–510
Lermontova I, Koroleva O, Rutten T, Fuchs J, Schubert V, Moraes I, Koszegi D, Schubert I (2011a) Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J 68:40–50
Lermontova I, Rutten T, Schubert I (2011b) Deposition, turnover, and release of CENH3 at Arabidopsis centromeres. Chromosoma 120:633–640
Lidsky PV, Sprenger F, Lehner CF (2013) Distinct modes of centromere protein dynamics during cell cycle progression in Drosophila S2R + cells. J Cell Sci 126:4782–4793
Maruyama S, Kuroiwa H, Miyagishima SY, Tanaka K, Kuroiwa T (2007) Centromere dynamics in the primitive red alga Cyanidioschyzon merolae. Plant J 49:1122–1129
Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH (2011) Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet 7:e1002068
Monen J, Maddox PS, Hyndman F, Oegema K, Desai A (2005) Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol 7:1248–1255
Moree B, Meyer CB, Fuller CJ, Straight AF (2011) CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194:855–871
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Nagaki K, Kashihara K, Murata M (2005) Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell 17:1886–1893
Padeganeh A, De Rop V, Maddox PS (2013a) Nucleosomal composition at the centromere: a numbers game. Chromosome Res 21:27–36
Padeganeh A, Ryan J, Boisvert J, Ladouceur AM, Dorn JF, Maddox PS (2013b) Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr Biol 23:764–769
Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K (2004) Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol 14:1962–1967
Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I (2004) Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113:258–269
Ravi M, Shibata F, Ramahi JS, Nagaki K, Chen C, Murata M, Chan SW (2011) Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet 7:e1002121
Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF (2012) Transgenerational propagation and quantitative maintenance of paternal centromeres depends on cid/cenp-a presence in Drosophila sperm. PLoS Biol 10:e1001434
Sanei M, Pickering R, Kumke K, Nasuda S, Houben A (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A 108:E498–E505
Schuh M, Lehner CF, Heidmann S (2007) Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17:237–243
She W, Grimanelli D, Rutowicz K, Whitehead MW, Puzio M, Kotlinski M, Jerzmanowski A, Baroux C (2013) Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development 140:4008–4019
Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE (2012) Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22:52–63
Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K (2008) Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell 19:682–690
Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053–1066
Watanabe Y (2012) Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev 13:370–382
Acknowledgments
We thank Jörg Fuchs for the flow sorting of nuclei, Andrea Kunze, Katrin Kumke and Joachim Bruder for their assistance, Armin Meister for the help with statistics and Andreas Houben for critically reading the manuscript. IS was partially supported by the European Social Fund (CZ.1.07/2.3.00/20.0189), IL by a DFG grant (LE2299/1-1).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
FACs profiles of sorted G1, S and G2 phase nuclei isolated and DAPI-stained from rye root tip meristems (GIF 19 kb)
Rights and permissions
About this article
Cite this article
Schubert, V., Lermontova, I. & Schubert, I. Loading of the centromeric histone H3 variant during meiosis–how does it differ from mitosis?. Chromosoma 123, 491–497 (2014). https://doi.org/10.1007/s00412-014-0466-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-014-0466-9