Abstract
All organisms sense and respond to conditions that stress their homeostasis by downregulating the synthesis of rRNA and ribosome biogenesis, thus designating the nucleolus as the central hub in coordinating the cellular stress response. One of the most intriguing roles of the nucleolus, long regarded as a mere ribosome-producing factory, is its participation in monitoring cellular stress signals and transmitting them to the RNA polymerase I (Pol I) transcription machinery. As rRNA synthesis is a most energy-consuming process, switching off transcription of rRNA genes is an effective way of saving the energy required to maintain cellular homeostasis during acute stress. The Pol I transcription machinery is the key convergence point that collects and integrates a vast array of information from cellular signaling cascades to regulate ribosome production which, in turn, guides cell growth and proliferation. This review focuses on the mechanisms that link cell physiology to rDNA silencing, a prerequisite for nucleolar integrity and cell survival.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The synthesis of rRNA, the first step in ribosome synthesis, determines the cell’s capacity to grow and proliferate. As ribosome biogenesis consumes a tremendous amount of cellular energy, rRNA synthesis and ribosome production are tightly linked to cell growth and proliferation to be responsive to general metabolism and specific environmental challenges. In fact, almost all signaling pathways that affect cell growth and proliferation directly regulate rRNA synthesis; their downstream effectors converging at the Pol I transcription machinery (Fig. 1). These topics have been reviewed in the past, and readers are referred to other articles for further reading (Grummt 2003; Russell and Zomerdijk 2005, 2006; Moss et al. 2007; McStay and Grummt 2008; Grummt 2010; Drygin et al. 2010; Grummt and Längst 2013).
Regulation of nucleolar transcription in response to external signals. The cartoon depicts some of the key signaling pathways and major enzymatic activities that modify basal Pol I-specific transcription factors, thereby adapting rDNA transcription to changes in extracellular conditions. The microscopic images show fluorouridine-labeled NIH3T3 cells that were cultured under normal conditions or exposed to nutritional stress (glucose deprivation). The red dots represent fluorouridine-labeled nascent nucleolar RNA visualized by immunofluorescence. A Miller spread of transcriptionally active rRNA genes showing the tandem arrangement of genes and the close spacing of nascent transcripts is shown below (from Scheer et al. 1997)
This review focuses on how stress responses in mammalian cells affect nucleolar function. The rationale for this is that all organisms sense and respond to conditions that stress their homeostasis by downregulating the synthesis of rRNA and ribosome biogenesis. Perturbation of any of the steps of ribosome biogenesis, including transcription, processing, and assembly of the 40S and 60S ribosomal subunits, causes nucleolar stress and inhibits cell cycle progression. Environmental cues, including virtually any type of stress, have been shown to feed into the tight regulation of rRNA synthesis as part of cellular quality control, thus placing the nucleolus as a central hub for coordination of cellular stress response. One of the strategies that cells use to preserve cellular energy homeostasis under stress conditions is the attenuation of ribosome biosynthesis. Cellular stress may be caused by a variety of unfavorable factors, including nutrient deprivation, transient exposure to high or low temperature, anaerobiosis, oxidative damage, heavy metals, and changes in osmolarity (Fig. 2). In view of the intricate networking within a cell, it is not surprising that attenuation of ribosome biosynthesis can occur at different levels, which include altered spatial distribution of nucle(ol)ar proteins, downregulation of RNA polymerase I (Pol I) transcription, precursor rRNA (pre-rRNA) processing and transport, or changes in chromatin structure. Genotoxic agents and UV irradiation induce an almost immediate shutdown of rDNA transcription by changing the frequency of transcription initiation or by adjusting the number of genes that are involved in transcription. Here, I will discuss the current understanding of the nucleolus and nucleolar protein pathways, both in terms of how they modulate the cellular stress response and how the integration of these pathways protects the genetic integrity of the cell.
Reorganization of nucleolar structure under stress
The nucleolus is composed of three structural compartments: the fibrillar centers (FC) and dense fibrillar centers (DFC), where transcription and processing take place, and the granular component (GC) where preribosomes are assembled (Scheer and Hock 1999). This spatial organization is disturbed if rDNA transcription is inhibited. Inhibition of rDNA transcription by drugs, commonly by low doses of actinomycin D, typically leads to rearrangement of nucleolar components, culminating in complete nucleolar disintegration. A similar segregation of nucleolar components, i.e., condensation and subsequent separation of FC, DFC, and GC and formation of “nucleolar caps,” is observed after UV irradiation or upon inhibition of topoisomerase II by drugs such as etoposide (Govoni et al. 1994). These dramatic changes in nucleolar morphology are caused by the simultaneous inhibition of major nuclear pathways and specific stress responses that affect transcription, replication, and DNA repair. The molecular mechanisms underlying the nucleolar stress response are complex and intertwined, underscoring the central role of the nucleolus in regulatory activities that extend beyond rRNA synthesis.
Basal factors required for Pol I transcription initiation
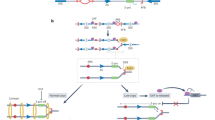
Mammalian rDNA clusters are characterized by multiple alternating modules of a long intergenic spacer (IGS) of approximately 30 kb and a pre-rRNA coding region of approximately 14 kb. Each active rRNA gene is transcribed by RNA Pol I to generate a 47S pre-rRNA. After synthesis, this primary transcript is subsequently processed and modified to generate one molecule each of mature 18S, 5.8S, and 28S rRNA that, together with 5S rRNA which is transcribed by Pol III, form the RNA backbone of the ribosome. Initiation of rDNA transcription requires the assembly of a specific multiprotein complex at the rDNA promoter containing Pol I and a surprising number of auxiliary proteins (Fig. 3). In mammals, the transcription initiation complex is assembled at the rDNA promoter by the synergistic action of two DNA binding factors, the upstream binding factor (UBF) (Jantzen et al. 1990) and the promoter selectivity factor (SL1/TIF-IB) (Learned et al. 1985; Comai et al. 1992; Clos et al. 1986). UBF is an abundant nucleolar protein that contains several high mobility group domains that activate rDNA transcription by stabilizing the binding of SL1/TIF-IB and Pol I at the rDNA promoter. Promoter specificity is brought about by SL1/TIF-IB, a multiprotein complex comprising the TATA-box binding protein (TBP) and five TBP-associated factors (TAFIs), i.e., TAFI110/95, TAFI68, TAFI48, TAFI41, and TAFI12 (Zomerdijk et al. 1994; Heix et al. 1997; Denissov et al. 2007; Gorski et al. 2007). The TAFI subunits perform important tasks in transcription complex assembly, mediating specific interactions between the rDNA promoter and Pol I, thereby recruiting Pol I, together with the essential transcription initiation factor TIF-IA and a collection of Pol I-associated factors, to rDNA.
Cartoon depicting the structural organization of mammalian rDNA repeats and the basal factors required for transcription initiation. The site of transcription initiation of 47S pre-rRNA (black arrow) and intergenic transcripts originating from the spacer promoter (red arrow) are indicated. The red boxes upstream and downstream of the pre-rRNA coding region represent binding sites for the transcription termination factor TTF-I. Repetitive enhancer elements (blue boxes) located between the spacer promoter and major gene promoter are also indicated. The ellipsoids show the factors that are associated with the rDNA promoter and Pol I, respectively. Synergistic binding of UBF and SL1/TIF-IB to the rDNA promoter is required for the recruitment of RNA polymerase I (Pol I) and multiple Pol I-associated factors, including the regulatory factor TIF-IA, to the transcription start site to initiate pre-rRNA synthesis (for details, see text)
Stress-dependent changes of the nucleolar proteome
Apart from its traditional role in ribosome biogenesis, the nucleolus is also involved in cellular functions not directly related to ribosome biogenesis. These include assembly of signal recognition particles, control of cell cycle progression, apoptosis, DNA replication, and repair, thus providing a link between ribosome biosynthesis, cell cycle progression, and stress signaling. Consistent with this functional diversity, the nucleolar proteome is likely to comprise more than 4,500 distinct proteins, including many chaperones or “nucleolar multitasking proteins,” i.e., proteins that contribute to other biological activities, which shuttle between the nucleolus, the nucleoplasm, and even the cytoplasm. Accordingly, the protein profile of nucleoli is not static but is modulated by changes in cell physiology, being altered by stress, tumor development, viral infections, and signaling events. High-throughput proteomics of mass spectrometry-based stable isotope labeling by amino acids in cell culture (SILAC) and live cell fluorescence microscopy of cells exposed to stress stimuli, e.g., actinomycin D treatment (Andersen et al. 2005), viral infection (Hiscox et al. 2010; Lam et al. 2010), or DNA damage (Boisvert and Lamond 2010), have revealed rapid changes in both the concentration and localization of proteins involved in rDNA transcription, pre-rRNA processing, DNA damage, and oxidative stress pathways (Banski et al. 2010; Ahmad et al. 2009). This dynamic network of chaperones, co-chaperones, and multitasking proteins is required to maintain nucleolar organization under normal growth conditions and to adapt nucleolar function to environmental changes elicited by stress or disease. As under conditions of cellular stress—for example, after heat shock, acidosis, or osmotic stress—the local network is reorganized, nucleolar chaperones, together with their cofactors, may provide “pillars” for the establishment of a compartment-specific homeostatic network (Banski et al. 2010).
Transcriptional inhibition by stress-dependent signaling pathways
Acute or adaptive response to unfavorable environmental conditions forces cells to regulate their energy expenses by focussing their metabolic activity on pro-survival tactics. Therefore, cells rapidly and efficiently shut down rRNA synthesis after exposure to extra- or intracellular stresses. One key nucleolar target of oxidative or ribotoxic stress is TIF-IA, a basal transcription factor for RNA polymerase I (Pol I) that regulates rRNA synthesis in response to external signals. Upon exposure to stress, c-jun N-terminal protein kinase 2 (JNK2), a ubiquitously expressed member of the JNK family, is activated by multiple cellular stresses (Schreiber et al. 1995; Chen et al. 1996; Davis et al. 2000). Stress-induced activation of JNK2 triggers phosphorylation of TIF-IA at a single threonine residue at position 200 (Thr200). Phosphorylation at Thr200 has two effects. First, it impairs the interaction of TIF-IA with Pol I and SL/TIF-IB, thus preventing transcription initiation complex formation at the rDNA promoter. Second, phosphorylation at Thr200 causes TIF-IA to move from the nucleolus to the nucleoplasm where it is sequestered from Pol I. Mutation of threonine 200 prevents inactivation of TIF-IA by JNK2-mediated phosphorylation and leads to stress resistance of Pol I transcription (Mayer et al. 2006). These findings highlight the important role of JNK2 in protecting rRNA synthesis against the harmful consequences of cellular stress, reinforcing that nucleoli orchestrate the chain of events the cell uses to effectively respond to stress signals.
Many of the pathways that convert stress signals into a cellular response link the nucleolus to stabilization and activation of the tumor suppressor p53 (Rubbi and Milner 2003). Although the signaling pathways rely on different mechanisms that can mediate the increase in p53 levels, it is likely that these pathways are interconnected, depending on the stress signal and cellular conditions. p53, widely dubbed as “the guardian of the genome”, is a transcription factor that triggers growth arrest, apoptosis, and cell senescence in response to DNA damage (Carson and Lois 1995). The levels of p53 are kept low under normal physiological conditions due to its interaction with the E3 ubiquitin ligase MDM2 (called HDM2 in human), marking p53 for proteasome-dependent proteolysis. Degradation of p53 by the ubiquitin pathway is intimately linked to nucleolar structure and function, which places nucleolar transcription at the center of control pathways that are influenced by the amount of p53. Under conditions of nucleolar stress, the p53–MDM2 complex is disrupted and p53-dependent pathways are activated. For example, abrogation of nucleolar activity by genetic inactivation of the TIF-IA gene leads to disintegration of the nucleolus, increased p53 levels, and p53-dependent apoptosis (Yuan et al. 2005). Regulation of p53 and induction of apoptosis in response to aberrant nucleolar activity is brought about by proteins that interact with MDM2, including the tumor suppressor protein ARF or several ribosomal proteins, such as L5, L11, or L23. In the absence of nascent pre-rRNA, these proteins bind to and inhibit the E3 ligase activity of MDM2, leading to the accumulation of p53 and induction of apoptosis (Sherr and Weber 2000; Zhang et al. 2003; Kurki et al. 2004). A recent study showed that inhibition of Pol I transcription by treatment with CX-5461, a specific small molecule inhibitor of Pol I transcription, induced cell death in malignant B cell lymphomas while allowing normal B cells to grow and proliferate (Bywater et al. 2012). CX-5461 treatment led to a rapid increase in p53 abundance caused by binding of ribosomal proteins L5 and L11 to MDM2. Together, the striking correlation between perturbation of nucleolar function, elevated levels of p53, and induction of cell suicide suggests that, depending on the gravity of the nucleolar stress, cells face the decision whether to arrest cell cycle progression and initiate repair mechanisms or to succumb to p53-dependent apoptosis. These results not only reinforce the central role of p53 in surveying cellular health and support the view that the nucleolus is a stress sensor that regulates the abundance of p53 (Rubbi and Milner 2003; Olson 2004; Mayer and Grummt 2005; Boulon et al. 2010), but also suggest that drugs targeting the rDNA transcription machinery may hold great promise for the treatment of cancer.
Changes in cellular energy levels modulate the activity of the transcription factor TIF-IA
As the availability of nutrients is one of the most important variables in maintaining cellular homeostasis and rRNA synthesis is a costly process, Pol I transcription is tightly linked to the metabolic state of a cell. It has been known for a long time that a given nutritional state gives rise to an equilibrium in which the synthesis of ATP and GTP is balanced by their use in protein synthesis (Grummt and Grummt 1976). Accordingly, rDNA promoter activity is regulated by intracellular levels of ATP, providing a molecular explanation for the growth-dependent control and homeostatic regulation of ribosome synthesis. The key enzyme that translates changes in energy levels into adaptive cellular responses is the AMP-activated protein kinase (AMPK). If energy levels are low and the intracellular AMP/ATP ratio is elevated, AMPK switches on energy-producing pathways and switches off energy-consuming pathways to restore cellular ATP levels. Therefore, under conditions of nutrient shortage, rDNA transcription is downregulated, linking nutrient availability to ribosome biogenesis (Fig. 4). In vitro and in vivo phosphorylation experiments combined with in vitro transcription assays revealed that the activation of AMPK triggers phosphorylation of TIF-IA at a single serine residue, Ser635, which leads to the inactivation of TIF-IA and inhibition of rRNA synthesis. AMPK-mediated phosphorylation of TIF-IA at Ser635 does not compromise binding of TIF-IA to Pol I, but abrogates the interaction between promoter-bound SL1/TIF-IB and TIF-IA, which in turn impairs transcription complex assembly (Hoppe et al. 2009). This observation reveals a sophisticated level of transcriptional regulation that converts changes in intracellular energy supply into specific phosphorylation of TIF-IA, thus leading to downregulation of Pol I transcription in response to nutrient shortage.
Glucose deprivation downregulates pre-rRNA synthesis. In glucose-rich medium, TIF-IA is phosphorylated by RSK, ERK, and mTOR at Ser649, Ser633, and Ser199, and these phosphorylations are required for Pol I transcription. Upon glucose deprivation, elevation of the cellular AMP/ATP ratio activates AMP-dependent protein kinase (AMPK). AMPK-dependent phosphorylation of TIF-IA at Ser635 prevents the interaction of TIF-IA with the pre-initiation complex comprising UBF and SL1. Consequently, recruitment of TIF-IA/Pol I to the rDNA promoter and formation of the transcription initiation complex are impaired
Sirtuins link the cellular energy status to rDNA transcription
Studies over the past decade have shown that the members of the sirtuin family are major players in sensing and coordinating the responses to environmental changes. Sirtuins share homology to the silencing factor Sir2 (silent information regulator 2) that regulates replicative lifespan and genomic stability in budding yeast (Imai and Guarente 2010; Haigis and Sinclair 2010). Sir2 plays an important role in rDNA stability and nucleolar activity links rRNA synthesis directly to the energy prosperity of the cell (Kobayashi and Ganley 2005). Sirtuins are NAD+-dependent deacetylases that sense oxidative stress conditions and promote a protective cellular response. Mammals have seven sirtuins, denoted SIRT1 to SIRT7; all of which have distinct cellular locations, target multiple substrates, and affect a broad range of cellular functions (Fig. 5). As sirtuins require NAD+ and the cellular redox state is paramount in cell metabolism, fluctuations of sirtuin activity play a central role in many processes, including gene expression, cell survival under stress, cell cycle control, metabolic homeostasis, etc. (Houtkooper et al. 2012). Several mammalian sirtuins have been shown to participate in gene expression through deacetylation of histone and non-histone proteins. SIRT1, the mammalian homolog of yeast Sir2, for example is conserved from yeast to humans and regulates a wide range of biological processes, including gene silencing, aging, differentiation, and cell metabolism. Regarding rDNA transcription, SIRT1 downregulates Pol I transcription by counteracting PCAF-dependent acetylation of TAFI68, a subunit of the promoter selectivity factor SL1/TIF-IB. As acetylation of TAFI68 is required for SL1/TIF-IB binding to the rDNA promoter and nucleation of the transcription initiation complex, deacetylation by SIRT1 downregulates transcription (Muth et al. 2001). Notably, this mechanism of SIRT1-dependent repression of Pol I transcription at “normal” energy levels is required to shut-off rRNA synthesis during mitosis (unpublished results).
Diet-induced changes in the NAD+/NADH ratio affect both SIRT1 and SIRT7, coupling changing energy levels to rRNA synthesis and ribosome production. However, in contrast to SIRT1, very little is known about the function of SIRT7. SIRT7 is localized in nucleoli being associated with both the promoter and the coding region of rDNA, interacts with UBF and Pol I, and promotes cell growth and proliferation in response to metabolic conditions by driving ribosome biogenesis (Ford et al. 2006). SIRT7 expression correlates with growth, being abundant in metabolically active cells and low or even absent in non-proliferating cells (Ford et al. 2006; Tsai et al. 2012; Grob et al. 2009). Depletion of SIRT7 or overexpression of a catalytically inactive point mutant leads to decreased rDNA transcription, inhibition of cell proliferation, and apoptosis, underscoring the pivotal role of SIRT7 in cell survival (Fig. 6). Furthermore, SIRT7-deficient mice suffer from reduced stress resistance, inflammatory cardiomyopathy, and premature aging (Vakrusheva et al. 2008). Consistent with its important role in cell proliferation and tumor initiation, SIRT7 was also found to be upregulated in human hepatocellular carcinoma patients, and suppression of SIRT7 reduced tumor growth in a mouse xenograft model (Kim et al. 2013). A recent study has shown that SIRT7 deacetylates histone H3 acetylated at lysine 18 (H3K18ac), hypoacetylation of H3K18 compromising transcription of genes that are linked to oncogenic transformation (Barber et al. 2012).
Opposing functions of SIRT1 and SIRT7 in rDNA transcription. Both SIRT1 and SIRT7 are localized in nucleoli and play an important role in the regulation of rDNA transcription. SIRT1 downregulates Pol I transcription by deacetylating TAFI68, a subunit of the basal transcription factor SL1. Deacetylation of TAFI68 impairs binding of SL1 to the rDNA promoter, thus preventing transcription complex assembly and transcription initiation (Muth et al. 2001). In contrast, SIRT7 activates Pol I transcription by deacetylating PAF53, a subunit of Pol I, which facilitates binding of PAF53/Pol I to rDNA and is necessary for Pol I transcription (Chen et al. 2013)
Although sirtuins were originally described as histone deacetylases, the range of deacetylase targets has broadened beyond histones and now includes transcription factors and enzymes (Feige and Auwerx 2007, 2008; Haigis and Sinclair 2010). We have recently shown that polymerase-associated factor 53 (PAF53), the mammalian homolog of the yeast Pol I subunit A49 (Hanada et al. 1996), is acetylated at lysine K373 (K373) by CBP and is deacetylated by SIRT7 (Chen et al. 2013). Hypoacetylation of PAF53 facilitates the association of PAF53/Pol I with rDNA and is required for Pol I transcription. The importance of reversible acetylation of PAF53 on Pol I transcription is most evident under conditions of cellular stress. Under normal conditions, SIRT7 is associated with elongating Pol I, contacting both PAF53 and nascent pre-rRNA (Fig. 7). Apparently, the dynamic interplay of CBP-dependent acetylation and SIRT7-dependent deacetylation of PAF53 is required for efficient transcription elongation, hypoacetylation of PAF53 promoting the association of Pol I with rDNA, and enhancing pre-rRNA synthesis. When cells are stressed, the assembly of transcription complexes is precluded and pre-rRNA synthesis is inhibited. The absence of nascent pre-rRNA chains in turn leads to the release of SIRT7 into the nucleoplasm. As a consequence, PAF53 is hyperacetylated and binding of Pol I to rDNA is reduced. These results uncover a novel mechanism of transcriptional regulation, acetylation, and deacetylation of PAF53 translating environmental changes into modulation of rRNA synthesis.
Reversible acetylation of PAF53 regulates rDNA transcription in response to stress signaling. Under normal growth conditions, elongating Pol I is associated with the histone acetyltransferase CBP and the deacetylase SIRT7. CBP acetylates the Pol I-associated factor PAF53 at lysine 373 (K373), whereas SIRT7 counteracts CBP-dependent acetylation, removing the acetyl group from K373. Under conditions of energetic stress, AMP-kinase (AMPK) phosphorylates TIF-IA at serine 635, which precludes transcription complex formation and inhibits transcription initiation (Hoppe et al. 2009). Binding to nascent RNA is essential for nucleolar retention of SIRT7. Impaired transcription and lack of nascent pre-rRNA (red line) lead to the release of SIRT7 from nucleoli and translocation into the nucleoplasm. As a consequence, PAF53 remains acetylated at K373, which in turn reduces rDNA occupancy of Pol I and reinforces transcriptional repression
Stress-induced relocalization of nucleolar proteins
An emerging field of nucleolar research involves subnuclear reorganization and detention of proteins in response to environmental stimuli (reviewed in Boulon et al. 2010). Nucleoli are very dynamic structures that differ, both in size and appearance, from one cell type to another depending on transcriptional activity. Growing evidence suggests that the nucleolus is used as a specific compartment where proteins can be sequestered to or excluded from, in order to transiently stabilize or destabilize them, or locally separate them from their interaction partners. This nucleolar confinement—like regulated nuclear import/export pathways—appears to play an important regulatory role in key cellular processes, such as cell cycle control and stress response. Relocalization of proteins and reorganization of nucleolar structure in response to environmental stimuli have often been observed, indicating that nucleolar proteins are in a constant influx and outflux (Dundr et al. 2002). For example, actinomycin D-mediated inhibition of Pol I transcription triggers the seclusion of various nucleolar components into peripheral caps, which is accompanied by an influx of extranucleolar proteins and drastic alterations of the nucleolar proteome (Andersen et al. 2005). Moreover, activation of JNK2 in response to oxidative or ribotoxic stress leads to phosphorylation of TIF-IA at a single threonine residue (Thr200) which is accompanied by translocation of TIF-IA from the nucleolus to the nucleoplasm (Mayer et al. 2006). Likewise, nutritional stress induced by treatment of cells with the mTOR inhibitor rapamycin leads to hyperphosphorylation of TIF-IA at serine 199 and to translocation to the cytoplasm (Mayer et al. 2004). Translocation is restricted to TIF-IA, whereas the nucleolar localization of Pol I and UBF remains unaffected (Fig. 8). Presumably, relocating just TIF-IA rather than the entire Pol I machinery is advantageous under stress conditions whereby transcription repression has to be both immediate and reversible.
Ribotoxic stress leads to the accumulation of TIF-IA in the nucleoplasm. Immunostaining of TIF-IA (red) and UBF (green) in untreated MEFs (mock) or MEFs treated with anisomycin (10 mM, 60 min). A merged image is shown on the right (from Mayer et al. 2006)
Recent SILAC-based and high throughput quantitative mass spectrometry approaches have identified global changes in the nucleolar proteome in stress-induced senescent fibroblasts (Kar et al. 2011). These studies revealed a distinct response of the nucleolar proteome to chemically induced senescence. In contrast to transcription inhibition by actinomycin D, an overall accumulation of proteins in the nucleolus was observed after butyrate-induced senescence in NIH3T3 cells, including histones and chromatin remodeling factors. Similar observations were made in other models of senescence, demonstrating that reorganization of nucleolar structure is not a specific cellular response to butyrate treatment, but more likely a general reflection of senescence.
Stress-induced intergenic transcripts immobilize proteins in the nucleolus
Numerous proteins with functions generally not ascribed to ribosome biogenesis are captured and immobilized in the nucleolus under certain cellular conditions, thus rendering them immobile and functionally inert (Scherl et al. 2002; Mekhail et al. 2005). For example, a recent study has demonstrated that RNA originating from the intergenic spacer (IGS) separating individual rDNA transcription units plays a key role in stimuli-specific nucleolar immobilization of proteins, including DNMT1, Hsp70, POLD1, and VH. The dynamic profile of these proteins changed from a high mobility state to a state of static detention under both heat shock and acidotic conditions (Audas et al. 2012). Sequestration of proteins in the nucleolus is mediated by several stimulus-specific RNAs that originate from the IGS. Extracellular cues, such as heat shock and acidosis, trigger transcription of IGS-RNAs to immobilize selected proteins and orchestrate rapid and drastic remodeling of the nucleolar structure. Condition-specific IGS-RNAs are capable of associating with a peptide code, referred to as the nucleolar detention sequence (NoDS), to target proteins for immobilization in the nucleolus and tether NoDS-containing proteins to the IGS. Knockdown of IGS-RNA enables proteins to retain their dynamic properties and prevents immobilization of NoDS-containing proteins. Thus, the intergenic spacer is a complex transcription unit from which RNA expression is induced in response to specific environmental cues. Given that a large number of proteins contain NoDS, nucleolar detention by inducible IGS RNAs is most likely a prevalent posttranslational regulatory mechanism.
NoRC—a chromatin remodeling complex that safeguards genome integrity
Although rRNA synthesis accounts for more than 50 % of cellular transcriptional activity, a significant fraction of rDNA repeats is constitutively silent (McStay and Grummt 2008). rDNA silencing plays an important role in genome stability, suppressing nonhomologous recombination pathways. Loss of silencing correlates with rDNA instability, nucleolar disintegration, and cellular senescence (Kobayashi 2008; Peng and Karpen 2007). Though the consensus is that stress-dependent regulation of pre-rRNA synthesis mainly occurs by influencing the transcription rate of already active genes, recent findings also point toward additional regulatory pathways, such as epigenetic regulation of rDNA transcription. Generally, transcriptionally active genes are characterized by an accessible “open” euchromatic structure, whereas silent ones exhibit a more compact heterochromatic structure. Specific histone modifications are associated with transcriptionally active and silent rDNA repeats, acetylation of histone H4 and H3K4me3 correlating with transcriptional activity, whereas histone H4 hypoacetylation and trimethylation of H3K9, H3K27, and H4K20 correlate with transcriptional silencing (Santoro et al. 2002; Zhou et al. 2002; Santoro and Grummt 2005; Peng and Karpen 2008; 2009). The evolutionarily conserved packaging of a fraction of rDNA into heterochromatin is not only important for nucleolar structure and function, but also protects the integrity of rDNA repeats and safeguards genomic stability by entailing the assembly of a generally repressive chromatin domain that is less accessible to the cellular recombination machinery. Switching between the active and silent state of rRNA genes is mediated by a chromatin remodeling complex, termed nucleolar remodeling complex (NoRC), a member of ATP-dependent chromatin remodeling machines comprising the ATPase SNF2h and a large subunit, termed TIP5 (Strohner et al. 2001). Targeting NoRC to rDNA leads to rewriting the histone code, increased DNA methylation, and ultimately heterochromatinization and transcriptional silencing of rRNA genes.
Like rRNA genes, centromeres and telomeres are organized in tandemly repeated gene clusters and exhibit a repressive heterochromatic structure. Maintenance of this heterochromatic structure at centromeres and telomeres is pivotal for kinetochore integrity and function as well as for protecting chromosome ends from exposure to the DNA damage response machinery, thus safeguarding chromosome stability. Considering the structural similarities and heterochromatic features of rRNA genes, centromeres, and telomeres and given that telomeres and centromeres are adjacent to rDNA at acrocentric chromosomes, it can be reasoned that NoRC function may not be restricted to silencing of a fraction of rRNA genes but may also be involved in heterochromatin formation at other repetitive sequences. Repetitive elements, comprising 30–50 % of mammalian genomes, are embedded in a compact heterochromatic structure, protecting genomic integrity by inhibiting DNA recombination and counteracting mutagenic rearrangement caused by uncontrolled transposition events (Peng and Karpen 2008, 2009). For this, specific histone-modifying enzymes are guided to these sequences to deacetylate and methylate specific lysine residues in histone tails, thus leading to chromatin compaction. Indeed, NoRC has been found to localize not only in nucleoli, but also at chromosome ends and centric repeats. This suggests that NoRC function is not restricted to heterochromatin formation and repression of rDNA transcription, but also plays a general role in the establishment of higher order chromatin structure at major clusters of repetitive sequences, including telomeres and centromeres. Consistent with NoRC sustaining the integrity and stability of telomeres and centromeres, enhanced recombination between telomeric repeats, chromosomal translocations, and mitotic defects has been observed after knockdown of TIP5, the large subunit of NoRC. Loss of chromatin compaction at pericentric regions leads to disorganized mitotic spindles, unaligned chromosomes in metaphase plates, and increased abundance of anaphase bridges (Postepska-Igielska et al. 2013). Such abnormal chromosome segregation during cell division has been shown to occur after disruption of pericentric heterochromatin and has been attributed to the increased rate of mutations in cancer cells (Ting et al. 2011). Although further analyses are required to explore the mechanism and consequences of NoRC function on regulation of chromatin structure and genomic stability, the finding that NoRC—probably among other players—is able to trigger the establishment and/or support the maintenance of compact chromatin conformation at major genomic clusters of tandemly repeated sequences may have wide-ranging implications for genomic structure and function.
A metabolic throttle regulates the epigenetic state of rDNA
Epigenetic control is not restricted to the formation of a stable, inheritable chromatin state but may also dynamically adapt the chromatin configuration to the physiological state of cells. However, the pathways that alter the chromatin structure in response to environmental or developmental cues remain largely unknown. Murayama et al. (2008) have uncovered an interesting interrelationship between the cellular energy status and rDNA transcription. They have shown that glucose starvation affects the epigenetic state of rRNA genes, suggesting a fine-tuned mechanism by which rDNA silencing may cut back energy expenditure and protect cells from energy deprivation-induced apoptosis. They identified a protein complex, dubbed energy-dependent nucleolar silencing complex (eNoSC), which represses rDNA transcription in response to the cellular energy status. eNoSC comprises three subunits, a nucleolar protein, termed nucleomethylin (NML), the protein deacetylase SIRT1, and the methyltransferase SUV39H1. NML binds to H3K9me2 throughout the rDNA transcription unit and affects rDNA transcription by modulating histone H3K9 methylation at the rDNA promoter. As a consequence, rDNA transcription is repressed. Silencing in response to glucose starvation requires the other eNoSC components, the NAD+-dependent deacetylase SIRT1, and the heterochromatic methyltransferase SUV39H1. Overexpression of SIRT1, the principal NAD+-dependent deacetylase for histones H4K16 and H3K9 and other proteins, augments the repressive effect of NML, while knockdown of SIRT1 or treatment of cells with nicotinamide (an inhibitor of NAD+-dependent deacetylases) prevents NML-dependent transcriptional repression and increases H3K9 acetylation. NML and SIRT1 interact with SUV39H1, which leads to elevated levels of H3K9 methylation. Significantly, the histone methyltransferase activity of SUV39H1 is strongly reduced by acetylation, deacetylation by SIRT1 relieving this inhibition, suggesting that a change in the NAD+/NADH ratio induced by reduction of the intracellular energy status activates SIRT1 which then leads to deacetylation of histone H3 and to dimethylation at Lys9 (H3K9me2) by SUV39H1 (Vaquero et al. 2007). Given that shortage of intracellular energy supply decreases the cellular ATP concentration and increases the NAD+/NADH ratio, activation of SIRT1 and SUV39H1 couples energy levels to ribosome production and links the cellular energy balance to epigenetic silencing of rDNA.
Perspectives
Recent years have seen remarkable progress in our understanding of traditional and nontraditional nucleolar functions. The nucleolus is a major target of signaling pathways that are activated by the cellular stress response network, leading to complex changes in nucleolar structure and protein content. Actually, the nucleolus plays a key role in sensing and responding to external signals, coordinating surveillance and stress control systems. As ribosome biogenesis is an extremely energy-consuming process, alteration of rRNA synthesis is one strategy that can preserve cellular homeostasis. Although the mechanisms controlling stress signaling pathways have been extensively analyzed, many details remain uncharacterized. This exciting and emerging field of research is of great importance not only to fundamental molecular and cellular biology, but also for the understanding of cellular aging and major diseases. Therefore, further characterization of nucleolar stress signaling pathways, identification of specific nucleolar biomarkers, and design of drugs that specifically inhibit components of the Pol I machinery will not only yield important insights into the mechanisms that regulate nucleolar function but may also provide tools to combat cancer and other stress-dependent diseases.
References
Ahmad Y, Boisvert FM, Gregor P, Lamond AI (2009) NOPdb: nucleolar proteome database—2008 update. Nucleic Acids Res 37:D181–D184
Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (2005) Nucleolar proteome dynamics. Nature 433(7021):77–83
Audas TE, Jacob MD, Lee S (2012) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45:147–157
Banski P, Kodiha M, Stochai U (2010) Chaperones and multitasking proteins in the nucleolus: networking together for survival? Trends Biochem Sci 35:361–367
Boisvert FM, Lamond AI (2010) p53-Dependent subcellular proteome localization following DNA damage. Proteomics 10:4087–4097
Barber M, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen R, Paredes S, Young N, Chen K et al (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487:114–118
Boulon S, Westman B, Hutten S, Boisvert SF-M, Lamond AI (2010) The nucleolus under stress. Mol Cell 40:216–227
Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L et al (2012) Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22:51–65
Carson DA, Lois A (1995) Cancer progression and p53. Lancet 346:1009–1011
Chen Y-R, Wang X, Templeton D, Davis RJ, Tan T-H (1996) The role of c-Jun N-terminal kinase (JNK) in apoptosis induced by ultraviolet C and radiation. J Biol Chem 271:31929–31936
Chen S, Seiler J, Santiago-Reichelt M, Felbel M, Grummt I, Voit R (2013) Interaction of SIRT7 with nascent RNA mediates deacetylation of PAF53 and confers stress response to RNA polymerase I transcription. Mol Cell, in press
Clos J, Buttgereit D, Grummt I (1986) A purified transcription factor (TIF-IB) binds to essential sequences of the mouse rDNA promoter. Proc Natl Acad Sci U S A 83:604–608
Comai L, Tanese N, Tjian R (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68:965–976
Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103:239–252
Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H (2007) Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J 26:944–954
Drygin D, Rice WG, Grummt I (2010) The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Ann Rev Pharm Tox 50:131–156
Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T (2002) A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623–1626
Feige J, Auwerx J (2007) Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol 17:292–300
Feige J, Auwerx (2008) Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol 20:303–309
Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20:1075–1080
Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JCBM (2007) A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J 26:1560–1568
Govoni M, Farabegoli F, Pession A, Novello F (1994) Inhibition of topoisomerase II activity and its effect on nucleolar structure and function. Exp Cell Res 211:36–41
Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V (2009) Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci 122:489–498
Grummt I (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17:1691–1702
Grummt I (2010) Wisely chosen paths: regulation of ribosomal RNA synthesis. FEBS J 277:4626–4639
Grummt I, Grummt F (1976) Control of nucleolar RNA synthesis by the intracellular pool sizes of ATP and GTP. Cell 7:447–453
Grummt I, Längst G (2013) Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta-Gene Regulatory Mechanisms 1829:393–404
Haigis MC, Sinclair DA (2010) Mammalian sirtuins: biological insights and disease relevance. Ann Rev Pathol 5:253–295
Hanada KI, Song CZ, Yamamoto K, Yano KI, Maedal Y, Yamaguchi K, Muramatsu M (1996) RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J 15:2217–2226
Heix J, Zomerdijk JCBM, Ravanpay A, Tjian R, Grummt I (1997) Cloning of murine RNA polymerase I-specific TAFs: conserved interactions between the four subunits of the species-specific transcription factor TIF-IB/SL1. Proc Natl Acad Sci U S A 94:1733–1738
Hiscox JA, Whitehouse A, Matthews DA (2010) Nucleolar proteomics and viral infection. Proteomics 10:4077–4086
Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, Voit R (2009) AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci U S A 106:17781–17786
Houtkooper RH, Pirinen E, Auwerx J (2012) Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13:225–238
Imai S, Guarente L (2010) Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci 31:212–220
Jantzen HM, Admon A, Bell SP, Tjian R (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344:830–836
Kar B, Liu B, Zhou Z, Lam YW (2011) Quantitative nucleolar proteomics reveals nuclear re-organization during stress-induced senescence in mouse fibroblasts. BMC Cell Biol 12:33
Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY, Borlak J, Nam SW (2013) Sirtuin oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125b. Hepatology 57:1055–1067
Kobayashi T, Ganley AR (2005) Recombination regulation by transcription-induced cohesion dissociation in rDNA repeats. Science 309:1581–1584
Kobayashi T (2008) A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. BioEssays 30:267–272
Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M (2004) Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5:465–475
Lam YW, Evans VC, Heesom KJ, Lamond AI, Matthews DA (2010) Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol Cell Proteomics 9:117–130
Learned RM, Cordes S, Tjian R (1985) Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol Cell Biol 5:1358–1369
Mayer C, Grummt I (2005) Cellular stress and nucleolar function. Cell Cycle 4:1036–1038
Mayer C, Zhao J, Yuan X, Grummt I (2004) mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev 18:423–434
Mayer C, Bierhoff H, Grummt I (2006) The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev 19:933–941
McStay B, Grummt I (2008) The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24:131–157
Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S (2005) Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol 170:733–744
Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V (2007) A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64:29–49
Muth V, Nadaud S, Grummt I, Voit R (2001) Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J 20:1353–1362
Murayama A, Ohmor K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, KimuraK K, Shimizu T, Yanagisawa J (2008) Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133:627–639
Olson MO (2004) Sensing cellular stress: another new function of the nucleolus? Sci STKE 224:pe10. doi:10.1126/stke.2242004pe10
Peng JC, Karpen GH (2007) H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9:25–35
Peng JC, Karpen GH (2008) Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev 18:204–211
Peng JC, Karpen GH (2009) Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet 5:1–14
Postepska-Igielska A, Krunic D, Greulich-Bode K, Boukamp P, Grummt I (2013) The chromatin remodeling complex NoRC safeguards genomic stability by heterochromatin formation at telomeres and centromeres. EMBO Rep, 14:704–710
Rubbi CP, Milner J (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22:6068–6077
Russell J, Zomerdijk JC (2005) RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 30:87–96
Russell J, Zomerdijk JC (2006) The RNA polymerase I transcription machinery. Biochem Soc Symp 73:203–216
Santoro R, Grummt I (2005) Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25:2539–2546
Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32:393–396
Scheer U, Hock R (1999) Structure and function of the nucleolus. Curr Opin Cell Biol 11:385–390
Scheer U, Xia B, Merkert H, Weisenberger D (1997) Looking at Christmas trees in the nucleolus. Chromosoma 105:470–480
Scherl A, Couté Y, Déon Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ (2002) Functional proteomic analysis of human nucleolus. Mol Biol Cell 13:4100–4109
Schreiber M, Baumann B, Cotten M, Angel P, Wagner ER (1995) Fos is an essential component of the mammalian UV response. EMBO J 14:5338–5349
Sherr CJ, Weber JD (2000) The ARF/p53 pathway. Curr Opin Genet Dev 10:94–99
Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I (2001) NoRC—a novel member of mammalian ISWI chromatin remodeling machines. EMBO J 20:4892–4900
Ting DT, Lipson D, Suchismita P, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, Rivera MN, Berdeesy N, Maheswaran S, Haber D (2011) Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331:593–596
Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM (2012) Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics 11:60–76
Vakrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 102:703–710
Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D (2007) SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450:440–444
Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Gröne HJ, Schütz G, Grummt I (2005) Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest and p53-mediated apoptosis. Mol Cell 19:77–89
Zhang Y, Wolf G, Bhat K, Jin A, Allio T, Burkhart W, Xiong Y (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23:8902–8912
Zhou Y, Santoro R, Grummt I (2002) The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21:4632–4640
Zomerdijk JC, Beckmann H, Comai L, Tjian R (1994) Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science 266:2015–2018
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grummt, I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma 122, 487–497 (2013). https://doi.org/10.1007/s00412-013-0430-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-013-0430-0