Abstract
In salivary gland polytene chromosomes of Drosophila melanogaster, the regions of intercalary heterochromatin are characterized by late replication, under-replication, and genetic silencing. Using Gal4/UAS system, we induced transcription of sequences adjacent to transgene insertions in the band 11A6-9. This activation resulted in a loss of “silent” and appearance of “active” epigenetic marks, recruitment of RNA polymerase II, and formation of a puff. The activated region is now early replicating and shows increased level of DNA polytenization. Notably, all these changes are restricted to the area around the inserts, whereas the rest of the band remains inactive and late replicating. Although only a short area near the insertion site is transcribed, it results in an “open” chromatin conformation in a much broader region. We conclude that regions of intercalary heterochromatin do not form stand-alone units of late replication and under-replication. Every part of such regions can be activated and polytenized independently of other parts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Finely coordinated spatial and temporal order of DNA replication events is one of the fundamental features of functional organization of eukaryotic genomes. Among cell types, replication patterns are fairly conserved and are inherited epigenetically. It was in 1960s that the concept relating replication timing and chromatin organization began to form, attracting further interest later on. Early replicating chromatin was typically observed to be “open,” transcriptionally active or poised for transcription, whereas transcriptionally inert, “closed” heterochromatic regions displayed late replication (Kalisch and Hägele 1976; Holmquist 1987; Aggarwal and Calvi 2004; Hiratani and Gilbert 2009; Gilbert 2010; Ryba et al. 2011; Schwaiger et al. 2009).
Replication patterns undergo significant changes throughout development. Not only is the replication timing locally altered but the affected chromosomal loci also change in size, as well as the positions they occupy in the nucleus change in space. In both mammals and fruit flies, such changes are known to involve about 20 % of genomic sequences, which encompass large domains 400–800 kb long in the latter and up to several megabases in the former (MacAlpine et al. 2004; White et al. 2004; Hiratani and Gilbert 2009; Schwaiger et al. 2009; Pope et al. 2010; Gilbert 2010). It was suggested that changes in replication patterns of such large domains are likely due to the general changes in chromatin organization rather than to the induction or repression of individual genes (MacAlpine and Bell 2005; Schwaiger and Schübeler 2006). In mammals and Drosophila, there are two basic types of replication domains: 0.2- to 2-Mb-long regions which harbor coordinately firing origins providing simultaneous replication within domains and 0.1–0.6 Mb origin-less or origin-depleted zones, whose replication occurs via unidirectional progression of a replication fork from an origin located outside of these zones (Norio et al. 2005; MacAlpine et al. 2010; Borowiec and Schildkraut 2011; Guilbaud et al. 2011; Sher et al. 2012).
Mechanisms that orchestrate cell replication timing are still poorly understood. Whereas genome-wide studies establish general correlations between chromatin state(s) and replication status, experiments focusing on individual domains help to uncover locus-specific replication effects (Eaton et al. 2011). Late-replicating domains of Drosophila melanogaster polytene chromosomes, also known as intercalary heterochromatin (IH) regions, represent one convenient model that helps to address these questions. As a term, IH was originally coined by Kaufmann 1939 and refers to the dense bands located in euchromatin, yet in many respects bearing similarity to pericentric heterochromatin (PH) (Kaufmann 1939). These features include transcriptional silencing, late replication, and under-replication, the latter causing formation of ectopic contacts and chromosome breaks. In contrast to PH, IH regions generally lack repeated DNA (reviewed in Belyaeva et al. 2008).
Taken together, IH regions occupy at least 14 % of the euchromatic portion of the genome; their positions have been molecularly mapped and were subsequently demonstrated to be enriched for SUUR, D1, and Lam proteins (Belyakin et al. 2005; Nordman et al. 2010; Belyaeva et al. 2012). Extensive overlap between IH regions and Lamin-associated DNA sequences suggests their role in the maintenance of spatial organization of a nucleus (Belyaeva et al. 2012). Furthermore, IH is characterized by low gene density, and IH-resident genes typically have narrow tissue- and stage-specific expression patterns, being particularly active in the male germ line (MacAlpine et al. 2004; Belyakin et al. 2010). Additionally, in different Drosophila species, the gene order in the late-replicating and SUUR-containing regions is well conserved throughout evolution (Ranz et al. 2012).
IH bands contain very few, if any, origins of replication (Sher et al. 2012; Belyaeva et al. 2012). Their replication is thought to occur by replication forks progressing from the flanks of IH bands inwards. By the end of S phase, converging forks often fail to meet within IH, thereby resulting in under-replication zones with minimal DNA copy number found in their centers (Belyaeva et al. 2012).
In order to explore how ectopically induced transcription might affect replication timing and DNA polytenization, we used Gal4-dependent activation of UAS-containing transgenes inserted in the IH region 11A6-9. We found that transgene activation causes this late-replicating region to become early replicating and increases its level of polytenization.
Materials and methods
Fly genetics and constructs
We used wild-type strain Oregon R and flystocks harboring unique insertions of EY transposons [P(EPgy2)] with known molecular localization: EY00353 (11A6-9), EY01976 (11A6-9), EY11998 (35B), EY13417 (59D), and EY10546 (75C1-2). Cytology mapping was done using Bridges map as a reference (Bridges 1935). EY transposons comprise 14 UAS copies that can be activated by Gal4 binding, and two visible markers, yellow + and mini-white +, whose transcription occurs upstream and antisense to UAS (Bellen et al. 2004) (Fig. 1a). The following stocks with Gal4 drivers were used: hsp70-Gal4 (heat shock inducible), y w; da-Gal4 (ubiquitous expression), y w; tub-Gal4/TM6 (ubiquitous expression), y w; AB1-Gal4 (salivary-gland-specific expression starting from early embryos), w; Sgs3-Gal4 (salivary-gland-specific expression starting from the mid-third instar stage), and w; GMR-Gal4/CyO (expression in the posterior part of the eye imaginal disk at the third instar larval stage). All Gal4 drivers except for tub-Gal4 and GMR-Gal4 are marked with mini-white +. Flystocks were obtained from Bloomington Drosophila Stock Center, except for tub-Gal4/TM6, which was kindly provided by Prof. P.G. Georgiev.
Structure of EY01976 and EY00353 transposons (from Bellen et al. 2004). a Arrows indicate the transcription orientation of yellow and white, as well as the induced transcription from UAS. Positions of the neighboring genes around transgene insertion sites are shown (b). Span of IH band 11A6-9 is from Belyaeva et al. (2012) (c) and under-replication zone borders are from Nordman et al. (2010) (d). Molecular map coordinates are as in Drosophila Genome Release 5.3 (e)
Stocks that were homozygous for both EY transgenes and Gal4 drivers were established by standard crosses: y w EY00353; da-Gal4, y w EY00353; hsp70-Gal4, y w EY01976; da-Gal4, and y w EY01976; hsp70-Gal4. Flies were kept on cornmeal medium at 18 and 22–25 °C.
Cytology
Preparations of salivary gland polytene chromosomes stained with acetic orcein were made by standard methods.
Immunostaining of polytene chromosomes
Immunostaining was performed as described in Kolesnikova et al. (2011). Primary antibody dilutions used were as follows: rabbit polyclonal anti-SUUR (E-45) (1:50), mouse anti-H1 (kindly provided by Prof. H. Saumweber, 1:100), anti-RNA PolII (specific for the Ser 5-phosphorylated form of Pol II CTD, Covance; 1:50), mouse monoclonal anti-PCNA (PC10, Abcam, ab29; 1:500), rabbit polyclonal anti-γ-H2Av (a gift from Prof. K. McKim, 1:200), mouse anti-D1 (a gift from Prof. E. Käs, 1:200), rabbit anti-H3K9ac (Abcam, 1:200), and goat anti-H3K4me2 (Abcam, 1:100).
The squashes were incubated with secondary FITC- or rhodamine-labeled goat anti-rabbit and anti-mouse IgG-specific conjugates (Abcam, 1:200), with anti-mouse IgM-FITC conjugates (Sigma, 1:250) or AF488 donkey anti-goat conjugates (Molecular Probes, 1:600). Chromosomes were examined using epifluorescence optics (Olympus BX50 microscope) and photographed with CCD Olympus DP50. For every staining class, at least 20 nuclei on several slides were analyzed.
Transcription measurements
Total RNA from 50 salivary glands was isolated using TRIzol (Invitrogen) according to the manufacturer’s protocol. RNA was used for reverse transcription with SuperScript III reverse transcriptase (Invitrogen). Obtained cDNA was used for the measurements of gene expression levels by standard curve qRT-PCR. Each experiment was repeated twice with three technical replicates each. qRT-PCR primers used are listed in Table 1, and their genomic positions are indicated in Fig. 4 and Suppl. Fig. 2.
To estimate the level of genomic DNA contamination in our qRT-PCR, we performed the exactly the same analysis on reverse transcriptase minus samples. We detected some amounts of genomic DNA in all samples, but they were negligible in comparison to the amounts of RNA.
Measurement of DNA polytenization
Genomic DNA was isolated from 50 salivary glands (dissected from female larvae). Polytenization levels were measured by standard curve RT-PCR. Each experiment had two biological replicates, three technical repeats each. The primers used are listed in Table 1 and indicated in Fig. 6 and Suppl. Fig. 3.
Results
Gal4/UAS system can be used to overcome inactivation of reporter genes
In order to make sure that proper transgene activation does occur in Gal4/UAS system, we analyzed several IH regions that harbored transgene insertions to the under-replication zone showing greatly suppressed marker expression. When on white-mutant background, these flies typically have pale-colored and mosaic eyes in both males and females (Babenko et al. 2010). We selected several stocks with single copy “repressed” insertions in the IH regions 59D1-2, 35B1-2, 75C1-2, and 11A6-9. Flies homozygous for these transgenes were crossed to tub-Gal4 flies, resulting in an offspring that was heterozygous for both the transgene and the driver. These flies showed strong mini-white expression, suggesting that Gal4 has overcome mini-white inactivation. Furthermore, IH band material was observed to decondense and form a puff on one of the homologs (data not shown). Thus, we established the functionality of the model system, i.e., that transgene activation does occur.
For a detailed follow-up analysis, we selected the region 11A6-9, which is a classical IH band. It is one of the latest to replicate in the genome, and its borders as well as under-replication profile have been molecularly mapped (Zhimulev et al. 2003; Belyaeva et al. 2006; 2012) (Fig. 1c, d). The region 11A6-9 spans 430 kb and encompasses 20 genes. Of these, 10 are male specific, and according to FlyBase, none of them are expressed in the salivary gland (Chintapalli et al. 2007; Graveley et al. 2011). According to the chromatin state classification made by Filion et al. using DamID approach, in Kc cells, this region is largely represented by BLACK, silent type of chromatin, and is enriched with typical BLACK-type proteins such as SUUR, Lam, and D1 (Filion et al. 2010). As a consequence, the transgenes inserted in this region are generally repressed (Babenko et al. 2010). Much like other IH regions, 11A6-9 shows no ORC binding in salivary glands (Belyaeva et al. 2012; Sher et al. 2012).
We analyzed two insertions in 11A6-9: EY00353, mapping to the center of the band and locating in an intron of Ten-a (insertion coordinate 12097557, genome release 5.3), and EY01976, mapping to the distal part of the band within the gene CG34323 (insertion coordinate 11964491) (Fig. 1). In both lines, homozygotes have eye pigmentation that is strongly different from wild type (Fig. 2a). In white-mutant background (Fig. 2b), the eyes of males and females appear poorly pigmented: they show groups of colored facets scattered over the pale-yellow eye (Fig. 2с). Pigmentation of testis sheath in these strains, which is dependent on white activity (Fig. 2l), is undetectable (Fig. 2m). Inactivation of the yellow reporter in the transgenes is manifested as a weak coloring of wing bristles (Fig. 2i). These appear much lighter than those in wild-type flies (Fig. 2g), yet darker than in the yellow mutants (Fig. 2h). In flies heterozygous for both EY00353 and tub-Gal4, eye (Fig. 2d), bristle (Fig. 2j), and testis sheath (Fig. 2n) pigmentation is restored and matches the phenotypes of w + and y + flies. In sibling heterozygous flies that inherited TM6 balancer instead of the tub-Gal4, chromosome pigmentation remains suppressed (Fig. 2e, k, o). Interestingly, when GMR-Gal4 driver is used (and GMR-Gal4 is known to be active upon cell differentiation in the posterior part of an eye (Ahmad and Henikoff 2001)), the heterozygotes that carry both the transgene and the driver consistently display restored pigmentation in exactly the posterior part of an eye (Fig. 2f). Similar results were obtained for the EY01976 insertion (data not shown). Taken together, these data indicate that Gal4 drivers activate IH-embedded insertions, which renders their reporter genes yellow and white accessible for tissue-specific factors that mediate expression of these genes.
Eye color in wild type (a) and in white mutant (b) is shown as a control. Eye pigmentation in flies with inactive EY00353 insertion (с) or activated with tub-Gal4 (d). In sibs that lack the driver, no increase in eye pigmentation is observed (e). When GMR-Gal4 driver is used, upregulation of white expression occurs only in the posterior part of an eye (f). Bristle color in wild type (g), yellow mutant (h), in EY00353 insert line (i), in EY00353 combined with tub-Gal4 (j), or in sibs lacking the driver (k). Testis sheath color in wild type (l), in males with inactive EY00353 (m), tub-Gal4-activated EY00353 (n), or in sibs without the driver (o)
Chromatin decondensation and changes in profile of epigenetic marks upon transgene activation
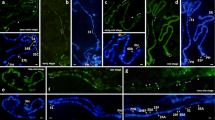
IH region 11A6-9 normally appears as a large dense band. It also shows the same morphology in chromosomes with inactive transgene insertions (Fig. 3a, left). We observed that the chromatin to decondense upon transgene activation, which was dependent on the “strength” of the specific Gal4 driver used. The weakest effect was seen with hsp70-Gal4 at 18 °С. The activated region appeared interband-like, i.e., a narrow stripe of decondensed material was observed within the band harboring activated EY00353 transgene (Fig. 3a, middle, red arrowheads), whereas flanking band shoulders remained dense. At 25 °С, the decondensed distal part of the band expands to form a puff, with proximal part remaining compact. The same was observed for ubiquitously expressed (da-Gal4 (Fig. 3a, right) and tub-Gal4) or salivary-gland-specific (AB1-Gal4 and Sgs3-Gal4) drivers. In the latter case, a much smaller puff was formed.
Chromatin changes observed upon activation of EY00353 insertion. Left column shows the fragment of X chromosome (regions 10 to 11) with an inactive transgene, column in the center shows the region morphology upon hsp70-Gal4 activation at 18 °С, and right column shows the chromosome region induced by da-Gal4. Phase contrast (a) and overlay of phase contrast and immunostaining (b–h). Lines connect homologous regions of the chromosomes. In the center column, red arrowheads point to the narrow decondensed region within 11A6-9, which resulted from Gal4/UAS activation. The images in the right column show that decondensation predominantly involves the distal part of 11A6-9, whereas its proximal part remains compacted (shown as black arrowheads). Images b–d in central and right columns show that silent chromatin proteins map to the dense and inactive parts of the band. Active chromatin marks, on the contrary, map to the decondensed parts (e–g). Asterisk indicates an ecdysone-induced puff 10EF. Bar, 5 μm
We observed that Gal4-driven puff formation was reversible. When hsp70-Gal4 is activated by heat shock at 37 °С for 40 min in the beginning of the third instar age, the puff is formed in 1 h. After 48–50 h at 18 °C, the puff shrinks back to a narrow stripe. The same pattern in the third instar larval polytene chromosomes is observed when single heat shock is applied during embryogenesis, first and second instar stages. Upon slight induction (hsp70-Gal4, 18 °C) of EY01976, the distal part of 11A6-9 separates from the band (Suppl. Fig. 1b, indicated by an arrowhead). Upon stronger induction (da-Gal4), the puff is formed which now occupies the entire distal part of the band (Suppl. Fig. 1c).
Band decondensation and puff formation are accompanied with changes in chromatin makeup. Chromatin marks specific for silent chromatin go away, and active marks appear instead. Specifically, reactivated chromatin now lacks SUUR and D1 (Fig. 3b, c; Suppl. Fig. 1d) and is significantly depleted for histone H1 (Fig. 3d). Instead, it becomes associated with RNA pol II, H3K4me2, and H3K9ac, the marks that were undetectable in this region prior to activation (Fig. 3e–g; Suppl. Fig. 1g, h; Suppl. Fig. 4). Thus, the activated zone acquires features of a typical developmental puff, which is best exemplified by its side-by-side comparison with ecdysone-inducible puff 10EF (indicated by an asterisk in Fig. 3). Importantly, the part of 11A6-9 that remains inactivated still shows major features of IH.
It has previously been demonstrated that phosphorylation of histone variant H2Av is immediately downstream of dsDNA break formation and that it shows extensive overlap with under-replicated regions of polytene chromosomes (Madigan et al. 2002; Andreyeva et al. 2008). This correlation is apparently attributable to the generation of free DNA ends when replication forks stall. In non-inducing conditions, γ-H2Av maps to IH regions, including 11A6-9 (Fig. 3h, left). However, it is no longer detectable in 11A6-9 when induced puff is formed (Fig. 3h, middle, right). This might serve as an indirect support to the idea that under-replication in this region can be suppressed by transcription.
Degree of chromatin decondensation correlates with the level of transcription
In order to confirm that decondensation, puff formation, and changes to protein composition of chromatin are caused by transcription, we analyzed how far the induced transcription proceeds and measured the RNA levels at several regions downstream of UAS using qRT-PCR. As a normalization control, RNA of a housekeeping gene Actin42A was used.
We observed that low transcription was detectable downstream of UAS even without activation, constituting about 1 % of Actin42A expression levels (Fig. 4, primers 353-7; Suppl. Fig. 2, primers 1976-6). Then, we proceeded to quantify RNA in salivary glands from da-Gal4; EY01976, da-Gal4; EY00353, and hsp70-Gal4; EY00353 larvae. We observed a pronounced increase in transcription level in 11A6-9, which varied depending on the driver and temperature used. When da-Gal4 and hsp70-Gal4 drivers were introduced at 22 °C, the transcript levels immediately downstream of UAS were 10–15 times higher than that of Actin42A. At a lower temperature (18 °C), hsp70-Gal4 activation resulted in just sixfold enrichment over Actin42A (Fig. 4; Suppl. Fig. 2). Transcription rapidly decreased with distance from UAS. In the case of EY01976, 2 kb away from the insertion, RNA levels were just 0.7-fold of the Actin42A level (Suppl. Fig. 2, primers 1976-7). EY00353 activation was observed to spread much farther, and even at 9 kb away from UAS, it was still significantly higher than Actin42A levels (Fig. 4, primers 353-8).
Gal4-induced activation of transcription is unidirectional and is known to proceed downstream of UAS, i.e., toward the 3′-end of the transgene, but not upstream. We measured the transcription levels immediately upstream of the transgene 5′-end. It corresponded to 1–10 % of Actin42A probe levels and was 100- to 1,000-fold less than those observed downstream of UAS (Fig. 4, primers 353-4; Suppl. Fig. 2, primers 1976-1).
Induced transcription switches replication timing from late to early within decondensed part of the IH band
To address the question whether replication timing in the band 11A6-9 is altered upon transcriptional activation of the inserted transgene, we analyzed at which stage of the S phase it replicates in wild-type case and upon transgene activation. S phase staging in the context of polytene chromosomes was previously carefully analyzed using radioautography (Arcos-Teran 1972; Hägele and Kalisch 1974; Zhimulev et al. 2003). At the very early stage, which is very short, the majority of the decondensed regions, such as interbands and puffs, replicate. Then, the bands become involved, and incorporation of a radiolabeled DNA precursor during DNA synthesis results in a pattern of continuous chromosome labeling. At the next stage of discontinuous labeling, the bands orderly complete replication, and finally, the last to replicate are the IH regions and the chromocenter. Using antibodies against PCNA, it was further shown that IH regions do not only complete replication late but they also enter replication the last (Belyaeva et al. 2012). It must be noted that PCNA staining pattern is indistinguishable from the pattern of pulse incorporation of labeled DNA precursor during DNA synthesis (Zhimulev et al. 2003). Hence, we used PCNA as a convenient replication marker (Belyaeva et al. 2012). Figure 5a shows that in the chromosomes of a driverless EY00353 stock at an early S phase stage, PCNA is detectable in all regions except for the bands 10A1-2, 10B1-2, 11A1-2, 11A6-9, and 11B1-2. Later, 10B1-2 and 11B1-2 are actively replicating, 10A1-2 and 11A1-2 only enter replication, whereas 11A6-9 still remains unreplicated (Fig. 5b). 11A6-9 can be observed to replicate, when most of other regions of the genome have already finished or about to finish replication (Fig. 5c).
EY00353 transgene activation alters replication timing at 11A6-9, which is best seen when induced with hsp70-Gal4 at 18 °C, when the decondensed zone is “sandwiched” between the fragments of condensed chromatin. At an early S phase stage, most of the regions actively replicate, including the ecdysone-induced puff 10EF. Bands 10A1-2 and 10B1-2 still remain replication negative, but the decondensed region within 11A6-9 has already started replication: PCNA is readily detectable in the middle of the region but is absent from the flanking condensed regions (Fig. 5d, puff is marked by an asterisk). Subsequently, when 10A1-2 and 10B1-2 acquire PCNA staining, replication is spread throughout the entire 11A6-9 (Fig. 5e). By the end of the S phase, replication is limited to the compact regions of 11A6-9 band (indicated by arrowheads in Fig. 5f). The proximal part of the band always remains condensed (arrowhead in Fig. 5i), and even when massive transcription is induced by da-Gal4, PCNA persists here longer as compared to the distal part (Fig. 5g–i). Thus, artificial induction of transcription results in a shift of replication timing from late to early in a decondensed zone within 11A6-9. Similar changes in replication timing were observed when genomic material close to EY01976 was induced by Gal4 (Suppl. Fig. 1e).
Transcription activation leads to increased DNA polytenization at 11A6-9
Since we observed the induced transcription to change the replication timing at 11A6-9, we asked whether the DNA polytenization level would also be altered. To address this question, we used qRT-PCR with primers positioned throughout the region as well as on the flanks of insertions (Fig. 6; Suppl. Fig. 3). Primer pair annealing in the region of a housekeeping gene Actin42A was used as a control showing 100 % polytenization.
Degree of polytenization within the region 11A6-9 harboring EY00353 transgene, activated with da-Gal4 (a) and hsp70-Gal4 (b) drivers. x-axis shows genomic coordinates; y-axis shows the level of polytenization. The y-axis was scaled so that Actin42A gene polytenization level was equal to 1. Red-shaded curves (red, orange, pink) correspond to the lines with inactive insertions or with Gal4 drivers. Blue-shaded lines (dark blue, light blue, purple) depict the polytenization profiles in stocks where transgenes are Gal4-activated. Arrows indicate insertion and PCR probe positions. Error bars represent standard deviation
First we plotted DNA polytenization profiles for 11A6-9 in the Gal4-only (da-Gal4 and hsp70-Gal4) and transgene-only stocks (Fig. 6a, b; Suppl. Fig. 3, red, orange, and pink curves). In all of these cases, replication profiles were highly consistent and matched the profiles previously established for wild-type chromosomes (Belyakin et al. 2005; Belyaeva et al. 2006). Minimum polytenization level is observed in the middle of the IH band, and it gradually increases toward its flanks. It must be noted, that even in the absence of activation, DNA polytenization level of a region around EY00353 is slightly higher than the level observed without an insert; this might be caused by the leaky background transcription of a transgene. This small difference is quite reproducible and is just below the significance level (p value = 7.84 × 10−2, t test).
Gal4-dependent transcriptional activation of a silenced transgene results in a significant increase in its polytenization level. Whereas in the absence of activation the polytenization levels of EY00353 and EY01976 are roughly 30 and 50 %, respectively, upon da-Gal4 activation, both insertions show replication levels close to that of Actin42A (dark blue curves in Fig. 6a, b; Suppl. Fig. 3). This increase is statistically significant (p value = 1.42 × 10−6 for EY00353 vs. EY00353; da-Gal4; p value = 1.80 × 10−7 for EY01976 vs. EY01976; da-Gal4; t test). Yet the effects of transcriptional activation on polytenization are local and do not encompass the entire 11A6-9. Replication profiles show that polytenization is the highest immediately adjacent to the insertion points and decreases outwards.
Polytenization levels were observed to vary depending on the transcription levels. The effect on polytenization was less pronounced when EY00353 is activated by hsp70-Gal4 at 18 °C (purple curve), rather than at 22 °C (light blue curve): at 18 °C, the maximum level of polytenization is lower than that observed at 22 °C, and it drops faster with the distance from the insertion (Fig. 6b). It is possible that transcription directly stimulates polytenization, as we saw that the genomic sequences transcribed from UAS displayed higher polytenization level than the non-transcribed sequences on the other side of the transgene. Figure 6 and Suppl. Fig. 3 show that in both insertion stocks, transcription is oriented distally. Accordingly, polytenization decreases in distal direction slower than it does proximally. Yet another argument supporting the direct effect of transcription on polytenization comes from a sharp peak around da-Gal4-induced EY00353 insertion (Fig. 6a, dark blue curve). Immediately downstream of UAS, polytenization level is first greatly increased, and then, it sharply drops. This polytenization peak is reproducible and is more or less pronounced depending on the conditions, although formally the difference is below the significance level (p value = 1.08 × 10−1, t test). Thus, induced transcription shifts replication timing of a region from late to early, which results in a significant increase in polytenization. The transcribed region (several kilobases, shown as yellow stripe in Fig. 6 and Suppl. Fig. 3) is much smaller than the region affected by the increased polytenization (several hundred kilobases, gray-shaded region in Fig. 6 and Suppl. Fig. 3).
Discussion
In the present work, we used different Gal4 drivers to induce transcription of constructs integrated in the IH regions of polytene chromosomes. This approach has already been applied to overcome position-effect variegation and PC-dependent silencing (Ahmad and Henikoff 2001; Cavalli and Paro 1998). In our experiments, transcription activation was relatively short and reversible, and its intensity and timing being dependent on the specific Gal4 driver used and position of the insert. When insertions in the distal part (EY01976) or in the center (EY00353) of 11A6-9 were compared side by side upon identical induction conditions, in the case of EY01976, RNA levels dropped 10-fold 4 kb away, whereas in the case of the EY00353, the decrease was just 3.5-fold about 10 kb from the transgene. This could be caused by the effects of the neighboring chromatin on transcription. EY00353 is located close to the exon of Ten-a gene and transcription proceeds through the introns and exons of this gene, whereas induced transcription from EY01976 faces an intergenic region.
Gal4-induced transcription is known to be unidirectional, yet chromatin decondensation spreads in both directions away from the transgene insertion. At a very approximate visual estimate, decondensation zone is roughly 200 kb, which is significantly more than the region actually being transcribed. Decondensed chromatin also includes reporter genes yellow and mini-white located upstream of UAS. These genes are probably expressed due to the more open chromatin structure, and so they are capable of attracting tissue-specific expression factors, which does not happen in the absence of Gal4-dependent induction. Such an explanation appears consistent with a well-established fact that chromatin decondensation and nucleosome remodeling may precede transcription and involve more material than that covered by transcription (Tumbar et al. 1999; Chambeyron and Bickmore 2004; Petesch and Lis 2008).
Transcription induction in 11A6-9 results in replication timing changes. Although transcribed area is short, timing changes span over a broader region. This is consistent with the data that not only transcription per se but also the transcription competence of the open chromatin correlates with early replication (Schwaiger et al. 2009; White et al. 2004). The decondensed region of the band becomes early replicating, whereas inactive parts stay late replicating. Thus, the region 11A6-9 does not function as a stand-alone unit of replication control; rather, it consists of separate fragments whose replication timing changes depend on the local chromatin state, transcription, decondensation, and binding of protein factors. It is possible that the changes in replication timing are due to the appearance of novel origins of replication. This is supported by the analysis of DNA copy number profiles across 11A6-9. Even upon minimal induction (hsp70-Gal4 at 18 °C), the polytenization degree in the activated zone is significantly increased compared to the inactive control (p value = 1.81 × 10−9, t test). This would be hard to explain without invoking the formation of a novel early origin of replication with bidirectional progression of replication forks.
One of the markers of late replication regions is SUUR protein (Makunin et al. 2002). SUUR is typically found in 11A6-9 but dissociates from this region upon decondensation. SUUR loss itself does not change replication timing. Even though in SuUR mutants replication of IH bands happens faster, these regions still enter and complete replication late, much like in wild-type chromosomes (Zhimulev et al. 2003; Belyaeva et al. 2012). Furthermore, in SuUR mutants, under-replication zones do not form novel origins (Sher et al. 2012). Thus, the most plausible explanation is that replication timing within the decondensed part of 11A6-9 switches from late to early due to the establishment of an open chromatin structure, but not due to the displacement of SUUR. This explanation is also consistent with the data on preferential association of ORC2 with an active chromatin (MacAlpine et al. 2010).
Thus, replication timing and polytenization degree of a region are coordinated by many factors, such as chromatin type, distribution and efficiency of origins, and speed of replication fork progression. These parameters define the order when genomic regions enter and exit replication. This agrees well with the idea that “high order of the chromatin structure is the most determinant of replication timing” (Donaldson 2005). Importantly, this structure is labile and can allow quick changes to the replication status of any chromosome region. Subdivision of chromosomes into replication domains thus reflects a general feature of eukaryotic genomes having open and closed chromatin regions. This in turn underlies the compartmentalization of transcription and replication in a cell nucleus (MacAlpine et al. 2004; Gierman et al. 2007; Gilbert 2010).
References
Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430:372–376
Ahmad K, Henikoff S (2001) Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104:839–847
Andreyeva EN, Kolesnikova TD, Belyaeva ES, Glaser RL, Zhimulev IF (2008) Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res 16:851–862
Arcos-Teran L (1972) DNA replication and the nature of late replicating loci in the X-chromosome of Drosophila melanogaster. Chromosoma 37:233–296
Babenko VN, Makunin IV, Brusentsova IV, Belyaeva ES, Maksimov DA, Belyakin SN, Maroy P, Vasil'eva LA, Zhimulev IF (2010) Paucity and preferential suppression of transgenes in late replication domains of the D. melanogaster genome. BMC Genomics 11:318
Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC (2004) The BDGP gene disruption project: Single transposon insertions associated with 40 % of Drosophila genes. Genetics 167:761–781
Belyaeva ES, Demakov SA, Pokholkova GV, Alekseyenko AA, Kolesnikova TD, Zhimulev IF (2006) DNA underreplication in intercalary heterochromatin regions in polytene chromosomes of Drosophila melanogaster correlates with the formation of partial chromosomal aberrations and ectopic pairing. Chromosoma 115:355–366
Belyaeva ES, Andreyeva EN, Belyakin SN, Volkova EI, Zhimulev IF (2008) Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster. Chromosoma 117:411–418
Belyaeva ES, Goncharov FP, Demakova OV, Kolesnikova TD, Boldyreva LV, Semeshin VF, Zhimulev IF (2012) Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS One 7(1):e30035
Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF (2005) Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc Natl Acad Sci USA 102:8269–8274
Belyakin SN, Babenko VN, Maksimov DA, Shloma VV, Kvon EZ, Belyaeva ES, Zhimulev IF (2010) Gene density profile reveals the marking of late replicated domains in the Drosophila melanogaster genome. Chromosoma 119:589–600
Borowiec JA, Schildkraut CL (2011) Open sesame: Activating dormant replication origins in the mouse immunoglobulin heavy chain (Igh) locus. Curr Opin Cell Biol 23:284–292
Bridges C (1935) Salivary chromosome map with a key to the banding of the chromosomes of Drosophila melanogaster. J Hered 26:60–64
Cavalli G, Paro R (1998) The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93:505–518
Chambeyron S, Bickmore WA (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18:1119–1130
Chintapalli VR, Wang J, Dow JAT (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature Genet 39:715–720
Donaldson AD (2005) Shaping time: Chromatin structure and the DNA replication programme. Trends Genet 21:444–449
Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM (2011) Chromatin signatures of the Drosophila replication program. Genome Res 21:164–174
Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143:212–224
Gierman HJ, Indemans MHG, Koster J, Goetze S, Seppen J, Geerts D, van Drie R, Versteeg R (2007) Domain-wide regulation of gene expression in the human genome. Genome Res 17:1286–1295
Gilbert DM (2010) Cell fate transitions and the replication timing decision point. J Cell Biol 191:899–903
Graveley BR, Brooks AN, Carlson JW et al (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471:473–479
Guilbaud G, Rappailles A, Baker A, Chen CL, Arneodo A, Goldar A, d’Aubenton-Carafa Y, Thermes C, Audit B, Hyrien O (2011) Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput Biol 7(12):e1002322
Hägele K, Kalisch WE (1974) Initial phases of DNA synthesis in Drosophila melanogaster. I. Differential participation in replication of the X chromosomes in males and females. Chromosoma 47:403–413
Hiratani I, Gilbert DM (2009) Replication timing as an epigenetic mark. Epigenetics 4:93–97
Holmquist GP (1987) Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet 40:151–173
Kalisch WE, Hägele K (1976) Correspondence of banding patterns to 3H-thymidine labeling patterns in polytene chromosomes. Chromosoma 57:19–23
Kaufmann BP (1939) Distribution of induced breaks along the X-chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA 25:571–577
Kolesnikova TD, Semeshin VF, Andreyeva EN, Zykov IA, Kokoza EB, Kalashnikova DA, Belyaeva ES, Zhimulev IF (2011) Induced decondensation of heterochromatin in Drosophila melanogaster polytene chromosomes under condition of ectopic expression of the Supressor of underreplication gene. Fly (Austin) 5:181–190
Madigan JP, Chotkowski HL, Glaser RL (2002) DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res 30:3698–3705
Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF (2002) The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160:1023–1034
MacAlpine DM, Bell SP (2005) A genomic view of eukaryotic DNA replication. Chromosome Res 13:309–326
MacAlpine DM, Rodríguez HK, Bell SP (2004) Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18:3094–3105
MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM (2010) Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20:201–211
Nordman J, Li S, Eng T, MacAlpine D, Orr-Weaver TL (2010) Developmental control of the DNA replication and transcription programs. Genome Res 21:175–181
Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL (2005) Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20:575–587
Petesch SJ, Lis JT (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74–84
Pope BD, Hiratani I, Gilbert DM (2010) Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Res 18:127–136
Ranz JM, Díaz-Castillo C, Petersen R (2012) Conserved gene order at the nuclear periphery in Drosophila. Mol Biol Evol 29:13–16
Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, Gilbert DM (2011) Replication timing: A fingerprint for cell identity and pluripotency. PLoS Comput Biol 7(10):e1002225
Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schübeler D (2009) Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23:589–601
Schwaiger M, Schübeler D (2006) A question of timing: Emerging links between transcription and replication. Curr Opin Genet Dev 16:177–183
Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL (2012) Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res 22:64–75
Tumbar T, Sudlow G, Belmont AS (1999) Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 145:1341–1354
White EJ, Emanuelsson O, Scalzo D, Royce T, Kosak S, Oakeley EJ, Weissman S, Gerstein M, Groudine M, Snyder M, Schübeler D (2004) DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc Natl Acad Sci USA 101:17771–17776
Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, Demakova OV (2003) Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 111:377–398
Acknowledgments
The authors are very grateful to Prof. K. McKim, Prof. E. Käs, and Prof. H. Saumweber for the antibodies; thank Bloomington Drosophila Stock Center and Prof. P.G. Georgiev for the flystocks; and thank Dr. A. Gorchakov for the discussion. This work was supported by the Program of Presidium of the Russian Academy of Sciences “Molecular and Cellular Biology” (grant no. 6.4) and by the grant of Russian Foundation of Basic Research 12-04-31128 (DAM).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Terry Orr-Weaver
Dmitry E. Koryakov and Galina V. Pokholkova contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Changes in chromatin make-up upon induced transcription of EY01976. Shown is a fragment of X chromosome (regions 10-11) with inactive insertion (a), insertion activated by hsp70-Gal4 at 18°С (b) and by da-Gal4 (c). Phase contrast (a-c) and overlay of phase contrast and immunostaining (d-h). Lines connect the same chromosome regions. Asterisk denotes a puff at 10EF. Bar, 5 μm (JPEG 59 kb)
Supplementary Fig. 2
Normalized RNA quantification around EY01976 insertion, shown as a log scale. qRT-PCR probes used and their positions on molecular map are indicated below the columns. Genotypes and induction conditions are color-coded. Error bars represent standard deviation (GIF 6 kb)
Supplementary Fig. 3
Degree of polytenization within the region 11A6-9 harboring EY01976 transgene, activated with da-Gal4 driver. x-axis shows genomic coordinates, y-axis shows the level of polytenization. The y-axis was scaled so that Actin42A gene polytenization level was equal 1. Red-shaded curves (red, orange, pink) correspond to the lines with inactive insertions or with Gal4-drivers. Blue-shaded lines (dark blue, light blue, purple) depict the polytenization profiles in stocks where transgenes are Gal4-activated. Arrows indicate insertion and PCR probes positions. Error bars represent standard deviation (GIF 32 kb)
Supplementary Fig. 4
Examples of typical immunostaining results with antibodies against RNA pol II, H3K9me2, and H3K9ac in the polytene sections 10-11. Fluorescent signal and phase contrast images are shown separately. The images with fluorescence clearly demonstrate the fact that prior to activation the region 11A6-9 (indicated by arrows) was not associated with these proteins. Bar, 5 μm (JPEG 99 kb)
Rights and permissions
About this article
Cite this article
Koryakov, D.E., Pokholkova, G.V., Maksimov, D.A. et al. Induced transcription results in local changes in chromatin structure, replication timing, and DNA polytenization in a site of intercalary heterochromatin. Chromosoma 121, 573–583 (2012). https://doi.org/10.1007/s00412-012-0382-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-012-0382-9