Abstract
Histone H3 variants play critical roles in the functional specialization of chromatin by epigenetically marking centromeric chromatin and transcriptionally active or silent genes. Specifically, the cenH3 histone variant acts as the primary epigenetic determinant of the site of kinetochore assembly at centromeres. Although the function of histone variants is well studied in plants, animals, and fungi, there is little knowledge of the evolutionary conservation of histone variants and their function in most protists. We find that Giardia intestinalis—a diplomonad parasite with two equivalent nuclei—has two phylogenetically distinct histone H3 variants with N-terminal extensions and nonconserved promoters. To determine their role in chromatin dynamics, conventional H3 and the two H3 variants were GFP-tagged, and their subcellular location was monitored during interphase and mitosis. We demonstrate that one cenH3-like variant has a conserved function in epigenetically marking centromeres. The other H3 variant (H3B) has a punctate distribution on chromosomes, but does not colocalize with active transcriptional regions as indicated by H3K4 methylation. We suggest that H3B could instead mark noncentromeric heterochromatin. Giardia is a member of the Diplomonads and represents an ancient divergence from metazoans and fungi. We confirm the ancient role of histone H3 variants in modulating chromatin architecture, and suggest that monocentric chromosomes represent an ancestral chromosome morphology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Both the assembly of the kinetochore on centromeric chromatin and the attachment of the kinetochore to the spindle microtubules are required for accurate chromosome segregation during mitosis. Although centromeric sequences vary greatly at the nucleotide level (Malik and Henikoff 2002), the cenH3 histone variant is an epigenetic mark of centromere identity. The cenH3 histone variant (also known as Cid in Drosophila, Cse4p in budding yeast, and CENP-A in metazoans and S. pombe) replaces the conventional H3 histone in centromeric nucleosomes (reviewed in Malik and Henikoff 2003). cenH3 plays an essential role in recruiting kinetochore proteins and is the primary determinant in establishing the site of kinetochore assembly (Malik and Henikoff 2003). Other constitutively expressed histone H3 variants, such as H3.3, are proposed to replace conventional H3 in nucleosomes during interphase, define active areas of transcription or mark the boundaries of euchromatin and heterochromatin (Malik and Henikoff 2003; Henikoff and Ahmad 2005).
Giardia intestinalis is a widespread parasitic protist and remains a leading cause of malabsorptive diarrhea worldwide (Adam 2001; Savioli and Smith et al. 2006). Giardia has two life cycle stages: the trophozoite or flagellate form that attaches to the intestinal microvilli and an encysted form that can persist in the environment (Adam 2001). As a member of the Diplomonads, Giardia has received some attention due to evolutionary controversies (Baldauf and Roger et al. 2000; Dacks and Marinets et al. 2002; Baldauf 2003; Knight 2004; Graczyk 2005; Ciccarelli and Doerks et al. 2006). However, in phylogenetic analyses with an archaeal outgroup, the Diplomonads do indeed represent the earliest diverging lineage of extant eukaryotes (Sogin and Gunderson et al. 1989; Baldauf and Roger et al. 2000; Van de Peer and Baldauf et al. 2000; Baldauf 2003; Best and Morrison et al. 2004; Ciccarelli and Doerks et al. 2006). Like other Diplomonads, Giardia possesses two transcriptionally equivalent diploid nuclei (2n = 10) and a complex microtubule cytoskeleton (Kabnick and Peattie 1990; Elmendorf and Dawson et al. 2003). In cell division, Giardia has two mitotic spindles and a semiopen mitosis (Sagolla et al. 2006). Both nuclei remain separate during mitosis and sister chromatids of each nucleus segregate equationally in anaphase to form four daughter nuclei (Yu and Birky et al. 2002; Sagolla et al. 2006). There is a growing interest in investigations of molecular aspects of the giardial cell cycle, principally the mechanism of chromosome segregation, with the aim of developing antigiardial compounds that could target cell division machinery. Studies of cell division in Giardia at the molecular level, however, have been complicated by the absence of cytological and molecular descriptions of intermediate stages of mitosis, including the mitotic spindle (Solari and Rahn et al. 2003) and chromosome behavior.
In eukaryotes, histone variants play a critical role in packaging functionally specialized chromatin (Henikoff and Ahmad 2005). Conventional histones (including H2A, H2B, H3, and H4) have been identified in Giardia both computationally and biochemically (Adam 2000; Wu and McArthur et al. 2000); in addition, two H3 variants have been identified in the Giardia genome (Malik and Henikoff 2003). To determine whether the site of attachment of chromosomes to the spindles is conserved in Giardia, and to distinguish whether either of these variant H3 histones defines giardial centromeric regions, we constructed C-terminal GFP fusions of the two H3 variants (H3B::GFP and cenH3::GFP) as well as the conventional H3 (H3::GFP). Based on its unique localization and its lack of colocalization with anti-H3K4 methylation antibodies that define active areas of transcription, we suggest that one H3 variant in Giardia, H3B, is likely associated with noncentromeric heterochromatin. Lastly, from the localization and behavior of the cenH3::GFP during interphase and mitosis (Sagolla et al. 2006), we confirm that cenH3 is the centromere-associated H3 variant, that giardial chromosomes are monocentric, and that centromeric regions containing cenH3::GFP serve to attach chromosomes to the spindle, as in other eukaryotes.
Materials and methods
Sequence and phylogenetic analyses
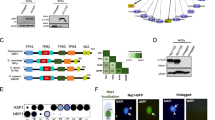
To determine evolutionary relationships among the H3 histone variants, we aligned both Giardia H3 histones and H3 variants with other eukaryotic H3 histone variants (Fig. 1a) using CLUSTALX (Aiyar 2000). We estimated the evolutionary relationships among the giardial and related H3 variants in other protists using the Bayesian method of phylogenetic inference within the Mr. Bayes 3.0 software package (Ronquist and Huelsenbeck 2003). Putative histone modification sites were identified in the N-terminal regions of H3 variants (shown in Fig. 1a). Posterior probabilities of trees were approximated with eight-chain Metropolis-coupled Markov chain Monte Carlo (MCMC) analysis, using a mixed amino acid frequency model. The chains were sampled every 1,000 generations, and inferences from each run were based upon a total of 100,000 sampled trees (Fig. 1b). A consensus of the stationary stage (following the subtraction of the first 10,000 generations as “burn-in”) of this phylogenetic analysis is presented in Fig. 1b.
Sequence analysis and evolutionary relationships of the histone H3 variants in Giardia. a Multiple sequence alignment of giardial histone H3 variants, including cenH3 histone variants and the giardial H3B variant (shaded in red). The lightly shaded red region corresponds to the highly conserved histone core motif; note the divergent N-terminal extensions of the cenH3 histone variants and the insertions in the loop 1 region. CenH3 variants typically lack a conserved glutamine residue (boxed) in the alpha-1 helix. The putative H3S10 phosphorylation site is marked with a carat. Asterisks mark conserved lysine residues (including K4, K9, K18, K23, and K27) that correspond to sites of epigenetic modification that are involved in both transcriptional silencing and activation in metazoans, fungi, and plants. Species abbreviations are: Dme Drosophila melanogaster, Sv Spironucleus vortens, Gi Giardia intestinalis, Lmaj Leishmania major, Ath Arabidopsis thaliana, Tth Tetrahymena thermophila, Ce Caenorhabditis elegans, Hs Homo sapiens, Spo Schizosaccharomyces pombe, and Sce Saccharomyces cerevisae. b Bayesian phylogenetic tree of histone H3 variants and conventional H3 histones demonstrating the evolutionary relationships of the giardial sequences (highlighted in red). Additional species abbreviations are: Ths Thalassosira pseudonana, Xla Xenopus laevis, Ehi Entamoeba histolytica, Cre Chlamydomonas reinhardtii, Dio Dictyostelium discoideum, Tbr Trypanosoma brucei, and Tcr Trypanosoma cruzi. Bayesian posterior probabilities are listed adjacent to branches when >0.50. Scale bar indicates 0.1 changes/site
C-terminal GFP tagging of conventional H3 histone and H3 variants
Histone::GFP fusion constructs were constructed by first modifying the GFPa.pac vector (Singer and Yee et al. 1998) by ligating complementary oligonucleotides (MCSA: 5′ AGCTTTGGCGCGCCGATATCCGGAT3′ and MCSB 5′ AGCTATCCGGATATCGGCGCGCCAA3′) into a HindIII restriction site, thereby creating novel AscI, BspEI, and EcoRV sites. The accession numbers of the giardial histones used in this study are: H3 (AF139875), H3B (XM_762300), and cenH3 (XM_766527). The following oligonucleotides were used in polymerase chain reaction (PCR) amplification of giardial genomic DNA to generate C-terminal histone:GFP constructs: gcenH3F 5′ AATTGGCGCGCCCCTGGGAACTGTCCCAGGCTTGTTAGCGAAGG3′ and gcenH3R 5′ ATCGAACCGGTAGCCGTAGTGAATTTAAGTTGCGCTGCTGAATTCT3′; gH3F: 5′ GGCGCGCCCGTTGTATTGACACTTTCGACGTATAGCAACTTATCACAGG 3′ and gH3R: 5′ ACCGGTAGCTTGCCCTTCCTGTACTCGGGCTCGATCCGCTTGCC 3′ and gH3BF:5′ GGCGCGCCTAGGGCGTGTCATCCCTTACTTAGGCC 3′ and gH3BR:5′ ACCGGTAGCAGCATACTTCCATGAATGCCGTTCA 3′. PCR products included terminal AscI/AgeI restrictions sites, and were initially cloned using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA, USA), and subsequently subcloned into Asc1/Age1 sites of the pMCS-GFP vector. Transformation and maintenance of GFP-tagged Giardia strains was performed as previously described (Singer and Yee et al. 1998).
Immunostaining of the microtubule cytoskeleton and the histone::GFP fusions
Histone::GFP strains were cultured in modified TYI-S-33 medium (Keister 1983) in either 13 ml or 8 ml plastic screw cap tubes (Fisher Scientific) at 37°C, maintained with 50 μg/ml puromycin (CalBioChem). GFP-tagged strains were imaged following direct fixation using either anti-alpha tubulin immunostaining (TAT1, Amersham) or anti-mono or anti-dimethylated H3K4 antibodies (Millipore) as previously described (Sagolla et al. 2006). Cultures enriched for mitotic cells were collected and processed for immunostaining at 1-h intervals over a 4-h period to ensure that a sufficient percentage of mitotic cells were observed (10–20% of total cells) (see Sagolla et al. 2006). Images were acquired using an Olympus IX70 wide-field inverted fluorescence microscope, captured using a Photometrics CCD CH350 camera (−35°C) (Roper Scientific, Tuscon, AZ, USA). Serial sections in the Z-axis were acquired at 0.2-μm intervals, data stacks were deconvolved, and 2D projections of 3D data sets were made using the SoftWoRx software (Applied Precision, Issaquah, WA, USA) for presentation purposes.
Results
We have found two copies of each core histone (H2A, H2B, and H3) and three copies of H4 in the Giardia genome, supporting the previous identification of histone sequences in the Giardia genome (Wu and McArthur et al. 2000). We also confirmed the absence of the H1 linker histone in the genome. Rather than tandem copies, the core histones were syntenic on chromosomes within an average of 3 kb of each other (data not shown). Each conventional histone retained a conserved 15 nucleotide element as previously reported (Wu and McArthur et al. 2000). In contrast, the H3 variants H3B and cenH3 did not contain this or any identifiable conserved upstream sequence element.
The alignment of H3 histones and histone variants (Fig. 1a) demonstrates that both the H3B and putative cenH3 variant in Giardia contain N-terminal extensions of roughly 21 and 44 amino acids, respectively, as compared to conventional H3 (Fig. 1a). We also have highlighted the putative H3S10 phosphorylation site in giardial H3 variants (H3:T9, H3B:S11, S13, cenH3:S1,S4,S6) that is conserved in other eukaryotes (Fig. 1a) and critical for the phosphoregulation of histones during mitosis (see Pascreau and Arlot-Bonnemains et al. 2003; Prigent and Dimitrov 2003). Histone lysine methylation occurs on both H3 and H4 histones in diverse organisms and the methylation of particular lysines correlates with either transcriptional activation or repression (Zhang and Reinberg 2001). Conserved lysine residues (H3K4, H3K9, H3K14, H3K18, H3K23, and H3K27) are also present in the N-terminal regions of the giardial H3 histone, as well as in the N-terminal extensions of H3B and cenH3 (Fig. 1a); the H3B sequence notably lacks the H3K9 position. These correspond to sites of epigenetic modification involved in transcriptional activation and/or silencing in metazoans, fungi, and plants.
Using Bayesian phylogenetic analysis, we determined the evolutionary relationships among the H3 variants. Based on minimal statistical support, we were unable to conclude that cenH3 variants are monophyletic with respect to conventional H3 histones. Furthermore, we find that other H3 variants, such as H3.3 and H3B, are not specifically related (Malik and Henikoff 2003). The relatively longer branch lengths associated with the cenH3 and H3B variants indicate they are undergoing a more rapid evolution (Fig. 1b).
To determine the specific localization patterns for the H3 variants, we imaged strains expressing individual GFP-tagged histone variants in conjunction with an anti-alpha tubulin antibody to highlight the microtubule cytoskeleton. We found that while the conventional histone (H3::GFP) localized along the entire length of chromosomes during interphase (colocalizing with DAPI-stained chromatin), both the H3B::GFP and cenH3::GFP localized to foci (Fig. 2, and Supplemental Fig. 1). As compared to H3B::GFP, the GFP-tagged H3 variant, cenH3::GFP localized to ten discrete foci in each interphase nucleus (Fig. 2i–l and Supplemental Fig. 1). However, there appear to be more than 10 H3B::GFP foci per interphase nucleus and many are smaller than the cenH3::GFP foci. In anaphase A, the cenH3::GFP localized to the leading edge (toward the spindle pole) of the segregating DNA; cenH3::GFP foci remained tightly clustered together at the spindle poles during anaphase B (Fig. 3i–l and see Sagolla et al. 2006). In contrast, during mitosis, H3::GFP remained localized along the length of chromosomes (Fig. 3a–d). H3B::GFP localized to many foci on chromatin, not just at the leading edge of the spindles, and some separating chromosomes appeared to have multiple H3B::GFP foci (Fig. 3e–h). Giardia has two presumably equivalent nuclei (Adam and Nash et al. 1988; Kabnick and Peattie 1990), and consistent with this supposition, we find no discernable differences in the localization patterns of the conventional H3 or the H3 variants between the two nuclei.
Localization of H3 and the H3B and cenH3 variants during interphase. Interphase localization of GFP-tagged H3 histone variants and the microtubule cytoskeleton in trophozoites. Conventional H3 histone colocalizes with chromatin (DAPI, blue) in interphase nuclei (H3::GFP in green, b–d). The giardial H3B variant also colocalizes with chromatin in interphase, but as many small foci (H3B::GFP in green, f–h). The centromere-specific histone variant, cenH3, defines centromeric chromatin in interphase forming 10 large spots/nucleus (cenH3::GFP foci in green, j–l). Anti-alpha tubulin labels the microtubule cytoskeleton (d, h, l in red) and DAPI labels chromatin (all cells, in blue). Images are representative of GFP localizations observed in over 250 cells per strain. Scale bar = 2 μm
Differential localization of GFP-tagged H3 and the H3B and cenH3 variants during mitotic anaphase. Immunolocalization of GFP-tagged H3 histone variants with the microtubule cytoskeleton in late mitosis. Conventional H3 histone colocalizes with chromatin in mitotic nuclei (DAPI in blue) (H3::GFP in green, b–d). The giardial H3B variant localizes to many foci (>10) in chromatin in mitosis (green, f–h). In anaphase, chromatin is segregated to spindle poles along the left–right axis and centromeres (cenH3::GFP foci) are clustered at the spindle poles (green, j–l). Anti-alpha tubulin defines the microtubule cytoskeleton and highlights the dual mitotic spindles, which are perpendicular to the long axis of the cell (d, h, l, in red). DAPI is used to mark chromatin (all cells, in blue). Images are representative of GFP localizations observed in over 250 cells per strain. Scale bar = 2 μm
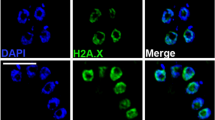
The H3B::GFP staining pattern could be localized to active areas of transcription or possibly to other noncentromeric heterochromatin. To identify active regions of transcription in the giardial nuclei, we used both anti-monomethyl or dimethyl H3K4 immunostaining in each of the three GFP-tagged histone H3 variants (Fig. 4). In general, we observed roughly 5–10 foci of anti-methyl H3K4 immunostaining in both nuclei as illustrated by H3::GFP staining using both antibodies (a–c, j–l). Foci from monomethyl H3K4 immunostaining do not colocalize with either H3B::GFP (Fig. 4m–o) or cenH3::GFP (Fig. 4p–r). In addition, dimethyl H3K4 foci also do not colocalize with H3B::GFP (Fig. 4d–f) or cenH3::GFP staining (Fig. 4g–i). Anti-H3K9 methylation (Millipore) antibodies have previously been shown to define areas of transcriptional gene silencing in other organisms (reviewed in Martin and Zhang 2005), but did not cross-react in Giardia (data not shown).
Active regions of transcription, as defined by mono- or dimethyl H3K4 immunostaining, do not colocalize with histone variant GFP fusions. Immunolocalization of anti-monomethyl H3K4 and anti-dimethyl H3K4 antibody to both giardial nuclei of interphase H3::GFP, H3B::GFP, and cenH3::GFP strains. The upper panel demonstrates that monomethyl H3K4 immunostaining does not colocalize with either H3B::GFP (d–f) or cenH3::GFP (g–i). Similarly, in the lower panel, dimethyl H3K4 immunostaining also does not colocalize with H3B::GFP (m–o) or cenH3::GFP staining (p–r). We generally observed 5–10 foci of antimethyl H3K4 immunostaining in all nuclei (see H3::GFP staining using both antibodies (a–c, j–l). Images are representative of GFP localizations observed in over 250 cells per strain. Scale bar = 1 μm
Discussion
cenH3 defines centromeric regions in Giardia
In many eukaryotes, histones are encoded by multiple sequence variants that are present either to increase gene dosage or to regulate the structural/functional heterogeneity of chromatin; different histone variants contribute to different chromosome architectures and functions. Histone H3 variants like cenH3 and H3.3 tend to lie outside of histone gene clusters and are usually transcribed independent of S-phase initiation, whereas core histones such as H3 are transcribed during S-phase. Centromeric proteins such as cenH3 are required to build the mitotic kinetochore by recruiting motors and other structural and regulatory proteins (Van Hooser and Ouspenski et al. 2001).
Although both H3 variants in Giardia possess N-terminal extensions, the localization patterns during both interphase and mitosis support the identification of a homolog of the centromeric histone variant cenH3 in Giardia. Specifically, we find that cenH3::GFP localizes to discrete foci on interphase chromosomes and to the spindle poles in anaphase (Figs. 2 and 3; Sagolla et al. 2006), indicating that cenH3 is an epigenetic marker of centromeres. Giardia has a haploid genome size of 12 Mb, with five chromosomes of sizes 1.4 to 3.4 Mb (Adam 2000). The presence of ten cenH3::GFP foci in each nucleus (see Supplemental Fig. 1) suggests that giardial chromosomes are monocentric like the majority of eukaryotic cell types, rather than holocentric, as is found in some eukaryotes such as C. elegans (Dernburg 2001; Maddox and Oegema et al. 2004). Although we have not identified homologs of other conserved kinetochore components, the presence of cenH3 at centromeres and the anaphase localization of cenH3::GFP imply that chromosome segregation is mediated by microtubule attachments at the kinetochore. Giardial centromeres presumably function as the site of kinetochore assembly and spindle microtubule attachment during chromosome segregation. Indeed, using the cenH3::GFP as a marker of centromeres, we recently provided the first comprehensive analysis of the complex nuclear and cytoskeletal behavior of mitosis in Giardia (Sagolla et al. 2006).
This analysis of cenH3 in Giardia confirms that the cenH3 histone variant represents a universal epigenetic marker of the eukaryotic centromere—that is independent of centromere sequence (Sullivan and Blower et al. 2001). Centromeres in plants and animals are maintained by highly repetitive satellite DNA or heterochromatin (Malik and Henikoff 2003), yet centromeric repeats differ considerably, even between closely related species (Malik and Henikoff 2001). In rare cases noncentromeric DNA sequences can acquire centromeric activity, and this change in function is correlated with the presence of cenH3, not a specific DNA sequence (Lo and Magliano et al. 2001). The emerging consensus from a variety of diverse organisms is that specific repetitive DNA sequences operate in cis to recruit centromere proteins (including the cenH3 histone) thus epigenetically imparting identity and function to centromeres (Malik and Henikoff 2002). In this way, the cenH3 histone variant functions to establish centromeric identity and the site of kinetochore assembly on chromosomes.
Although the conventional H3 histone is highly conserved, the cenH3 genes are rapidly evolving (Fig. 1b and Malik and Henikoff 2003)—chiefly in terms of the length and sequence divergence of the N-terminal region. Even in Giardia, it appears that both cenH3 and centromeric chromatin rapidly coevolved and diverged in sequence, but maintained the same conserved function. The giardial cenH3 N-terminal region—like that of other cenH3s—lacks sequence conservation, with the exception of the putatively conserved H3S10 histone modification site (Fig. 1a). As in other eukaryotes, we find putative serine phosphorylation sites in the H3B and cenH3 variants, suggesting that these variants could share the same mechanism of phosphoregulation as other organisms (Prigent and Dimitrov 2003). For instance, the phosphorylation of S10 in H3 histones begins in mitotic prophase, after centromere duplication, but before the attachment of chromosomes to the spindle. Phosphorylation decreases during anaphase, but may regulate the function of the kinetochore (Prigent and Dimitrov 2003). The subsequent analysis of such modifications in Giardia could provide further insight into centromere function and kinetochore assembly.
Unique localization and putative function of the H3B histone variant
Histone H3 variants, such as cenH3, play a key role in the functional specialization of chromatin (Henikoff and Ahmad 2005). Besides the cenH3 variant, other H3 variants have evolved multiple times; these variants, such as the H3.3 variant in Drosophila, are often associated with high levels of transcription, particularly at ribosomal gene arrays (Ahmad and Henikoff 2002a,b). Alternatively, noncentromeric H3 variants may also define boundaries of euchromatin and heterochromatin or noncentromeric regions of heterochromatin (Henikoff and Ahmad 2005). The unique punctate localization of the giardial H3B histone variant suggested that it could have a more specialized function in nucleosomes than the core histone H3, possibly analogous to H3.3 histone variants in other eukaryotes (Brown 2001; Smith 2002). In Drosophila, the H3.3 variant has a replication-independent deposition in nucleosomes and is a marker of activated chromatin (Ahmad and Henikoff 2002a,b). Furthermore, H3.3 accounts for roughly 25% of the total H3 histone in bulk chromatin (McKittrick and Gafken et al. 2004) and can establish heritable epigenetic distinctions affecting chromatin function. Because the many H3B::GFP foci produced a fine punctate pattern on chromatin in interphase and mitosis—as compared to that of conventional H3::GFP and the cenH3::GFP (see Figs. 2 and 3)—H3B could define either active areas of transcription or noncentromeric regions of heterochromatin.
Core nucleosomal histones, such as H3, are transcribed coincident with DNA replication during S-phase (Smith 2002), whereas H3 variants such as H3.3 and Cid (a CenH3 homolog) are deposited throughout the cell cycle (Ahmad and Henikoff 2002a,b). There appears to be a correlation between the regulation of cenH3 expression and the timing of centromere replication, distinct from when the bulk of histone H3 nucleosome deposition occurs in S-phase (Smith 2002). For example, in metazoans, the expression of the cenH3 variant, CENP-A, under control of an H3 promoter (active early in S-phase) results in its mislocalization (Van Hooser and Ouspenski et al. 2001). The upstream regions (around −35) of core Giardia histones contain a 15 nucleotide element, which has been proposed to be either a histone-specific or perhaps an S-phase-specific promoter (Wu and McArthur et al. 2000). We have found that both the H3B and cenH3 variants lack this upstream sequence element. The absence of the histone-specific sequence element in H3B and cenH3 promoter regions implies that their expression is regulated differently from the core nucleosomal histones, most likely in a DNA replication-independent manner.
Currently, there is no method of cell synchronization in Giardia, and so we were unable to define transcriptional regions during the cell cycle. Histone lysine methylation of specific sites within the conventional H3 histone (monomethyl-, dimethyl-, or trimethylation) is known to indicate either activation or repression of transcription: the main sites of activation include K4, K36, and K79 of histone H3 (Martin and Zhang 2005). H3K4 methylation acts as a binding site to recruit the SAGA (Spt-Ada-Gcn5-acetyltransferase) complex, leading to transcriptional activation in yeast (Pray-Grant and Daniel et al. 2005). H3K4 di- and trimethylation modifications are particularly enriched at actively transcribed genes; however, while the monomethyl and dimethyl modifications are generally localized to actively transcribed genes, the trimethyl modification is restricted to the 5′ region of these genes (reviewed recently in Martin and Zhang 2005).
To determine whether the giardial histone variant H3B defines actively transcribed regions (analogous to H3.3) we used both anti-monomethyl and anti-dimethyl H3K4 immunostaining to mark active areas of transcription in giardial chromatin (Fig. 4). We found that either monomethyl or dimethyl modification of H3K4 localized to discrete foci that did not overlap with either H3B::GFP or cenH3::GFP foci during interphase. Thus, we conclude that H3B does not function in defining actively transcribed genes, but possibly has a role in defining noncentromeric heterochromatin.
Insights into evolution of heterochromatin from Giardia
A crucial event in the cellular evolution of eukaryotic genomes was the invention of a mechanism to segregate the genome using the cytoskeleton. Both Archaea and eukaryotes package DNA using histones (reviewed in Malik and Henikoff 2003), yet only eukaryotes segregate DNA using the microtubule cytoskeleton. How did mitosis evolve? It is apparent that the evolution of the kinetochore and centromere was integral to this novel mechanism of chromosome segregation. Because of Giardia’s divergent evolutionary position, we predict that monocentric chromosomes are the ancestral state, in contrast to proposals that holocentric chromosomes reflect an ancestral state of chromosome structure (Dernburg 2001). We further hypothesize that additional functions of the kinetochore—such as the maintenance of sister chromatid cohesion and the monitoring of incomplete or inappropriate microtubule attachments (Bernard and Maure et al. 2001)—are also ancient functions. Understanding the molecular mechanisms and dynamics of the mitotic spindles in Giardia provide a unique perspective on mitosis in other eukaryotes, and may illuminate ancient components of this mechanism.
References
Adam RD (2000) The Giardia lamblia genome. Int J Parasitol 30(4):475–484
Adam RD (2001) Biology of Giardia lamblia. Clin Microbiol Rev 14(3):447–475
Adam R, Nash T et al (1988) The Giardia lamblia trophozoite contains sets of closely related chromosomes. Nucleic Acids Res 16(10):4555–4567
Ahmad K, Henikoff S (2002a) Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci USA 99(Suppl 4):16477–16484
Ahmad K, Henikoff S (2002b) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9(6):1191–1200
Aiyar A (2000) The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol Biol 132:221–241
Baldauf SL (2003) The deep roots of eukaryotes. Science 300(5626):1703–1706
Baldauf SL, Roger AJ et al (2000) A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290(5493):972–977
Bernard P, Maure JF et al (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294(5551):2539–2542
Best AA, Morrison HG et al (2004) Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia. Genome Res 14(8):1537–1547
Brown DT (2001) Histone variants: are they functionally heterogeneous? Genome Biol 2(7):REVIEWS0006
Ciccarelli FD, Doerks T et al (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311(5765):1283–1287
Dacks JB, Marinets A et al (2002) Analyses of RNA Polymerase II genes from free-living protists: phylogeny, long branch attraction, and the eukaryotic big bang. Mol Biol Evol 19(6):830–840
Dernburg AF (2001) Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol 153(6):F33–F38
Elmendorf HG, Dawson SC et al (2003) The cytoskeleton of Giardia lamblia. Int J Parasitol 33(1):3–28
Graczyk TK (2005) Is Giardia a living fossil? Trends Parasitol 21(3):104–107
Henikoff S, Ahmad K (2005) Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21:133–153
Kabnick KS, Peattie DA (1990) In situ analyses reveal that the two nuclei of Giardia lamblia are equivalent. J Cell Sci 95(Pt 3):353–360
Keister DB (1983) Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg 77:487–488
Knight J (2004) Giardia: not so special, after all? Nature 429(6989):236–237
Lo AW, Magliano DJ et al (2001) A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res 11(3):448–457
Maddox PS, Oegema K et al (2004) “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res 12(6):641–653
Malik HS, Henikoff S (2001) Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157(3):1293–1298
Malik HS, Henikoff S (2002) Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev 12(6):711–718
Malik HS, Henikoff S (2003) Phylogenomics of the nucleosome. Nat Struct Biol 10(11):882–891
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6(11):838–849
McKittrick E, Gafken PR et al (2004) Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA 101(6):1525–1530
Pascreau G, Arlot-Bonnemains Y et al (2003) Phosphorylation of histone and histone-like proteins by aurora kinases during mitosis. Prog Cell Cycle Res 5:369–374
Pray-Grant MG, Daniel JA et al (2005) Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433(7024):434–438
Prigent C, Dimitrov S (2003) Phosphorylation of serine 10 in histone H3, what for? J Cell Sci 116(Pt 18):3677–3685
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574
Sagolla MS, Dawson SC, Mancuso JJ, Cande WZ (2006) Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite Giardia intestinalis. J Cell Sci 119 (23):4889–4900
Savioli L, Smith H et al (2006) Giardia and Cryptosporidium join the ‘Neglected Diseases Initiative’. Trends Parasitol 22(5):203–208
Singer SM, Yee J et al (1998) Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol Biochem Parasitol 92(1):59–69
Smith MM (2002) Centromeres and histone variant: what, where, when and why? Curr Opin Cell Biol 14(3):279–285
Sogin ML, Gunderson JH et al (1989) Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science 243(4887):75–77
Solari AJ, Rahn MI et al (2003) A unique mechanism of nuclear division in Giardia lamblia involves components of the ventral disk and the nuclear envelope. Biocell 27(3):329–346
Sullivan BA, Blower MD et al (2001) Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet 2(8):584–596
Van de Peer Y, Baldauf SL et al (2000) An updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances. J Mol Evol 51(6):565–576
Van Hooser AA, Ouspenski II et al (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114(Pt 19):3529–3542
Wu G, McArthur AG et al (2000) Core histones of the amitochondriate protist, Giardia lamblia. Mol Biol Evol 17(8):1156–1163
Yu LZ, Birky CW Jr et al (2002) The two nuclei of Giardia each have complete copies of the genome and are partitioned equationally at cytokinesis. Eukaryot Cell 1(2):191–199
Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15(18):2343–2360
Acknowledgements
We would like to acknowledge Heidi Elmendorf (Georgetown University), C.C. Wang and colleagues (UCSF), and Keith Gull (Oxford University, UK) for plasmids, reagents, and methodologies. Additionally, we thank Hillary Morrison and other members of the Giardia Genome Project for access to genomic data. We also thank Elizabeth Slawson and members of the Cande lab for the helpful discussions. This work was supported by an NIH grant A1054693 and grant from the University of California Cancer Research Coordinating Committee (CRCC) to WZC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Choo
Appendix
Below is the link to the electronic supplementary material.
Fig. S1
cenH3::GFP foci are visible in 3D stack. This movie steps through individual sections in a 3D stack of images taken at 0.2 μm depth using a cenH3::GFP strain. Individual foci are visible (MOV 2.6 MB).
Rights and permissions
About this article
Cite this article
Dawson, S.C., Sagolla, M.S. & Cande, W.Z. The cenH3 histone variant defines centromeres in Giardia intestinalis . Chromosoma 116, 175–184 (2007). https://doi.org/10.1007/s00412-006-0091-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-006-0091-3