Abstract
Heterochromatin is composed of tightly condensed chromatin in which the histones are deacetylated and methylated, and specific nonhistone proteins are bound. Additionally, in vertebrates and plants, the DNA within heterochromatin is methylated. As the heterochromatic state is stably inherited, replication of heterochromatin requires not only duplication of the DNA but also a reinstallment of the appropriate protein and DNA modifications. Thus replication of heterochromatin provides a framework for understanding mechanisms of epigenetic inheritance. In recent studies, roles have been identified for replication factors in reinstating heterochromatin, particularly functions for origin recognition complex, proliferating cell nuclear antigen, and chromatin-assembly factor 1 in recruiting the heterochromatin binding protein HP1, a histone methyltransferase, a DNA methyltransferase, and a chromatin remodeling complex. Potential mechanistic links between these factors are discussed. In some cells, replication of the heterochromatin is blocked, and in Drosophila this inhibition is mediated by a chromatin binding protein SuUR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Overview
In recent years the crucial role of epigenetics has become increasingly apparent, as many human diseases have been linked to epigenetic defects (for review, see Jiang et al. 2004). Gene expression is controlled not only by DNA sequence elements but also by the configuration of proteins in the chromatin and by methylation of the DNA itself. In mammals some genes are imprinted such that expression of the paternal or maternal alleles is blocked; in mammalian females one X chromosome is inactivated for expression, and in a variety of organisms genes in proximity to heterochromatin are repressed. The epigenetic as well as genetic states are inherited, making it important to decipher mechanisms for the establishment and maintenance of the epigenetic state. In this review we discuss recent advances in our understanding of replication of heterochromatin, an extreme epigenetic state that serves as an excellent model for elucidating how chromatin structure and DNA methylation are regulated.
Heterochromatin was first recognized and defined cytologically as regions of the genome that were highly condensed throughout the cell cycle, as distinguished from euchromatin in which condensation was visible only during mitosis (for reviews, see Dillon and Festenstein 2002; Henikoff 2000). As these regions remain condensed in interphase, heterochromatin is frequently associated with silenced regions of the genome in which genes are not expressed. Facultative heterochromatin refers to regions that can be transiently condensed and silenced during development of an organism. These regions, therefore, are capable of transitioning between heterochromatin and euchromatin. Constitutive heterochromatin, including pericentric and telomeric heterochromatin, refers to chromatin that remains condensed and silenced throughout development. Heterochromatin plays critical roles in chromosome structure and transmission, and most eukaryotic centromeres are surrounded by blocks of heterochromatin. In Drosophila, up to 30% of the chromosome is heterochromatin, and in fission yeast it is clearly established that centric heterochromatin blocks transcription across the centromere that could cripple its function in chromosome segregation (Ekwall et al. 1997). Similarly, the heterochromatic nature of telomeres is important for their function.
There are several key molecular characteristics of both the DNA content and chromatin organization of heterochromatin, although these are general characteristics and certainly there are exceptions. Many heterochromatic regions consist mainly of highly repetitive satellite DNA and moderately repetitive elements like transposable elements (Dillon and Festenstein 2002; Henikoff 2000). Transposable elements tend to accumulate in heterochromatin where reduced expression may limit their mobility and restrict accumulation (reviewed in Henikoff 2000; Schramke and Allshire 2004). There is also a sparse distribution of single copy genes in heterochromatin. Although the expression of most genes is repressed by heterochromatin, there are essential genes such as the Drosophila light gene that can be expressed only in a heterochromatic environment (Wakimoto and Hearn 1990).
Nucleosomes in heterochromatin tend to display a regular, ordered spacing, and heterochromatic DNA is not accessible to nucleases (Dillon and Festenstein 2002). Heterochromatin is typically associated with hypermethylated and hypoacetylated histones. Histone H3 is often methylated at lysine 9, which generates a binding site for the heterochromatic protein HP1 (Lachner et al. 2001; Bannister et al. 2001). By its ability to dimerize, HP1 has been suggested to promote the formation of higher-order chromatin structure of heterochromatin (Nielsen et al. 2001), and localization of this protein has been shown to be sufficient to promote chromatin condensation (Verschure et al. 2005). Based on these characteristics, heterochromatin is assumed to be a highly organized, compacted chromatin structure, and the details of this structure are being elucidated. This definition of heterochromatin, however, was recently challenged by observations of transcription in heterochromatin and exchange of heterochromatin proteins, suggesting that the heterochromatic state may be more pliable than originally thought (Volpe et al. 2002; Cheutin et al. 2003; Greil et al. 2003; Martens et al. 2005).
The ability of heterochromatin to repress gene expression is exemplified by situations in which the expression of genes normally located in euchromatic regions is reduced or abolished if they are translocated next to heterochromatin (Dillon and Festenstein 2002; Henikoff 2000). This transcriptional repression is seen in cases in which chromosomal rearrangements, such as inversions, place previously expressed euchromatic genes adjacent to heterochromatin. Even on normally configured chromosomes, the expression of euchromatic genes adjacent to the heterochromatin is repressed. This occurs for genes adjacent to centromeres, and in yeast repression it also occurs next to the silenced mating-type loci (Dillon and Festenstein 2002; Henikoff 2000). This type of positional repression is often unstable, with the genes being expressed in some clonal cells but not in others, an epigenetic phenomenon that has been termed position effect variegation (PEV). PEV is a powerful phenotype for genetic studies in yeasts and Drosophila, and key proteins controlling chromatin have been identified by the ability of mutations in the genes that encode them either to suppress or enhance PEV (reviewed in Schotta et al. 2003a). In particular, roles in promoting heterochromatin were confirmed for a conserved heterochromatin binding protein, HP1, and a histone methyl-transferase enzyme, SU(VAR)3-9, by their identification as suppressors of PEV in Drosophila (Schotta et al. 2003b). In addition to genetics, the size of heterochromatic blocks and stability of heterochromatin permit biochemical studies and cytological visualization both of chromatin-bound proteins and chromatin modifications (Dillon and Festenstein 2002; Maison and Almouzni 2004).
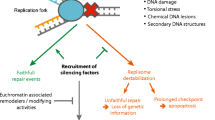
There are several challenges to faithfully duplicating heterochromatin in each cell cycle. The first concerns the replication of the DNA itself, given the highly condensed state of the chromatin. Most heterochromatin is replicated late in S phase, but the significance of this is unknown (reviewed in Gilbert 2002; McNairn and Gilbert 2003). It is possible that it takes longer for replication origins to fire within the heterochromatin, but it is also possible that the timing of replication is actively regulated. For example, limiting heterochromatic replication until late in S phase could facilitate reassembly of the epigenetic state of the heterochromatin if heterochromatin binding proteins were not present early in S phase or were prevented from binding until late S phase. In polyploid cells, the heterochromatin frequently is not replicated, such that these regions are underrepresented (Rudkin 1969; Leach et al. 2000). This may be a mechanism to optimize the metabolic state of polyploid cells by dispensing with gene-poor regions of the genome. It is important to emphasize that although DNA replication necessitates a mechanism to maintain heterochromatin, it has been shown in yeast that it is possible to establish heterochromatin without DNA replication (Kirchmaier and Rine 2001; Li et al. 2001). The second aspect of heterochromatin duplication concerns how the chromatin is assembled into a heterochromatic state with the appropriate positioning of the nucleosomes, histone modifications, and binding of heterochromatin proteins following replication. A third aspect involves the modification of the DNA itself, the methylation of DNA sequences.
Because our focus is on the replication of heterochromatin, much of the recent literature on the increasing list of regulators required for the maintenance of heterochromatin is not discussed here (see Craig 2005 for review). Factors needed to maintain heterochromatin are likely to act both during and following S phase, but in most examples the time of action with respect to replication has not been established. It is possible, therefore, that these factors play a role in heterochromatin duplication that will be elucidated in future studies. This is true for the exciting finding on the role of noncoding RNAs in heterochromatin. Noncoding RNAs play crucial roles in the establishment of heterochromatin to inactivate the mammalian X chromosome, and the RNAi pathway is important for H3K9 methylation and HP1 localization in the centric heterochromatin. To date, these RNA-mediated mechanisms have not been shown to participate in the replication of heterochromatin, and thus we refer readers to several recent reviews for a full discussion of this topic (Lippman and Martienssen 2004; Schramke and Allshire 2004; Matzke and Birchler 2005). Histone protein variants play a critical role in epigenetic regulation, with some predominating in, and others excluded from, heterochromatin (as reviewed in Kamakaka and Biggins 2005; Ahmad and Henikoff 2002); although some variants can be incorporated during S phase, it remains to be determined how they impact DNA replication. Consequently, the role of these variants in chromatin structure and gene expression is not covered in this review.
In this work we discuss the following issues unique to propagation of heterochromatin: (1) the role of replication proteins in the replication of heterochromatic DNA and the recruitment of heterochromatin binding proteins; (2) a Drosophila protein, SuUR, that specifically controls replication of the heterochromatin; (3) the chromatin assembly factors, specifically CAF1, that act to maintain heterochromatin following DNA replication; (4) the link between DNA replication and DNA methylation; and (5) evidence for roles of chromatin remodeling complexes in the replication of heterochromatin (see Table 1 for factors discussed in this review).
The replication machinery and replication of heterochromatin
Role of the origin recognition complex in heterochromatin
One key concept to emerge from the analysis of the replication machinery and heterochromatin is that replication proteins can act both to replicate the DNA and to recruit the heterochromatin binding proteins that epigenetically confer the heterochromatic state. This is most clear for the origin recognition complex (ORC), an evolutionarily conserved complex consisting of six subunits (for reviews, see Bell and Dutta 2002; Leatherwood and Vas 2003). Studies in many organisms have demonstrated that ORC is a link between the processes of DNA replication and heterochromatin maintenance. ORC was originally identified by the role of the complex in the initiation of DNA replication. In budding yeast, mutations in the subunits of the ORC also disrupt silencing of the mating-type loci. Surprisingly, studies have demonstrated that the replication and silencing functions of ORC are genetically separable (Bell et al. 1995; Dillin and Rine 1997). Bell et al. found that the N terminus of ORC1 in Saccharomyces cerevisiae is specifically required for mating-type repression, but is dispensable for normal growth and, therefore, DNA replication. Dillin and Rine isolated mutants of orc5 specifically defective in either DNA replication or mating-type silencing. These mutations are able to complement each other, suggesting that different domains of the protein acted in the two processes, and furthering a model where ORC has two domains that confer separate functions. ORC's role in silencing involves interaction with Sir1, the S. cerevisiae functional homolog of HP1, and the subsequent recruitment of Sir proteins to specific loci (Triolo and Sternglanz 1996).
Although the relationship between ORC and heterochromatin in higher eukaryotes is less clear, a role for ORC both in DNA replication and in recruitment of heterochromatin proteins has been described. Analyses of ORC localization during the cell cycle provide evidence for ORC in heterochromatin replication in mammalian cells. Prasanth et al. documented cell-cycle changes in ORC2 localization in MCF7 cells; ORC2 generally localizes with heterochromatic foci, marked by the presence of HP1α and β, during G1 and early S phase. However, as the cells progress further into S phase, ORC2 localizes to punctate foci that are characteristic of late-replicating pericentric regions (Prasanth et al. 2004). Lidonnici et al. (2004) examined the localization of tagged, ectopic human ORC1 in mammalian cells and also noted that ORC1 preferentially localizes to the pericentric heterochromatin foci that colocalize with HP1. The localization of ORC to heterochromatic foci when they are likely to be replicating in late S phase suggests that ORC is involved in the replication of heterochromatin in higher eukaryotes.
In addition, the phenotype of Drosophila orc2 mutants indicates an important role for ORC in the proper timing of replication. Generally, euchromatic regions of the genome are replicated prior to heterochromatic regions in S phase. In orc2 mutants, however, replication of some euchromatic regions is delayed and these regions are inappropriately replicated after heterochromatic regions (Loupart et al. 2000). The authors suggest this intriguing possibility for the phenotype: ORC may have a higher affinity for heterochromatin, and the limited, functional ORC complexes in this mutant are recruited more efficiently to heterochromatin and enable replication of these regions. The euchromatic regions then are less likely to recruit ORC and display delayed replication initiation. Euchromatic and heterochromatic regions may, therefore, require ORC for replication and for coordination of their replication timing.
A role for ORC in the formation of heterochromatin is supported by physical interaction between ORC and the HP1 protein (Table 1). Drosophila ORC2 localizes in vivo to heterochromatin and colocalizes with HP1 on mitotic chromosome spreads (Pak et al. 1997). Immunoprecipitation experiments from Drosophila embryo extracts confirm a physical interaction with HP1 and the ORC complex (Pak et al. 1997; Huang et al. 1998). This direct interaction was also demonstrated in Xenopus (Pak et al. 1997) and in mammalian cell lines (Lidonnici et al. 2004; Prasanth et al. 2004). Lidonnici et al. also used fluorescence resonant energy transfer (FRET) to demonstrate an in vivo interaction between ORC1 and HP1α.
A second protein, HOAP (HP1/ORC-associated protein), present in heterochromatin not only interacts with ORC but also recruits HP1/ORC to heterochromatin. Drosophila HOAP copurifies with ORC subunits and HP1α and has also been shown to colocalize with ORC and HP1α (Shareef et al. 2001). By disrupting the HP1/HOAP interaction, Badugu et al. revealed that this interaction is required for the proper localization of HP1, but HOAP localization is not disrupted (Badugu et al. 2003). Consistent with a role for the HOAP/ORC complex in recruiting HP1 to heterochromatin and promoting heterochromatin, a mutant in hoap suppresses PEV (Shareef et al. 2001). This effect on PEV confirms that HOAP has a functional role in heterochromatin architecture in vivo.
Other experiments also suggest that ORC promotes the assembly of heterochromatin in metazoans and that, as in S. cerevisiae, ORC's main function in heterochromatin may be to recruit HP1. Like mutants in hoap, Drosophila orc2 mutants also suppress PEV (Pak et al. 1997) and HP1 localization is disrupted in orc2 mutants (Huang et al. 1998). In mammalian cells, depletion of ORC2 by siRNA resulted in a disruption of HP1α and HP1β foci, although the HP1 binding sites (trimethylated lysine 9 on histone H3) remained intact (Prasanth et al. 2004). These results imply that ORC is necessary to recruit HP1 and that this interaction promotes the formation of heterochromatin.
Given that ORC is likely involved in the replication of heterochromatin sequences, a model can be envisioned in which ORC localizes to heterochromatic sites for DNA replication, recruiting HP1 to those sites to reestablish the heterochromatic state after passage of the replication fork. This simplistic model remains to be demonstrated, and it will be important to determine how ORC recruits HP1 only to heterochromatin. It is conceivable that HP1 is available for loading onto newly duplicated chromatin only late in S phase, but the mechanism by which it would be precluded earlier is not clear. Another complexity is that some observations lead to the conclusion that HP1 localizes ORC rather than the opposite (Leatherwood and Vas 2003). Lidonnici et al. observed that in synchronized mammalian cells in early G1, HP1β was found at heterochromatic loci, but tagged ORC1 was not. As the cells progressed through G1, ORC1increasingly colocalized with HP1β at heterochromatic loci. Furthermore, an ORC1 mutant lacking the HP1 binding domain did not localize to heterochromatin (Lidonnici et al. 2004). HP1 is not, however, required to maintain ORC localization because disruption of HP1 localization by treatment with either trichostatin A (TSA), an inhibitor of a subset of known histone-deacetylases, or RNAse A does not disrupt ORC1 localization (Lidonnici et al. 2004). It seems, therefore, that the HP1–ORC1 interaction could have two functions during the cell cycle: to recruit ORC1 to sites of heterochromatin in G1 and to recruit HP1 to sites of heterochromatic replication in late S phase.
Roles of other replication proteins in heterochromatin
The majority of eukaryotic DNA replication is catalyzed by polymerases α, δ, and ɛ (POL α, δ, and ɛ). POL α associates with primase to synthesize and extend RNA primers in initiation. POL α and δ do lagging strand synthesis, and synthesis of the leading strand at the replication fork is achieved by POL ɛ. Does the replication of heterochromatin require different replication machinery or mechanics? Two studies in S. pombe link POL α to the establishment of heterochromatin. Mutations in pol α suppress PEV at the mating-type loci, centromeres, and telomeres, and POL α directly interacts with Swi6, a protein known to be important in the silencing of mating-type loci and the S. pombe HP1 homolog (Table 1) (Ahmed et al. 2001; Nakayama et al. 2001). Additionally, mutations in pol α affect the localization of Swi6 to mating-type loci and to heterochromatic loci (Ahmed et al. 2001; Nakayama et al. 2001). The mutation used for the studies by Nakayama et al. is not located in a conserved region required for polymerase activity, nor does it increase UV sensitivity as expected if the catalytic activity were reduced. This raises the possibility that the mutant effects on Swi6 are not simply due to DNA replication failure. These observations suggest a model in which Pol α, at replication forks, is able to recruit and maintain Swi6 to reestablish heterochromatin following replication.
In S. cerevisiae, establishment of silencing can occur without DNA replication, in particular, independently of passage of the replication fork (Kirchmaier and Rine 2001; Li et al. 2001). Nevertheless, mutations in many replication factors, including proliferating cell nuclear antigen (PCNA), replication factor C (RF-C), the replication initiation factor Cdc45, POL α, and POL ɛ affect silencing either by disrupting it or by suppressing silencing defects (Huang 2002). At present, it is not clear whether these replication factors are required only for replication of heterochromatic regions or whether they participate in the formation of heterochromatin independently of, and in addition to, their actions in DNA replication. Given that silencing can be established independently of passage of the replication fork, definitive tests of whether replication factors have a replication-independent role in heterochromatin will require the recovery of mutants that affect silencing but not replication.
In human cells, DNA polymerase ɛ may assist in the replication of heterochromatin. In addition to its role in chromosomal DNA replication, POL ɛ is involved in DNA repair and the S-phase DNA damage checkpoint (Hubscher et al. 2002). Analysis of the subcellular localization of the POL ɛ subunit p261 (the catalytic subunit) in human fibroblasts revealed that PCNA, BrdU, and POL ɛ colocalize in late S phase specifically to the large foci that are characteristic of heterochromatic DNA replication (Fuss and Linn 2002). Interestingly, in early S phase, PCNA and p261 do not colocalize, but are adjacent. The specific colocalization in late S phase could mean that POL ɛ synthesizes DNA only at late-replicating heterochromatic loci or may be specifically suited for replication at these foci. This could reflect a need for a different replication machinery for heterochromatin (Fuss and Linn 2002).
Studies on PCNA also suggest a link between DNA replication and epigenetic inheritance. PCNA is a member of the DNA sliding clamp family that increases DNA polymerase processivity (for review, see Majka and Burgers 2004). In addition to its role in DNA replication, PCNA interacts with a wide variety of cell factors and may be the major scaffold for recruiting and directing chromatin enzymes (Table 1) (for review, see Maga and Hubscher 2003). In S. cerevisiae, mutations in the pcna gene increase expression of genes near the telomere and at mating-type loci, linking PCNA to silencing (Zhang et al. 2000). PCNA mutants in Drosophila suppress PEV, indicating that PCNA participates in chromatin assembly in higher eukaryotes (Henderson et al. 1994). PCNA localizes to mammalian heterochromatic loci where it interacts with CAF1, a chromatin-assembly factor, and chromatin-remodeling enzymes, both of which are discussed below (Fig. 1).
Model of protein–protein interactions at the replication fork that are relevant to the heterochromatic state. Many characteristics of heterochromatin, like histone modifications, nucleosome positioning and bound proteins, are likely displaced as the replication fork passes through the DNA sequences. The depicted factors are speculated to assist in the reestablishment of the heterochromatic state after the DNA has been replicated. Their interaction with proliferating cell nuclear antigen (PCNA) suggests that they may travel with the progressing replication fork. a PCNA acts as a scaffold for nucleosome processes, bringing the chromatin-assembly factor, CAF1, and the chromatin-remodeling factor, Imitation Switch (ISWI), to nascent DNA. CAF1 deposits histone H3/H4 tetramers on newly replicated DNA, which are joined by two H2A/H2B dimers to form the full nucleosome. ISWI alters the spacing of these nucleosomes on the DNA, forming a regularly spaced array. Additionally, CAF1 binds the heterochromatin protein, HP1, likely keeping the local concentration of HP1 high so that it can quickly bind modified histone H3. b DNA methylation and DNA methyl binding proteins must also be reestablished after progression of the replication fork. Again, PCNA is speculated to act as a scaffold, recruiting the DNA methyltransferase, DNMT1, which in turn recruits the MBD2a-3 methyl binding proteins. PCNA and CAF1 also bind MBD1, another methyl binding protein, and SETDB1, a histone H3 methyl transferase. This coordination between DNA and histone modification enzymes is speculated to rapidly promote heterochromatin formation after DNA replication
These studies demonstrate a requirement for ORC and the DNA replication machinery in heterochromatin but illustrate the complexities in deciphering the exact role of DNA replication factors, particularly whether they play roles independent of their replication activities in the establishment of epigenetic state. If indeed the replication factors have roles in maintaining heterochromatin that are independent of DNA synthesis, there must be a means by which these actions are restricted to the heterochromatin, unless this restriction is an outcome of heterochromatin replication being limited to late S phase. The ability to genetically separate the activities of ORC in replication and silencing demonstrates that ORC has independent activities, and such genetic analyses on other replication factors is likely to be informative. The question as to whether replication of DNA in heterochromatin requires distinct functions from the replication of DNA in euchromatin also merits further investigation.
A specialized trans regulator of heterochromatin replication
The Drosophila SuUR (suppressor of underreplication) gene encodes an intriguing chromosomal protein that specifically affects the replication of heterochromatin (Belyaeva et al. 1998; Makunin et al. 2002). It is the sole protein identified to date that is uniquely responsible for the replicative properties of heterochromatin. Understanding the role of the SuUR protein requires an appreciation of the parameters of heterochromatin replication during a variant cell cycle, the endo cycle, that gives rise to polyploid or polytene cells (Edgar and Orr-Weaver 2001). In the endo cycle there are repeated rounds of S phase, punctuated by gap phases during which gene expression and cell growth occur, but mitosis does not take place. Endo cycles produce either polyploid or polytene chromosomes, in which the replicated sister chromatid copies are held in physical register. Polyploid and polytene cells are found throughout the plant and animal kingdoms, most commonly associated with cell types that are highly metabolically active.
Consistent with the implementation of the endo cycle as a means to produce a “factory” cell, in many endo cycling cells S phase is cut short and heterochromatin is not replicated (Edgar and Orr-Weaver 2001). This is evident in Drosophila polytene cells, in particular the larval salivary glands. The approximately 1000 copies of each chromosome pair are aligned to produce a distinctive banding pattern. This banding pattern, however, is present only in the euchromatin; the 20–30% of each chromosome arm adjacent to the centromere that is composed of heterochromatin is not visible in salivary gland chromosomes, and neither is the heterochromatic Y chromosome. Quantitation of DNA doublings in the endo cycle indicates that approximately 20% of the genome is not replicated in each endo cycle S phase (Rudkin 1969; Smith and Orr-Weaver 1991). In addition to the centric heterochromatin and Y chromosome, there are regions throughout the euchromatin with constrictions and fragile sites, known as intercalary heterochromatin, that are also underreplicated (Zhimulev and Belyaeva 2003).
Cell cycle regulators controlling the G1–S transition and transcription of genes necessary for S phase have been found to affect underreplication of heterochromatin in the endo cycle. Decreased function of cyclin E or of either subunit of the E2F1 transcription factor results in increased replication of centric heterochromatin in the polyploid nurse cells of the adult ovary (Lilly and Spradling 1996; Royzman et al. 2002). It has been proposed that in the endo cycle S phase is truncated such that late-replicating heterochromatin is not copied (Lilly and Spradling 1996). The cyclin E and dE2F1 mutant phenotypes are explained as the consequence of a slowed S phase resulting in the replication of late-replicating heterochromatin. By pulse labeling replicating salivary gland DNA and then cytologically examining the pattern of nucleotide incorporation on polytene chromosomes, it was confirmed that the regions adjacent to the centromeres and the constrictions replicate late in the endo cycle S phase (Zhimulev et al. 2003a). Not all late-replicating regions are underreplicated, however; only 60 out of 156 late-replicating sites correspond to weak constriction points on the polytene chromosomes (Zhimulev et al. 2003a).
The SuUR mutant arose spontaneously and was identified because it eliminated the constrictions at intercalary heterochromatin and restored replication to parts of the centric heterochromatin in salivary glands (Fig. 2). Quantitation of DNA copy number for several of these intervals demonstrated that in SuUR mutants the regions are less underreplicated, i.e., they have increased DNA copy number (Belyaeva et al. 1998). Pulse labeling of mutant cells indicated that normally late-replicating regions are replicated earlier with the bulk of euchromatic DNA (Zhimulev et al. 2003a). There is suppression of PEV at several loci in the SuUR mutant, implying that the wild-type protein is needed for heterochromatin structure (Belyaeva et al. 2003).
Dosage effects of the Drosophila SuUR gene on heterochromatin replication in polytene chromosomes. a In larval salivary gland chromosomes the centric heterochromatin comprising the proximal 20–30% of each chromosome arm is so severely underreplicated that these segments of the chromosomes are not visible following orcein staining. Region 80 on chromosome 3L and region 81 on chromosome 3R are indicated. b Mutation of the SuUR gene results in replication of the centric heterochromatin such that banded regions become visible, shown here for cytological intervals 80 and 81. c In addition to the blocks of heterochromatin flanking the centromeres, underreplication of intercalary heterochromatin can be visualized by missing or thin bands, chromosome constrictions and breaks. These sites also frequently attach ectopically to other chromosome regions. Two sites of intercalary heterochromatin at 75A and 75C1-2 on chromosome 3L are shown. d In the SuUR mutant, sites of intercalary heterochromatin become more fully replicated. e Overexpression of the SuUR protein from extra copies of the gene results in many new sites of intercalary heterochromatin. Two of the sites with pronounced breaks are highlighted by arrows, but there are many visible regions in which the bands are partially missing. Panels a and b are from Belyaeva et al. (1998), panels c and d are from Semeshin et al. (2001), and panel e is from Zhimulev et al. (2003a); reproduced with permission
The effects of the SuUR protein on heterochromatin structure and replication are dose-specific (Fig. 2). In the presence of extra copies of the wild-type gene, the number of constrictions and weak points on salivary gland chromosomes increases, and these correspond to late-replicating regions (Zhimulev et al. 2003a). Copy number of the DNA decreases at the new sites and is further decreased at the normal constriction points (Zhimulev et al. 2003a; Moshkin et al. 2001). Thus it appears that the SuUR protein leads to underreplication by further delaying the replication of late-replicating genes such that they fail to replicate at all during the endo cycle. Increased levels of the protein can dramatically alter polytene chromosome structure, leading to swellings that resemble DNA puffs (Zhimulev et al. 2003b). Extra copies of the wild-type SuUR gene enhance PEV, which also argues that the protein promotes heterochromatin formation (Belyaeva et al. 2003). Recently, the response of underreplicated regions to changes in dosage of the SuUR gene was exploited in microarray studies that identified approximately 1000 genes clustered in 52 regions whose copy number is affected by SuUR (Belyakin et al. 2005).
The SuUR gene encodes a protein of 962 amino acids whose N terminus has some similarity to the conserved motifs in the SNF2/SWI2 chromatin remodeling proteins (Makunin et al. 2002). The N-terminal half of SuUR is 42% identical to the bromodomain of the Brahma trxG transcriptional activator (Tchurikov et al. 2004). The spontaneous mutation, discussed above, is attributable to an insertion that leads to loss of the single transcript from the gene, which is normally particularly abundant in females and embryos. As expected from the homology motifs, the SuUR protein binds to chromosomes and is observed at heterochromatin regions of polytene chromosomes (Makunin et al. 2002). It localizes to 113 bands, and 108 are sites of late replication. When overexpressed, it localizes to 280 sites. The binding of the protein to affected regions argues that SuUR directly promotes the heterochromatin state and restricts DNA replication. Its mechanism of action remains to be deciphered at a molecular level, particularly whether the primary effect is via heterochromatin structure or via perturbation of the replication machinery. SuUR colocalizes with HP1 at a cytological level, but the relationship between these proteins has not been investigated (Zhimulev and Belyaeva 2003).
Given the dramatic effects of SuUR mutants on PEV and underreplication, it is puzzling that the mutant is fully viable and fertile (Belyaeva et al. 1998). Increased levels of protein, however, are deleterious. Continuous overexpression of the protein in the salivary gland results in a small gland, and ubiquitous overexpression results in lethality (Volkova et al. 2003). Overexpression in the follicle cells is capable of repressing replication at specific sites during the amplification of the chorion eggshell genes (Volkova et al. 2003). Thus the organism can survive without this protein and the resulting increased copy number of heterochromatin regions, but increasing the number of underreplicated domains lead to lethality.
Reestablishment of heterochromatin after DNA replication
As DNA replication requires the ability of the polymerase to directly contact the nucleotide sequence and move processively along the DNA, higher-order chromatin would need to be disassembled and then reassembled following the replication fork. Indeed, it has been demonstrated in vitro that nucleosomes, the basic unit of chromatin, are disrupted at the replication fork (Gruss et al. 1993). Both euchromatin and heterochromatin, therefore, require factors to recruit and deliver nucleosomes to newly replicated DNA. Although the nucleosome deposition function of these chromatin-assembly enzymes is likely similar in both euchromatin and heterochromatin, it is possible that these enzymes have additional roles in reestablishing heterochromatin after the replication fork. Here we address evidence for such a role for the chromatin-assembly factor 1 (CAF1).
Chromatin-assembly factor 1 is a multi-subunit complex that assists in loading newly synthesized H3–H4 tetramers onto chromatin, preferentially after DNA replication and DNA repair (for review, see Ridgway and Almouzni 2000; Mello and Almouzni 2001). As the replication fork progresses, the parental nucleosomes are transiently disrupted and H3/H4 tetramers bind H2A/H2B dimers to reconstitute the octamer core. In addition to the transfer of parental nucleosomes to two daughter DNA duplexes, nucleosomes can be assembled de novo by the initial recruitment of the H3/H4 tetramers followed by H2A/H2B dimers (for review, see Krude 1999). CAF1 was previously observed to localize to mammalian euchromatic DNA replication foci first, and later to associate with heterochromatic replication foci as soon as the euchromatic replication is completed (Krude 1995). Additionally, CAF1 physically associates with PCNA, implying that incorporation of new histones directly follows the DNA polymerase (Fig. 1a) (Shibahara and Stillman 1999). In S. cerevisiae, silencing at the HML locus can be restored in cac1 sir3 mutants by expression of SIR3, indicating that CAF1 is not required for the establishment of silencing. However, the presence of silencing defects in cac1 mutants suggests a role for CAF1 in the maintenance and transmission of heterochromatin (Enomoto and Berman 1998).
Studies on the largest CAF1 subunit in S. cerevisiae, Cac1/Rfl2, have suggested a model in which CAF1 plays an integral role in incorporating “heterochromatin competent” H3–H4 tetramers, by virtue of their acetylation pattern (Enomoto and Berman 1998). In mammalian cells, acetylated H4K5 and H4K12 are incorporated at late-replicating foci and colocalize with HP1α, HP1β, and CAF1 (Taddei et al. 1999). Although heterochromatic histone H4 is characteristically underacetylated, newly synthesized histone H4 is acetylated at lysine 5 and lysine 12 regardless of the previous chromatin state. Taddei et al. (1999) also found that the association of acetylated H4K5, H4K12, and CAF1 with late-replicating foci is related to DNA synthesis, as BrdU labeling in a pulse-chase experiment colocalizes with CAF1 at these foci. Thus, the default chromatic state postreplication may be more euchromatic or “open,” and the reestablishment of heterochromatin is likely to be an active process. Interestingly, at mammalian late-replicating foci, the hyperacetylated H4 and CAF1 remain associated with the heterochromatic foci for 20 min postreplication, revealing a window in which heterochromatin begins its reestablishment. CAF1 continues its association with heterochromatin at least until late G2 (Murzina et al. 1999). It is tantalizing to speculate that the lingering presence of acetylated H4K5, H4K12, and CAF1 at newly replicated heterochromatic foci may act as a particular mark for heterochromatin and recruit heterochromatin factors to stimulate heterochromatin formation.
Like ORC and POL α, CAF1 also physically interacts with HP1 (Table 1, Fig. 1a). Murzina et al. (1999) demonstrated that the largest subunit of CAF1, p150, and several isoforms of HP1 associate through the MOD1 interacting region (MIR) of p150 in mammalian cells. However, mutations in MIR that disrupt the CAF1–HP1 interaction did not affect the recruitment of CAF1 to either euchromatic or heterochromatic replication foci (Murzina et al. 1999). Additionally, HP1 localization to heterochromatin does not appear to require heterochromatic replication, implying that HP1 can localize to heterochromatin by means independent of CAF1's link with fork progression (Murzina et al. 1999). Thus the function of the CAF1–HP1 interaction remains unclear.
Visualization of replicating heterochromatin enables a better understanding of the spatial and temporal relationship between DNA replication and heterochromatin assembly. Quivy et al. (2004) recently reported that pulse–chase–pulse experiments, in conjunction with high-resolution microscopy and 3D modeling, reveal the nuclear positioning and architecture of replicating mammalian pericentric heterochromatin domains. DNA synthesis, based on colocalization of a BrdU pulse and PCNA, occurs at the periphery of the pericentric domain. As heterochromatin is replicated and the chromatin configuration disrupted, it may be displaced to the exterior of the pericentric domain. The newly replicated DNA is observed to then move into the interior of the pericentric domain.
Whereas the architecture of replicating pericentric domains may be specific to pericentric heterochromatin and/or mammalian cells, Quivy et al. (2004) reveal details of heterochromatin reassembly that may be more universal. These experiments demonstrate the presence of two pools of nuclear HP1 in these cells: a replication-associated pool and an independent pool. The replication-associated pool of HP1 is characterized by its colocalization with PCNA, CAF1, and acetylated H4K5 at the periphery, but not methylated H3K9, which is found in the core of the pericentric heterochromatin domain. This pool is resistant to RNAse treatment and is detected in knockout cells of Suv39h, a histone methyltransferase, whereas as knockdown of CAF1 by siRNA to the p150 subunit leads to a loss of HP1 staining at the periphery. In contrast, the independent pool of HP1 requires Suv39h for localization. These data add to a model in which PCNA recruits CAF1 to loci and CAF1 assists in reestablishing heterochromatin after passage of the replication fork, by recruiting HP1 to newly replicated foci (Fig. 1a). Once assembled, HP1 is retained by interactions with methylated H3 and RNA.
Reestablishment of DNA Methylation Patterns
Hypermethylation of cytosine bases is another characteristic of silenced chromatin, most prominently in vertebrates. DNA replication and methylation appear to occur concurrently; by isolating newly synthesized DNA (containing origins of replication) from mammalian cells, it was demonstrated that levels of cytosine methylation were equal in the parental and daughter DNAs (Araujo et al. 1998). The methyltransferase DNMT1 has been linked to maintenance of this epigenetic state due to its association with hemimethylated DNA and its interaction with the replication machinery at late-replicating foci (Fig. 1b). DNMT1 has been demonstrated to copurify with in vitro DNA replication activity and to coelute with POL α activity, supporting a model in which methylation occurs concomitant with DNA replication (Vertino et al. 2002). Consistent with these observations, DNMT1, BrdU, and PCNA colocalize at the sites of mammalian pericentric heterochromatin replication; (Leonhardt et al. 1992; Chuang et al. 1997), and DNMT1 and PCNA physically interact by GST pulldown (Table 1) (Chuang et al. 1997). This interaction supports a model in which PCNA, traveling with the replication fork, acts as a scaffold to recruit a number of chromatin-modifying enzymes (Fig. 1b). Indeed, DNMT1 is recruited to DNA more efficiently if the DNA is bound by PCNA, and PCNA-bound DNA is methylated more efficiently by DNMT1 than a PCNA-free control (Iida et al. 2002).
In addition to the reestablishment of the DNA methylation pattern, specific methyl-binding proteins that contribute to the silenced state of chromatin must also rebind following replication. A family of proteins consisting of MeCP2 and MBD1, -2, -3, and -4 binds methylated CpG sequences in vertebrates. Importantly, these methyl-binding proteins are commonly found in complexes with histone deacetylases and chromatin remodeling enzymes, suggesting that these proteins assist in the recruitment of factors that reestablish the heterochromatic state (for review, see Wade 2001). Methyl-binding proteins, particularly the MBD2a–MBD3 complex, also colocalize with DNMT1 in late S phase at mammalian pericentric heterochromatin, but not before (Tatematsu et al. 2000). This suggests that both methylation of the DNA and binding of this mark by methyl-binding proteins occur quickly after replication, although it remains to be demonstrated that these methyl-binding proteins are present on nascent DNA. A link between silencing and replication is also suggested by the fact that MBD1 physically interacts with CAF1 by immunoprecipitation and yeast two-hybrid, and they colocalize to mammalian pericentric heterochromatin domains (Table 1, Fig. 1b) (Reese et al. 2003). It has not been tested whether PCNA is involved in the MBD1–CAF1 interaction or whether CAF1 may act as a second scaffold behind the fork. The notion that CAF1 can act as a scaffold is supported by the fact that the MBD1/CAF1 complex associates with HP1, but that HP1 has not been demonstrated to interact physically with PCNA (Fig. 1b) (Reese et al. 2003).
Reese et al. also examined the effects of disrupting CAF1 p150 on CAF1–MBD1 localization and on several heterochromatin markers. Overexpression of the C terminus of CAF1 p150, the domain required for the MBD1 interaction, disrupted localization of CAF1 to pericentric heterochromatin foci (Reese et al. 2003). In addition, this overexpression prevented localization of MBD1 to the heterochromatin foci, but did not seem to disrupt other markers of heterochromatin such as MeCP2 or HP1α. This experiment suggests that CAF1 mediates MBD1's localization to pericentric heterochromatin. Additionally, it suggests that other factors assist in recruiting HP1α to heterochromatin in the absence of proper CAF1 localization. It may be that the methylation of histone H3 can recruit HP1 on its own postreplication, and that this is facilitated by HP1's interaction with CAF1. It is also possible that ORC and replication proteins could recruit HP1 or that unidentified factors assist in recruiting HP1 (Fig. 1a). Whether or not these factors normally assist in recruiting HP1α or only in this aberrant state remains to be elucidated.
As previously noted, in heterochromatin lysine 9 of histone H3 (H3K9) is often methylated and this site is bound by HP1 (Lachner et al. 2001; Bannister et al. 2001). Reestablishment of histone methylation following replication is linked to establishment of methylated DNA. DNMT1 and DNMT3a, a de novo DNA methyltransferase, bind to SUV39H1, a known H3K9 methyltransferase, and HP1β and SUV39H1 associate with DNA methyltransferase activity (Fuks et al. 2003a). Additionally, DNMT3b, another de novo DNA methyltransferase, fails to localize in Suv39h null cells, and these cells display an altered DNA methylation status at particular sequences, highlighting the importance of the DNA methylation–histone methylation relationship (Lehnertz et al. 2003). Methyl-binding proteins, specifically MeCP2, were previously shown to recruit H3K9 histone methyltransferase activity in mammalian cells (Fuks et al. 2003b).
Sarraf and Stancheva (2004) demonstrated a physical interaction between MBD1 and SETDB1, another H3K9 histone methyltransferase. In addition, MBD1/SETDB1 associates with CAF1 and PCNA specifically in S phase, and the formation of this complex requires ongoing DNA replication (Table 1, Fig. 1b). By using RNAi to MBD1, Sarraf and Stancheva found that MBD1 is required to recruit SETDB1 to CAF1 during DNA replication (Sarraf and Stancheva 2004). The interaction between DNA methylation and histone methylation is intriguing, as it may facilitate the rapid transition from newly synthesized chromatin to heterochromatin.
Although heterochromatic factors must be synthesized to meet the demands of the daughter genomes, it seems unlikely that the old factors are discarded and fresh factors are incorporated at each round of replication. How then does the passing replication fork keep track of “old” factors and ensure that the proper epigenetic state is reestablishment? Although many details of these questions remain, experiments by Sarraf and Stancheva imply that the fork may transiently displace MBD1 from methylated DNA, but keeps MBD1 close by to incorporate the factor postreplication. MBD1 binding to methylated DNA and CAF1 are mutually exclusive, suggesting that replication forks may generate a transient CAF1/MBD1/SETDB1 complex by displacing MBD1 from methylated DNA (Sarraf and Stancheva 2004). The CAF1/MBD1/SETDB1 complex also associates with histones H3 and H4 in S phase, hinting that methylation of H3K9 occurs during chromatin assembly (Sarraf and Stancheva 2004). These studies also suggest that passage of the replication fork is necessary to reestablish a heterochromatic state at a promoter site in mammals, in contrast to yeast. An intriguing explanation is proposed: DNA methylation directs H3K9 methylation by SETDB1 at MBD1-bound loci. If DNA methylation is removed, the recruitment of MBD1/SETDB1 complex to CAF1 is disrupted and results in a gradual loss of methylation following rounds of replication. It will be interesting to determine the details of the DNA methylation and histone methylation relationship: the number and importance of histone methylases acting to restore the heterochromatic state, the importance of replication in recruiting these factors, and whether or not every histone methylase is dependent upon DNA methyl-binding proteins.
Higher-order chromatin structure and the role of chromatin-remodeling complexes
Multiple chromatin-remodeling enzymes, which alter the positioning and spacing of nucleosomes without removal from DNA, were identified in eukaryotes and shown to play a role in the formation of heterochromatin. Heterochromatin is characterized by regular spacing of nucleosomes and tight compaction, restricting accessibility of the DNA (Wallrath and Elgin 1995; Sun et al. 2001). Chromatin-remodeling enzymes utilize ATP to shift nucleosomes into equally spaced positions and to remove them from promoter regions. Complexes containing Imitation Switch (ISWI) have been implicated in replication and maintenance of heterochromatin (for review, see Corona and Tamkun 2004; de la Serna and Imbalzano 2002). Defining the time of action of these complexes will be complicated, however, as chromatin-remodeling enzymes may be involved in moving nucleosomes to open DNA for replication and/or to reestablish the nucleosome pattern of heterochromatin. In addition, disruption of these enzymes likely affects all chromatin, complicating dissection of a specific role in heterochromatin. Current research on the role of ISWI complexes reveals roles in regulating replication and heterochromatin, although at present it is not clear whether they act primarily to open heterochromatin to promote replication or restrict replication through heterochromatin by maintaining a closed configuration, as detailed below.
Studies in human cells demonstrate a requirement for the ACF1 (ATP-utilizing chromatin assembly and remodeling factor 1)–ISWI complex in replication of heterochromatin. Prior to late S phase, ACF1 and ISWI exhibit general nuclear staining. At late S phase, these factors colocalize with BrdU and HP1β at the characteristic pericentric heterochromatin foci (Collins et al. 2002). Although ACF1 can localize to pericentric heterochromatin without its interaction with ISWI, the function of ACF requires ISWI (Collins et al. 2002). RNAi to ACF1 decreases the number of cells incorporating BrdU at pericentric heterochromatin, but does not alter HP1β localization. In addition, these ACF1 depleted cells show a delay in late S phase, which the authors interpret as a delay in the replication of heterochromatin. These phenotypes could be reversed by treatment of the ACF1-depleted cells with a DNA methylation inhibitor that leads to the decondensation of heterochromatin. This suggests that the delay in S phase is a result of impairment in opening heterochromatin for replication.
Studies on ACF1 in Drosophila, however, suggest a different role for the ACF1–ISWI complex in heterochromatin. Extracts made from acf1 null mutants assemble nucleosomes arrays less efficiently than wild-type extracts and show a decrease in the periodicity of these arrays on isolated chromatin (Fyodorov et al. 2004). Mutations in acf1, however, act as strong suppressors of PEV, suggesting that ACF1 contributes to the formation of heterochromatin rather than the opening of heterochromatin (Fyodorov et al. 2004). Observations on DNA replication in acf1 mutant embryos and larval neuroblasts also indicate that the functions of ACF1 in Drosophila differ from those observed in human cells. Drosophila acf1 mutant tissues spend less time in S phase and DNA replication appears normal. Thus these tissues appear to progress more rapidly through late S phase (Fyodorov et al. 2004). An accelerated S phase is also observed in mutants with decreased levels of histones, which further suggests that the repressive nature of heterochromatic DNA replication is relieved by poor chromatin assembly in acf1 mutants (Fyodorov et al. 2004).
Is it possible to reconcile the observations in human cells and Drosophila? As Fyodorov et al. (2004) note, the behavior of cultured mammalian cells and acf1 mutant embryos and larvae may not be identical. The ACF1–ISWI may perform slightly different roles in different organisms or at different developmental stages. Another possibility may be that ACF1–ISWI is involved in both roles, opening heterochromatin for replication and arranging nucleosome arrays for the heterochromatic state. Perhaps each system or experimental technique is particularly suited to observe predominantly one role over the other. Nevertheless, it is apparent that the ACF1–ISWI complex is important in the propagation of heterochromatin after DNA replication.
Another ISWI-containing complex, Williams syndrome transcription factor (WSTF)–ISWI chromatin remodeling complex, has been linked to maintenance of chromatin state in mammalian cells. WSTF and ISWI form a complex in vertebrates that, in vitro, can reconfigure disorganized nucleosomal arrays into more regularly spaced and organized configurations in an ATP-dependent manner (Bozhenok et al. 2002). A role for the WSTF–ISWI complex in heterochromatin maintenance is suggested by its localization to mammalian pericentric heterochromatin in late S phase, where it colocalizes in large, distinct foci with HP1β (Bozhenok et al. 2002). Based on the localization of WSTF in late S phase, the authors suggest that the WSTF–ISWI complex either facilitates DNA replication through heterochromatin or has a role in the assembly of heterochromatin reestablishment postreplication. A recent paper from the same laboratory probed the role of WSTF–ISWI further and revealed that WSTF–ISWI may have a role earlier in S phase (Poot et al. 2004). Importantly, in mid, late and very late S phase, WSTF nearly always colocalized with sites of active DNA replication. Additionally, WSTF and ISWI physically interact with PCNA and are retained at replication foci via their interaction with PCNA (Table 1, Fig. 1a) (Poot et al. 2004). This interaction with PCNA is consistent with the observation that WSTF–ISWI is retained at foci postreplication, because PCNA can persist at replication foci after DNA synthesis is complete.
Experiments in which WSTF has been depleted from cells provide evidence for a different role for WSTF–ISWI: WSTF acts to maintain open chromatin structures (Poot et al. 2004). WSTF-depleted cells have small nuclei that are more resistant to DNase I and micrococcal nuclease digestion, indicative of the chromatin in these cells being closed and more packaged. In addition, these cells demonstrated an increase in heterochromatic markers; levels of HP1α and -β, and histone H3 lysine 9 trimethylation and lysine 27 dimethylation were increased and this increase was not due to increased transcription of these factors (Poot et al. 2004). Interestingly, the increase in HP1β levels can be prevented if the cells are blocked in G1, indicating that passage through S phase is required for the observed increase in HP1β protein levels (Poot et al. 2004). The authors present two possible interpretations for the role of WSTF–ISWI. Nucleosomes may be less mobile in the absence of WSTF–ISWI, thereby promoting formation of heterochromatin. It is also possible that WSTF–ISWI may directly prevent HP1 binding to newly replicated DNA and actively maintain euchromatic structure. These interpretations suggest that heterochromatin is the default state for newly replicated DNA or that newly replicated DNA is highly susceptible to heterochromatin assembly in the absence of an active inhibition factor. If heterochromatin is indeed a default state, then keeping an organism's genome open would require a high level of heterochromatin-inhibition factors and substantial energy. Nevertheless, a precedence exists in the requirement of Dot1 in S. cerevisiae to actively block the spread of heterochromatin (Katan-Khaykovich and Struhl 2005). It is also not intuitive why a factor that inhibits the formation of heterochromatin would localize to pericentric heterochromatin while it replicates. Much remains to be deciphered about the role of chromatin-remodeling factors in the replication and regulation of heterochromatin.
Conclusions and perspectives
We have presented the evidence for roles of replication proteins, histone modification enzymes, DNA methyltransferase, and chromatin remodeling complexes in the reinstatement of heterochromatin at the replication fork in S phase. Many of these functions are likely to be required outside of S phase for the maintenance of heterochromatin and to be critical for the establishment of heterochromatin at new genomic locations in response to developmental cues such as position-effect variegation or X chromosome inactivation. Even within S phase, precise evaluation of the mechanism by which these proteins contribute to heterochromatin replication is impeded by the complexities of distinguishing their roles in DNA replication versus reestablishment of the chromatin. Use of mutations that dissociate DNA replication and chromatin requirements will be a powerful means to decipher these roles.
Conversely, new factors required for the maintenance of heterochromatin now need to be analyzed for roles in the replication of heterochromatin within the S phase. Among the most exciting new activities needed for heterochromatin are the RNAi machinery and the retinoblastoma (Rb) tumor suppressor protein. In fission yeast, Drosophila, and mammalian cells, the RNAi machinery is required for heterochromatin protein binding, heterochromatic silencing, and centromere function (Pal-Bhadra et al. 2004; Verdel et al. 2004; Huertas et al. 2004; Kanellopoulou et al. 2005; Motamedi et al. 2004; Fukagawa et al. 2004). The Rb protein family was also shown to be required for DNA methylation, hypoacetylation of histone H3, and trimethylation of histone H4, most likely via a direct interaction with the histone H4 lysine 20 trimethyl transferase Suv4-20 (Gonzalo et al. 2005). Given that Rb is known to be present and to act within the S phase (Bosco et al. 2001; Avni et al. 2003), it is likely that Rb has a function in reinstating heterochromatin during DNA replication. In addition to the predominant histone proteins, there are histone variants that contribute both to the formation of heterochromatin and protection against the spread of heterochromatin into euchromatic regions (for review, see Kamakaka and Biggins 2005). Although some of these histone variants such as H3.3 do not require DNA replication for their assembly into nucleosomes, the assembly requirements of other variants and potential roles in replication are in the early stages of investigation.
A crucial issue is how the histone modifications and associated chromatin proteins are templated onto the daughter duplex after replication. Given the interdependency of histone modifications (Czermin and Imhof 2003; Fischle et al. 2003), semiconservative reassembly of the nucleosome could provide a means to reestablish histone modifications that could then promote proper protein association (Tagami et al. 2004). The evidence to date, however, suggests conservative assembly of the nucleosome (for review, see Henikoff et al. 2004). The relationship between DNA methylation and histone modification provides an additional template mechanism. Although the problem of templating chromatin architecture is common to both euchromatin and heterochromatin, in the case of heterochromatin it has the increased complexity of requiring the reassociation of heterochromatin binding proteins. Further investigation of the regulation of heterochromatin replication will produce insights into how these modifications and protein associations are templated.
The timing of heterochromatin replication within S phase and the mechanism by which it is delayed until late in the S phase is another issue that remains to be unraveled. This is of biological significance in that this delayed timing facilitates the underreplication of heterochromatin in endo cycles. At present, it is not clear whether heterochromatin replicates late in the S phase because the chromatin structure restricts access to replication factors or whether the restriction of replication to late S phase helps form heterochromatin due to delayed availability of heterochromatin proteins to assemble. As proposed by McNairn and Gilbert (2003), both may be possible: late replication of heterochromatin initially contributes to formation of heterochromatin structure and that thereafter this restricts replication until late S phase. Future analysis on the timing of synthesis and assembly of heterochromatin proteins within S phase will be informative. Defining the means by which the intriguing SuUR protein both affects the timing and extent of heterochromatin replication in endo cycles is likely to provide crucial insights into how heterochromatin replication can be restricted until late in S phase.
References
Ahmad K, Henikoff S (2002) Epigenetic consequences of nucleosome dynamics. Cell 111:281–284
Ahmed S, Saini S, Arora S, Singh J (2001) Chromodomain protein Swi6-mediated role of DNA polymerase alpha in establishment of silencing in fission yeast. J Biol Chem 276:47814–47821
Araujo FD, Knox JD, Szyf M, Price GB, Zannis-Hadjopoulos M (1998) Concurrent replication and methylation at mammalian origins of replication. Mol Cell Biol 18:3475–3482
Avni D, Yang H, Martelli F, Hofmann F, ElShamy WM, Ganesan G, Scully R, Livingston DM (2003) Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol Cell 12:735–746
Badugu R, Shareef MM, Kellum R (2003) Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J Biol Chem 278:34491–34498
Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124
Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71:333–374
Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B (1995) The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83:563–568
Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE (1998) Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci U S A 95:7532–7537
Belyaeva ES, Boldyreva LV, Volkova EI, Nanayev RA, Alekseyenko AA, Zhimulev IF (2003) Effect of the Suppressor of Underreplication (SuUR) gene on position-effect variegation silencing in Drosophila melanogaster. Genetics 165:1209–1220
Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanyev RA, Makunin IV, Kafatos FC, Zhimulev IF, Heidelberg Fly Array Consortium (2005) Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc Natl Acad Sci U S A 102:8269–8274
Bosco G, Du W, Orr-Weaver TL (2001) DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat Cell Biol 3:289–295
Bozhenok L, Wade PA, Varga-Weisz P (2002) WSTF–ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J 21:2231–2241
Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721–725
Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF (1997) Human DNA-(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science 277:1996–2000
Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD (2002) An ACF1–ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet 32:627–632
Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677:113–119
Craig JM (2005) Heterochromatin—many flavours, common themes. BioEssays 27:17–28
Czermin B, Imhof A (2003) The sounds of silence—histone deacetylation meets histone methylation. Genetica 117:159–164
de la Serna IL, Imbalzano AN (2002) Unfolding heterochromatin for replication. Nat Genet 32:560–562
Dillin A, Rine J (1997) Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics 147:1053–1062
Dillon N, Festenstein R (2002) Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet 18:252–258
Edgar BA, Orr-Weaver TL (2001) Endoreplication cell cycles: more for less. Cell 105:297–306
Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC (1997) Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021–1032
Enomoto S, Berman J (1998) Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev 12:219–232
Fischle W, Wang Y, Allis CD (2003) Histone and chromatin cross-talk. Curr Opin Cell Biol 15:172–183
Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6:784–791
Fuks F, Hurd PJ, Deplus R, Kouzarides T (2003a) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31:2305–2312
Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T (2003b) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem 278:4035–4040
Fuss J, Linn S (2002) Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J Biol Chem 277:8658–8666
Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT (2004) Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev 18:170–183
Gilbert DM (2002) Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol 14:377–383
Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, Blasco MA (2005) Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 7:420–428
Greil F, van der Kraan I, Delrow J, Smothers JE, de Wit E, Bussemaker HJ, van Driel R, Henikoff S, van Steensel B (2003) Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev 17:2825–2838
Gruss C, Wu J, Koller T, Sogo JM (1993) Disruption of the nucleosomes at the replication fork. EMBO J 12:4533–4545
Henderson DS, Banga SS, Grigliatti TA, Boyd JB (1994) Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J 13:1450–1459
Henikoff S (2000) Heterochromatin function in complex genomes. Biochim Biophys Acta 1470:1–8
Henikoff S, Furuyama T, Ahmad K (2004) Histone variants, nucleosomal assembly and epigenetic inheritance. Trends Genet 20:320–326
Huang Y (2002) Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res 30:1465–1482
Huang DW, Fanti L, Pak DT, Botchan MR, Pimpinelli S, Kellum R (1998) Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol 142:307–318
Hubscher U, Maga G, Spadari S (2002) Eukaryotic DNA polymerases. Annu Rev Biochem 71:133–163
Huertas D, Cortes A, Casanova J, Azorin F (2004) Drosophila DDP1, a multi-KH-domain protein, contributes to centromeric silencing and chromosome segregation. Curr Biol 14:1611–1620
Iida T, Suetake I, Tajima S, Morioka H, Ohta S, Obuse C, Tsurimoto T (2002) PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells 7:997–1007
Iizuka M, Stillman B (1999) Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem 274:23027–23034
Jiang YH, Bressler J, Beaudet AL (2004) Epigenetics and human disease. Annu Rev Genomics Hum Genet 5:479–510
Kamakaka RT, Biggins S (2005) Histone variants: deviants? Genes Dev 19:295–310
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Katan-Khaykovich Y, Struhl K (2005) Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J 24:2138–2149
Kirchmaier AL, Rine J (2001) DNA replication-independent silencing in S. cerevisiae. Science 291:646–650
Krude T (1995) Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res 220:304–311
Krude T (1999) Chromatin assembly during DNA replication in somatic cells. Eur J Biochem 263
Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120
Leach TJ, Chotkowski HL, Wotring MG, Dilwith RL, Glaser RL (2000) Replication of heterochromatin and structure of polytene chromosomes. Mol Cell Biol 20:6308–6316
Leatherwood J, Vas A (2003) Connecting ORC and heterochromatin: why? Cell Cycle 2:573–575
Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13:1192–1200
Leonhardt H, Page AW, Weier HU, Bestor TH (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865–873
Li YC, Cheng TH, Gartenberg MR (2001) Establishment of transcriptional silencing in the absence of DNA replication. Science 291:650–653
Lidonnici MR, Rossi R, Paixao S, Mendoza-Maldonado R, Paolinelli R, Arcangeli C, Giacca M, Biamonti G, Montecucco A (2004) Subnuclear distribution of the largest subunit of the human origin recognition complex during the cell cycle. J Cell Sci 117:5221–5231
Lilly MA, Spradling AC (1996) The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev 10:2514–2526
Lippman Z, Martienssen R (2004) The role of RNA interference in heterochromatic silencing. Nature 431:364–370
Loupart M-L, Krause SA, Heck MMS (2000) Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol 10:1547–1556
Maga G, Hubscher U (2003) Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci 116:3051–3060
Maison C, Almouzni G (2004) HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5:296–304
Majka J, Burgers PM (2004) The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 78:227–260
Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF (2002) The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160:1023–1034
Martens JHA, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24:800–812
Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6:24–35
McNairn AJ, Gilbert DM (2003) Epigenomic replication: linking epigenetics to DNA replication. BioEssays 25:647–656
Mello JA, Almouzni G (2001) The ins and outs of nucleosome assembly. Curr Opin Genet Dev 11:136–141
Moshkin YM, Alekseyenko AA, Semeshin VF, Spierer A, Spierer P, Makarevich GF, Belyaeva ES, Zhimulev IF (2001) The bithorax complex of Drosophila melanogaster: underreplication and morphology in polytene chromosomes. Proc Natl Acad Sci U S A 98:570–574
Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119:789–802
Murzina N, Verreault A, Laue E, Stillman B (1999) Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell 4:529–540
Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113
Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R (2001) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell 7:729–739
Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311–323
Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC (2004) Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303:669–672
Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD (2004) The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol 6:1236–1244
Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B (2004) Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 23:2651–2663
Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G (2004) A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J 23:3516–3526
Reese BE, Bachman KE, Baylin SB, Rountree MR (2003) The methyl-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol Cell Biol 23:3226–3236
Ridgway P, Almouzni G (2000) CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J Cell Sci 113:2647–2658
Royzman I, Hayashi-Hagihara A, Dej KJ, Bosco G, Lee JY, Orr-Weaver TL (2002) The E2F cell cycle regulator is required for Drosophila nurse cell DNA replication and apoptosis. Mech Dev 119:225–237
Rudkin GT (1969) Non replicating DNA in Drosophila. Genetics 61(Suppl):227–238
Sarraf SA, Stancheva I (2004) Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 15:595–605
Schotta G, Ebert A, Dorn R, Reuter G (2003a) Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 14:67–75
Schotta G, Ebert A, Reuter G (2003b) SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing. Genetica 117:149–158
Schramke V, Allshire R (2004) Those interfering little RNAs! Silencing and eliminating chromatin. Curr Opin Genet Dev 14:174–180
Semeshin F, Belyaeva S, Zhimulev F (2001) Electron microscope mapping of the pericentric and intercalary heterochromatic regions of the polytene chromosomes of the mutant Suppressor of underreplication in Drosophila melanogaster. Chromosoma 110:487–500
Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R (2001) Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell 12:1671–1685
Shibahara K, Stillman B (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575–585
Smith AV, Orr-Weaver TL (1991) The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112:997–1008
Sun FL, Cuaycong MH, Elgin SC (2001) Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol Cell Biol 21:2867–2879
Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G (1999) Duplication and maintenance of heterochromatin domains. J Cell Biol 147:1153–1166
Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51–61
Tatematsu KI, Yamazaki T, Ishikawa F (2000) MBD2–MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 5:677–688
Tchurikov NA, Kretova OV, Chernov BK, Golova YB, Zhimulev IF, Zykov IA (2004) SuUR protein binds to the boundary regions separating forum domains in Drosophila melanogaster. J Biol Chem 279:11705–11710
Triolo T, Sternglanz R (1996) Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381:251–253
Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676
Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont, AS, van Driel R (2005) In vivo HP1 targetting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol 25:4552–4564
Vertino PM, Sekowski JA, Coll JM, Applegren N, Han S, Hickey RJ, Malkas LH (2002) DNMT1 is a component of a multiprotein DNA replication complex. Cell Cycle 1:416–423
Volkova EI, Yurlova AA, Kolesnikova TD, Makunin IV, Zhimulev IF (2003) Ectopic expression of the Suppressor of Underreplication gene inhibits endocycles but not the mitotic cell cycle in Drosophila melanogaster. Mol Genet Genomics 270:387–393
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA (2002) Regulation of heterochormatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837
Wade PA (2001) Methyl CpG-binding proteins and transcriptional repression. Bioessays 23:1131–1137
Wakimoto BT, Hearn MG (1990) The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125:141–154
Wallrath LL, Elgin SC (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9:1263–1277
Zhang Z, Shibahara K, Stillman B (2000) PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408:221–225
Zhimulev IF, Belyaeva ES (2003) Intercalary heterochromatin and genetic silencing. BioEssays 25:1040–1051
Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, Demakova OV (2003a) Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 111:377–398
Zhimulev IF, Belyaeva ES, Semeshin VF, Shloma VV, Makunin IV, Volkova EI (2003b) Overexpression of the SuUR gene induces reversible modifications at pericentric, telomeric and intercalary heterochromatin of Drosophila melanogaster polytene chromosomes. J Cell Sci 116:169–176
Acknowledgements
We thank A. Aslanian, S. Bell, H. Blitzblau, S. Bumgarner, J. Pesin, and E. Rogakou for helpful comments on the manuscript. We are grateful to I. Zhimulev for permission to reproduce the micrographs in Fig. 2. The authors were supported by grants from the NIH (GM57541) and the Harold G. and Leila Y. Mathers Charitable Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.A. Nigg.
Rights and permissions
About this article
Cite this article
Wallace, J.A., Orr-Weaver, T.L. Replication of heterochromatin: insights into mechanisms of epigenetic inheritance. Chromosoma 114, 389–402 (2005). https://doi.org/10.1007/s00412-005-0024-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-005-0024-6