Abstract
Like various other diurnal birds of prey, the world's largest eagle, the Harpy (Harpia harpyja), presents an atypical bird karyotype with 2n=58 chromosomes. There is little knowledge about the dramatic changes in the genomic reorganization of these species compared to other birds. Since recently, the chicken provides a “default map” for various birds including the first genomic DNA sequence of a bird species. Obviously, the gross division of the chicken genome into relatively gene-poor macrochromosomes and predominantly gene-rich microchromosomes has been conserved for more than 150 million years in most bird species. Here, we present classical features of the Harpy eagle karyotype but also chromosomal homologies between H. harpyja and the chicken by chromosome painting and comparison to the chicken genome map. We used two different sets of painting probes: (1) chicken chromosomes were divided into three size categories: (a) macrochromosomes 1–5 and Z, (b) medium-sized chromosomes 6–10, and (c) 19 microchromosomes; (2) combinatorially labeled chicken chromosome paints 1–6 and Z. Both probe sets were visualized on H. harpyja chromosomes by multicolor fluorescence in situ hybridization (FISH). Our data show how the organization into micro- and macrochromosomes has been lost in the Harpy eagle, seemingly without any preference or constraints.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The typical avian karyotype with 2n=78 chromosomes is widely conserved among very divergent orders of birds (Burt 2002; Schmid et al. 2000). It is composed of few large macrochromosomes that are comparable in size with mammalian chromosomes and numerous morphologically indistinguishable microchromosomes ranging in size from some 23 to less than 5 Mbp. Some birds of prey of the order Falconiformes, including hawks, buzzards, and eagles, however, show strikingly different karyotypes with lower numbers of mostly medium-sized chromosomes and only few chromosomes of the size of typical avian microchromosomes (Bed'Hom et al. 2003).
Because of its economic impact and its importance in biomedical research as a model organism for developmental biology and neurobiology, the genome of the domestic chicken Gallus gallus domesticus generally serves as the starting point and reference for the investigation of other avian genomes. Its physiology is well understood, and various genomic resources are available, including a recent first draft of the genome sequence (International Chicken Genome Sequencing Consortium 2004) (http://www.ensembl.org/Gallus_gallus). The chicken shares the typical avian karyotype with 2n=78 and micro- and macrochromosomes. Chicken macrochromosomes (1–9, Z, W) comprise about 75% of the genome, which mainly consists of AT-rich, relatively gene-poor, and late replicating DNA, whereas microchromosomes show converse features (recently summarized in Schmid et al. 2000 and International Chicken Genome Sequencing Consortium 2004).
Lately, chromosome-specific painting probes for the chicken macrochromosomes 1–9, for several of the larger microchromosomes and for fractions of the smaller microchromosomes, became available (Griffin et al. 1999; Habermann et al. 2001). More recently, the entire chromosome set of the chicken was defined by fluorescence in situ hybridization (FISH) with DNA probes (Masabanda et al. 2004). Cross-species chromosome painting studies between the chicken and the emu (Dromaius novaehollandiae) demonstrated that macrochromosomes are well conserved for more than 80 million years of bird evolution (Shetty et al. 1999). These data suggest that the typical avian karyotype with macro- and microchromosomes may be the ancestral state for all birds. Further, microchromosomes are also present in some primitive amphibians and most reptiles, suggesting that some, if not most avian microchromosomes, represent ancestral vertebrate syntenies (Burt 2002).
Thus, in birds of prey, numerous chromosome rearrangements must have occurred, including fusions of microchromosomes and/or their translocation onto larger chromosomes (Bed'Hom 1999; de Lucca and Rocha 1992; Rodionov 1996; Tegelstrom et al. 1983). Like other raptors, the Harpy eagle (Harpia harpyja, Family Accipitridae), which inhabits neotropical rainforests from southern Mexico to northern Argentina, is a representative example for these atypical bird karyotypes. H. harpyja presents a karyotype of 2n=58 (Hoffmann et al. 1976), consisting of largely medium-sized chromosomes that gradually decrease in size, and of which only four chromosome pairs are comparable in size to chicken microchromosomes.
We performed an initial characterization of the chromosome complement of the Harpy eagle employing differential staining techniques and FISH with a telomeric repeat probe conserved in vertebrates [(TTAGGG)n; Meyne et al. 1989]. Further, we delineated chromosomal homologies between the Harpy eagle and the chicken by cross-species chromosome painting of various chicken chromosome-specific probes. These experiments provide initial insight into the extent and the evolutionary direction of chromosomal rearrangements that changed the karyotype of the Harpy eagle and probably those of other raptors.
Materials and methods
Cell samples
Blood samples were collected from two specimens (male and female) of Harpy eagle kept at the Zoológico Municipal de Curitiba (Curitiba, PR, Brazil). Blood cells were grown and harvested as described by Moorhead et al. (1960), with minor modifications. Metaphase preparations followed standard procedures. Chicken metaphase spreads were prepared from a fibroblast culture (Habermann et al. 2001).
Banding techniques
Metaphase chromosomes of H. harpyja were studied by C- and nucleolar organizer region (NOR) banding techniques according to Sumner (1972) and Howell and Black (1980). G-banding technique followed Seabright (1971) with minor modifications. 7-Aminoactinomycin-D (7-AAD; emission maximum 647 nm, red fluorescence) differential DNA staining was performed essentially according to Gill et al. (1975), but in combination with a subsequent 4′,6-diamidino-2-phenylindole (DAPI) counterstain (emission maximum 461 nm, blue fluorescence).
Probe composition, in situ hybridization, and detection
The biotinylated “All Human Telomere Probe” (ONCOR) was applied in FISH experiments according to the manufacturer's protocol. For multicolor cross-species chromosome painting, two independent experiments using different multiplex probe sets were performed. The probe sets were the same as previously described (Habermann et al. 2001): (1) chicken chromosome 1–6 and Z paint probes were combinatorially labeled with three fluorochromes and hybridized to eagle chromosomes; (2) chromosome paint probes of closely the entire chicken karyotype were divided into three differentially labeled probe pools according to their size: (a) macrochromosomes 1–5 and Z, (b) medium-sized chromosomes 6–10, and (c) 19 different microchromosomes. Both probe sets were labeled with biotin–dUTP, digoxigenin–dUTP, or TAMRA–dUTP, respectively, and mixed with tenfold excess of chicken Cot-1 DNA. Cross-species hybridizations to eagle chromosomes were performed for 72 h, followed by stringency washes for 2×5 min in 50% formamide/2×SSC (37°C) and 2×5 min in 2×SSC (37°C). Biotinylated probes were detected with avidin-Cy3 or avidin-Cy5 (Amersham), digoxigenin-labeled probes with sheep anti-digoxigenin FITC coupled antibody (Roche). Chromosomes were counterstained with DAPI and embedded with Vectashield mounting medium (Vector Laboratories).
Microscopy and image analysis
Metaphase images were captured with a cooled CCD camera (Photometrics C250/A equipped with a Kodak KAF1400 chip) coupled to a Zeiss Axiophot microscope. Camera control and digital image acquisition was performed using SmartCapture VP software (Digital Scientific, Cambridge, UK).
Results
Classical cytogenetics, differential DNA staining, and telomeric repeats
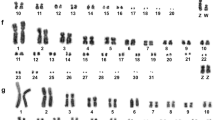
The karyotype of H. harpyja (2n=58 chromosomes) has 18 biarmed and 10 acrocentric autosomal pairs. The sex chromosomes are ZZ in the male and ZW in the female. The Z is a large submetacentric, whereas the W is a medium-sized submetacentric chromosome. Constitutive heterochromatin blocks visualized by C-banding were located in the pericentromeric region of most chromosomes. The W chromosome showed a large heterochromatic block, corresponding to roughly two thirds of its total length (Fig. 1a). NORs were located by silver staining on chromosome 8 and on a microchromosome-sized pair, most likely chromosome 25 (Fig. 1b).
a C-banded metaphase of a female H. harpyja. The arrow marks the W chromosome, of which a large region is heterochromatic. b Sequential G-banding and silver staining reveal H. harpyja NOR-bearing chromosomes 8 and 25. Only one of the two chromosomes 8 is stained in this metaphase. Differential DNA staining of c a chicken and d a H. harpyja metaphase with DAPI/7-AAD. GC-rich DNA is shown in red, AT-rich DNA in green. e In situ hybridization of a telomere-specific (TTAGGG)n probe (green) to a H. harpyja metaphase (red) produced hybridization signals on chromosome ends, but not in interstitial chromosome regions
Differential DNA staining of chicken and H. harpyja metaphase chromosomes was performed using a combination of DAPI and 7-AAD (Fig. 1c,d). DAPI preferentially binds AT-rich DNA, whereas 7-AAD is a GC-selective DNA stain. GC-rich chicken microchromosomes showed intense 7-AAD fluorescence, compared to the macrochromosomes. In the Harpy eagle, bright 7-AAD fluorescence was observed on the microchromosomes but also in several blocks up to the size of chromosome arms on medium-sized and large chromosomes. In situ hybridization of a biotinylated telomere-specific (TTAGGG)n probe to metaphase chromosomes from H. harpyja (Fig. 1e) provided no evidence for the existence of interstitial telomeric sequences.
Cross-species chromosome painting
Chromosome painting with combinatorially labeled chicken (GGA) macrochromosome probes 1–6 and Z (Fig. 2a) revealed 16 homologous chromosome regions on 15 different autosome pairs of H. harpyja (HHA). The chicken Z painting probe hybridized to the entire H. harpyja Z chromosome and showed cross-hybridization to the entire W chromosome (Fig. 2b). The assignment of chicken homologous regions to G-banded H. harpyja chromosomes is summarized in Fig. 2d.
a In situ hybridization of combinatorially labeled chicken (GGA) macrochromosome paint probes 1–6 and Z on a H. harpyja metaphase. GGA 1 is shown in blue, GGA 2 in green, GGA 3 in red, GGA 4 in magenta, GGA 5 in cyan, GGA 6 in white, and GGA Z in yellow. a′ Chromosomal counterstain (inverted DAPI) of the metaphase depicted in a. b Summary of the hybridization results with chicken (GGA) chromosome 1–6 and Z paint probes on H. harpyja chromosomes. c Hybridization of pooled paint probes of chicken macrochromosomes 1–5 (blue), medium-sized chromosomes 6–10 (green), and 19 different microchromosomes (red) to H. harpyja metaphase chromosomes (inset: partial chicken metaphase hybridized with the same probe). c′ Chromosomal counterstain (inverted DAPI) of the metaphase depicted in c. d G-banded karyotype of a male H. harpyja (inset: sex chromosome complement of a female individual), together with the assignment of chicken chromosome homologies delineated by cross-species chromosome painting. Homologies to individual chicken chromosomes 1–6 and Z (compare with Fig. 2a) are indicated by numbers to the right of each chromosome; homologies delineated by pooled chicken paint probes (compare with Fig. 2c) are depicted by color-coded vertical bars on the left (GGA 1–5 in blue, GGA 6–10 in green, and 19 different microchromosomes in red). Asterisks indicate NOR-bearing chromosomes
The set of pooled and differentially labeled chicken macrochromosomes 1–5, medium-sized chromosomes 6–10, and 19 different microchromosomes also delivered reproducible hybridization signals when hybridized to H. harpyja chromosomes (Fig. 2c). The probe pool of chicken macrochromosomes hybridized to 14 different H. harpyja autosomes (Fig. 2d). The probe pool representing the five medium-sized chicken chromosomes labeled five homologous chromosomes in H. harpyja. Finally, the probe pool representing 19 out of the 29 chicken microchromosomes delineated 16 regions on 15 different chromosomes of the Harpy eagle. Nine of these Harpy eagle chromosomes exclusively showed homologies to chicken microchromosomes. The remaining seven hybridization signals of the microchromosome probe pool were observed on segments of six different Harpy eagle chromosomes.
Discussion
To date, the karyotype of the Harpy eagle (H. harpyja) has only been analyzed by classical Giemsa staining (Hoffmann et al. 1976). The present study, using different banding techniques, has permitted the conventional characterization of the karyotype. The use of chromosome-specific probes from G. gallus in “painting” experiments, however, allowed the delineation of chromosomal homologies for an initial analysis of the mechanisms that led to the differentiation of Accipitridae karyotypes from the typical avian chromosome complement.
H. harpyja shows a karyotype of 2n=58 chromosomes with features similar to other Accipitridae species. C-banding demonstrated that most of the W chromosome is heterochromatic (Fig. 1a). Moreover, the chicken Z painting probe showed cross-hybridization to the entire H. harpyja W chromosome, indicating that similar repetitive DNA sequences are found in the W and Z.
The location of NORs on a pair of large chromosomes as found in the Harpy eagle is also shared by some other Accipitridae species. In addition, however, in H. harpyja, a pair of microchromosomes showed positive Ag–NORs, whereas in most other Accipitridae, either a pair of macro- or a pair of microchromosomes bear NORs (de Boer 1980). The most interesting observation from the classical cytogenetic characterization of the karyotype of the Harpy eagle compared to the chicken comes from DAPI/7-AAD staining (Fig. 1c,d). In the chicken, the staining pattern follows recent DNA sequencing results of the chicken genome (International Chicken Genome Sequencing Consortium 2004) (http://www.ensembl.org/Gallus_gallus). Microchromosomes were more intensively stained by the GC-selective fluorochrome 7-AAD, whereas macrochromosomes were highlighted by more AT-specific DAPI staining (Fig. 1c). This differentiation is lost in the Harpy eagle, where several medium-sized chromosomes show strong 7-AAD positive segments (Fig. 1d).
The chromosome painting analysis between the Harpy eagle and the chicken revealed that both species differ by at least 20 chromosome rearrangements (Fig. 3). The direction of change becomes evident when comparing karyotypes and genomes between the chicken and other birds, between birds and reptiles, and finally between the chicken and mammals. Recent cross-species chromosome painting studies suggest that between chicken and emu, all macrochromosomes are conserved, except for chicken chromosome 4 (Shetty et al. 1999). The comparison of chicken chromosome 4 and its homologs in other birds and mammals suggests its origin by fusion of an ancestral avian microchromosome (GGA 4p), with an ancestral avian macrochromosome (GGA 4q) (International Chicken Genome Sequencing Consortium 2004) (Fig. 3). Except for chromosome 4, the chicken appears to closely reflect the ancestral karyotype of Galliformes, Anseriformes, Columbiformes, Ciconiformes, Passeriformes, Strigiformes, and Struthioniformes, with chromosome numbers ranging from 2n=78 to 82 (Derjusheva et al. 2004; Guttenbach et al. 2003; Kasai et al. 2003; Raudsepp et al. 2002; Schmid et al. 2000; Shibusawa et al. 2004a,b). Moreover, recent gene mapping in reptiles identified the same gene order on reptile macrochromosomes as in birds (Matsuda et al. 2004), suggesting that both reptiles and birds share the same ancestral chromosome organization for more than 250 million years of evolution. As a consequence, the karyotype of the Harpy eagle should be considered as highly derived.
Chromosomal rearrangements in the Harpy eagle. Compared to the putative ancestral avian karyotype, the chicken karyotype is derived by the fusion of a macrochromosome (blue) with a microchromosome (brown). In the Harpy eagle, chicken macrochromosome homologs are further derived by ten fissions and one fusion. Harpy eagle microchromosome homologs (red) were involved in at least 10 fusions (note: only 20 of 29 microchromosome homologs could be analyzed, because probes for the remaining nine chicken chromosomes were not available in this study)
Further evidence in support of this conclusion is provided by the chicken–human genome alignment (http://www.ensembl.org). For example, chicken chromosome 3 is largely homologous to human 6p21.2-qter. Taking humans as the ultimate “outgroup” for birds, the contiguous human 6p21.2-qter and its chicken homolog should represent a shared ancestral chromosomal entity for both birds and mammals. In H. harpyja, however, this region is found fragmented into four homologous segments.
The homologs of chicken macrochromosomes 1–5 are split in H. harpyja as a consequence of ten fissions and one fusion (Fig. 3), whereas the five medium-sized chicken chromosomes 6–10 may have conserved large-scale homology between the two species. Most notably, a minimum of four fusions between homologs to chicken micro- and macrochromosomes, at least three further fusions between chicken microchromosome and medium-sized homologs, and another three fusions among microchromosomes occurred in the Harpy eagle (Fig. 3). Apparently, the homologs to chicken microchromosomes are fused to other chromosomes without any preference or constraints. A probe specific for telomeric DNA common to most vertebrates (Meyne et al. 1989), however, did not reveal any interstitial telomeric sites at fusion points of chromosomes (Fig. 1c).
As a consequence of this dramatic karyotype reorganization, the gross division of the genome into relatively gene-poor macrochromosomes and gene-rich microchromosomes, as observed in chicken and in many other bird species, has been lost in the Harpy eagle and most probably in other birds of prey as well. Traditional analyses of chromosomes already identified various highly conserved “flavors” of chromosomes or chromosome regions in evolution regarding gene density and replication timing (Holmquist 1992). This has been confirmed and dramatically extended by the initial results from the chicken genome project. In addition, a distinct arrangement of macro- and microchromosomes has been observed in three-dimensionally preserved chicken cell nuclei as well as in mitotic rosettes. Microchromosomes are preferentially clustered in the center of interphase nuclei and mitotic rosettes, whereas macrochromosomes are consistently located in the nuclear periphery (Habermann et al. 2001). Birds of prey may provide a model for cell biologists to unravel the gross genome organization and genomic “flavors” in an evolutionary framework.
References
Bed'Hom B (1999) Etude des Caryotypes Atypiques des Accipitridae (Aves, Falconiformes) par Cytogenetique Classique et Molécular, et Modélisation de leur Èvolution. Ph.D. Thesis, University of Tours, France
Bed'Hom B, Coullin P, Guillier-Gencik Z, Moulin S, Bernheim A, Volobouev V (2003) Characterization of the atypical karyotype of the black-winged kite Elanus caeruleus (Falconiformes: Accipitridae) by means of classical and molecular cytogenetic techniques. Chromosome Res 11:335–343
Burt DW (2002) Origin and evolution of avian microchromosomes. Cytogenet Genome Res 96:97–112
de Boer LE (1980) Do the chromosomes of the kiwi provide evidence for a monophyletic origin of the ratites? Nature 287:84–85
de Lucca EJ, Rocha GT (1992) Citogenética de Aves. Bol Mus Para Emilio Goeldi, Zool 8:33–68
Derjusheva S, Kurganova A, Habermann F, Gaginskaya E (2004) High chromosome conservation detected by comparative chromosome painting in chicken, pigeon and passerine birds. Chromosome Res 12:715–723
Gill JE, Jotz MM, Young SG, Modest EJ, Sengupta SK (1975) 7-Amino-actinomycin D as a cytochemical probe. I. Spectral properties. J Histochem Cytochem 23:793–799
Griffin DK, Habermann F, Masabanda J, O'Brien P, Bagga M, Sazanov A, Smith J, Burt DW, Ferguson-Smith M, Wienberg J (1999) Micro- and macrochromosome paints generated by flow cytometry and microdissection: tools for mapping the chicken genome. Cytogenet Cell Genet 87:278–281
Guttenbach M, Nanda I, Feichtinger W, Masabanda JS, Griffin DK, Schmid M (2003) Comparative chromosome painting of chicken autosomal paints 1–9 in nine different bird species. Cytogenet Genome Res 103:173–184
Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I (2001) Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res 9:569–584
Hoffmann R, Faust R, Weinand U, Hoffmann-Fezer G (1976) Chromosomenuntersuchungen an fünf Spezies der Ordnung Falconiformes. Zool Gart (Jena) 46:99–107
Holmquist GP (1992) Chromosome bands, their chromatin flavors, and their functional features. Am J Hum Genet 51:17–37
Howell WM, Black DA (1980) Controlled silver-staining of nucleolar organizer regions with protective colloidal developer: a 1-step method. Experientia 36:1014
International Chicken Genome Sequencing Consortium (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Kasai F, Garcia C, Arruga MV, Ferguson-Smith MA (2003) Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa): evidence of the occurrence of a neocentromere during evolution. Cytogenet Genome Res 102:326–330
Masabanda JS, Burt DW, O'Brien PC, Vignal A, Fillon V, Walsh PS, Cox H, Tempest HG, Smith J, Habermann F, Schmid M, Matsuda Y, Ferguson-Smith MA, Crooijmans RP, Groenen MA, Griffin DK (2004) Molecular cytogenetic definition of the chicken genome: the first complete avian karyotype. Genetics 166:1367–1373
Matsuda Y, Nishida-Umehara C, Tarui H, Kuroiwa A, Yamada K, Isobe T, Ando J, Saito T, Agata K (2004) Karyological relationships between birds and reptiles inferred from comparative gene mapping. 15th International Chromosome Conference, Brunel University, UK. Chromosome Res, pp 45
Meyne J, Ratliff RL, Moyzis RK (1989) Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A 86:7049–7053
Moorhead PS, Nowell PC, Mellman WJ, Battips DM (1960) Chromosome preparations of leucocytes cultured from human peripheral blood. Exp Cell Res 20:613–616
Raudsepp T, Houck ML, O'Brien PC, Ferguson-Smith MA, Ryder OA, Chowdhary BP (2002) Cytogenetic analysis of California condor (Gymnogyps californianus) chromosomes: comparison with chicken (Gallus gallus) macrochromosomes. Cytogenet Genome Res 98:54–60
Rodionov AV (1996) Micro vs. macro: structural–functional organization of avian micro-and macrochromosomes. Genetika 32:597–608
Schmid M, Nanda I, Guttenbach M, Steinlein C, Hoehn M, Schartl M, Haaf T, Weigend S, Fries R, Buerstedde JM, Wimmers K, Burt DW, Smith J, A'Hara S, Law A, Griffin DK, Bumstead N, Kaufman J, Thomson PA, Burke T, Groenen MA, Crooijmans RP, Vignal A, Fillon V, Morisson M, Pitel F, Tixier-Boichard M, Ladjali-Mohammedi K, Hillel J, Maki-Tanila A, Cheng HH, Delany ME, Burnside J, Mizuno S (2000) First report on chicken genes and chromosomes 2000. Cytogenet Cell Genet 90:169–218
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2:971–972
Shetty S, Griffin DK, Graves JA (1999) Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res 7:289–295
Shibusawa M, Nishibori M, Nishida-Umehara C, Tsudzuki M, Masabanda J, Griffin DK, Matsuda Y (2004a) Karyotypic evolution in the Galliformes: an examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenet Genome Res 106:111–119
Shibusawa M, Nishida-Umehara C, Tsudzuki M, Masabanda J, Griffin DK, Matsuda Y (2004b) A comparative karyological study of the blue-breasted quail (Coturnix chinensis, Phasianidae) and California quail (Callipepla californica, Odontophoridae). Cytogenet Genome Res 106:82–90
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306
Tegelstrom H, Ebenhard T, Ryttman H (1983) Rate of karyotype evolution and speciation in birds. Hereditas 98:235–239
Acknowledgements
The authors would like to thank Thomas Cremer for his continuous support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.A. Nigg
Rights and permissions
About this article
Cite this article
de Oliveira, E.H.C., Habermann, F.A., Lacerda, O. et al. Chromosome reshuffling in birds of prey: the karyotype of the world's largest eagle (Harpy eagle, Harpia harpyja) compared to that of the chicken (Gallus gallus). Chromosoma 114, 338–343 (2005). https://doi.org/10.1007/s00412-005-0009-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-005-0009-5