Abstract
The centromere-specific histone H3 variant CENP-A plays a crucial role in kinetochore specification and assembly. We chose a genetic approach to identify interactors of the Drosophila CENP-A homolog CID. Overexpression of cid in the proliferating eye imaginal disk results in a rough eye phenotype, which is dependent on the ability of the overexpressed protein to localize to the kinetochore. A screen for modifiers of the rough eye phenotype identified mutations in the Drosophila condensin subunit gene Cap-G as interactors. Yeast two-hybrid experiments also reveal an interaction between CID and Cap-G. While chromosome condensation in Cap-G mutant embryos appears largely unaffected, massive defects in sister chromatid segregation occur during mitosis. Taken together, our results suggest a link between the chromatin condensation machinery and kinetochore structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Accurate segregation of sister chromatids in mitosis is a prerequisite for maintenance of genetic stability. Missegregation of chromatids results in the formation of aneuploid daughter cells that are either non-viable or may progress to malignant growth (for a recent review see Draviam et al. 2004). Proper segregation is ensured by establishment, and meticulous control of amphitelic attachment of the replicated chromosomes within the mitotic spindle apparatus. The microtubules of the mitotic spindle attach with the chromosomes via the kinetochores, which are large proteinaceous structures that assemble at the centromeres of each chromatid.
In budding yeast, a short centromeric DNA sequence of 125 bp is required and sufficient for determining the assembly site of the kinetochore, which in mitosis is composed of at least 50 different proteins (for review see McAinsh et al. 2003). In contrast to these point centromeres of Saccharomyces cerevisiae, the regional centromeres in metazoans are much larger and the DNA sequence elements found in these regions appear to be neither required nor sufficient for nucleation of functional kinetochores. Thus, it is believed that an epigenetic mechanism ensures the faithful propagation of a centromeric locus in these species. One epigenetic mark may be represented by the histone H3 variant CENP-A, which is found exclusively in active centromeres throughout the cell cycle (Cleveland et al. 2003; Sullivan et al. 2001). Several studies suggest that CENP-A stands at the base of the kinetochore assembly pathway, as almost all other kinetochore components are mislocalized in the absence of CENP-A (Blower and Karpen 2001; Howman et al. 2000; Moore and Roth 2001; Oegema et al. 2001). However, CENP-A is not sufficient for assembly of fully functional kinetochores, since mistargeted CENP-A recruits some, but not all, essential kinetochore components (Van Hooser et al. 2001). In vertebrates, CENP-A forms together with the proteins CENP-C, CENP-H, and CENP-I a prekinetochore that constitutively localizes to centromeres throughout the cell cycle (for review see Fukagawa, 2004). While homologs for all these constitutive components could be identified in yeast (Meluh and Koshland 1995; Nishihashi et al. 2002; Westermann et al. 2003), in Drosophila the CENP-A homolog CID (Henikoff et al. 2000) is presently the only described kinetochore component localizing to the centromere throughout the cell cycle.
For proper segregation, the chromosomes also need to be converted into a compacted transport form during mitosis. This compaction is in part brought about by action of the conserved 13S condensin complex, which is composed of five subunits (for review see Swedlow and Hirano 2003). Two subunits, SMC2 and SMC4, belong to the family of structural maintenance of chromosome proteins. SMC2 and SMC4 associate with three non-SMC subunits, originally identified in Xenopus egg extracts as XCAP-D2, XCAP-H, and XCAP-G. Homologs of the condensin subunits could be identified in a wide range of eukaryotes (for review see Hagstrom and Meyer 2003). Intriguingly, condensin subunits are not only involved in general chromosome condensation, but have also been implicated in X-chromosome dosage compensation in Caenorhabditis elegans, in epigenetic control of homeotic gene expression in Drosophila, and in DNA repair in Saccharomyces pombe (Aono et al. 2002; Chuang et al. 1994; Lupo et al. 2001). Furthermore, in C. elegans, the condensin subunits SMC-4, SMC-2/MIX-1, and CAP-D2/HCP-6 were shown to be involved in centromere organization (Hagstrom et al. 2002; Stear and Roth 2002). C. elegans features holocentric chromosomes, in which the poleward facing entire lengths of the condensed metaphase chromosomes are attached with kinetochore microtubules of the spindle apparatus. Thus, disturbing overall chromosome structure in C. elegans might be expected to affect centromere structure and function.
Here, we present results of a genetic screen aimed at identifying interactors of the Drosophila kinetochore component CID. We found that the condensin subunit Cap-G interacts genetically and physically with CID, expanding the evidence of a link between condensin and kinetochore structure to organisms with monocentric chromosomes with regional centromeres, similar to the situation found in vertebrates. Moreover, the cytological phenotype of Cap-G mutations reveals apparent defects in chromatid segregation rather than chromosome condensation.
Materials and methods
Fly strains
For expression of UAS transgenes, we used ey-GAL4 (Hazelett et al. 1998), GMR-GAL4 (Freeman 1996), da-GAL4 (Wodarz et al. 1995), F4-GAL4 (Weiss et al. 1998), and prd-GAL4 (Brand and Perrimon 1993). The strains carrying the deficiencies collectively removing most of the Drosophila melanogaster euchromatin (“deficiency kit”) including the deficiencies Df(2R)CX1 and Df(2R)vgD as well as strains carrying the alleles gluk08819, barrL305, l(2)49Ff1 [also called vr911], and l(2)49Ff6 [also called vr943] were obtained from the Drosophila stock center (Bloomington, Indiana). The alleles gluk08819 and barrL305 have been described as severe hypomorphs (Bhat et al. 1996; Steffensen et al. 2001). The deficiency Df(2R)vg56 and the allele l(2)49Ff3 [also called vr923] were a gift from C.-ting Wu. The EP(2)2346 line was obtained from Exelixis. Stocks carrying cid mutant chromosomes (cid12-1 and cid22-4) were generously provided by Thom Kaufman (Indiana University). UAS-cid, UAS-cid-myc, and UAS-cid(hx2H3)-myc lines were obtained after P-element-mediated germline transformation using constructs in pUAST (Brand and Perrimon 1993) or in a modified pUASP vector (Rørth 1998) following standard procedures. This modified pUASP vector (pUASP1) harbors a more extended polylinker region than the original pUASP. We consistently found that overexpression phenotypes caused by transgenes cloned in pUASP1 were weaker when compared with pUAST. The detailed cloning procedures are available upon request. Briefly, the cid open reading frame was amplified by the polymerase chain reaction (PCR) with genomic DNA as template and then cloned into pUAST or pUASP1. UAS-cid-myc constructs were generated by cloning in-frame restriction fragments encoding ten copies of the myc epitope tag immediately downstream of the last codon of cid. For construction of the cid version containing helix 2 of histone H3, three overlapping fragments were generated by PCR. The first fragment encodes the N-terminal region of CID (aa 1–172) up to helix 2 in the histone fold domain (HFD), the second fragment encodes the corresponding helix 2 of H3, and the third fragment encodes the part of CID C-terminal to helix 2 (aa 201–225). The three PCR products were combined and a final PCR with the terminal flanking primers was performed. The resulting recombinant molecule was ligated into pUAST and a pUASP1 vector containing the fragment encoding the ten myc epitope tags.
A Cap-G genomic rescue fragment was assembled in pSLfa1180fa (Horn and Wimmer 2000) by combining a 7.5 kb NheI fragment encompassing the region 900 bp upstream of the translational start codon and the adjacent 5′-terminal 6.6 kb of the Cap-G gene (based on the Cap-G-RB annotation) with a 3.7 kb NheI/ MluI fragment of the 3′-terminal part of Cap-G including 1.2 kb downstream of the translational stop codon. These restriction fragments were isolated from a 16 kb BamHI subclone of the BAC 48I13 (Berkeley Drosophila Genome Project; BDGP). The assembled genomic fragment containing the complete Cap-G gene according to the Cap-G-RB annotation was excised as an 11.2 kb AscI/FseI fragment and cloned into the transformation vector pBac 3xP3-EGFP (Horn and Wimmer 2000). For the construction of UAS-Cap-G, the Cap-G coding region was isolated from the expressed sequence tag (EST) clone SD10043 (BDGP) and cloned into pUASP1. For the rescue experiment with UAS-Cap-G, males of the genotype l(2)49Ff6/CyO, P[w+, ftz-lacZ]; UAS-Cap-G III.1 were crossed with females of the genotype l(2)49Ff1/CyO, P[w+, ftz-lacZ]; da-GAL4. For the rescue experiment with the genomic Cap-G construct, males of the genotype l(2)49Ff3 /CyO, P[w+, ftz-lacZ]; gCap-G III.2/+ were crossed with females of the genotype l(2)49Ff6 /CyO, P[w+, ftz-lacZ]. Progeny were allowed to develop at 22°C. Rescued transheterozygous individuals could be readily identified by the recessive markers al, b, c and sp present on the l(2)49Ff chromosomes (Lasko and Pardue 1988).
Antibodies and immunolabeling
The antibodies against Drosophila cyclin B (Knoblich and Lehner 1993) and the human c-myc epitope (Evan et al. 1985) have been described previously. An antibody against CID was raised in rabbits using bacterially expressed, affinity-purified full-length protein as immunogen. The antiserum was affinity purified using standard procedures. For immunolabeling, the anti-cyclin B, anti-CID, and anti-myc antibodies were used at 1:3, 1:500, and 1:10 dilutions, respectively. Secondary antibodies were obtained from Jackson Laboratories. DNA was stained with Hoechst 33258.
Yeast two-hybrid experiments
Protein–protein interactions were analyzed using the yeast two-hybrid system as described previously (Jäger et al. 2001). The complete cid coding region as well as cid fragments encoding the N-terminus or the HFD were cloned as fusions with the Gal4 DNA-binding domain in-frame into the vector pGBKT7 (Clontech). DNA fragments encoding parts of the Cap-G protein were PCR amplified using the EST SD10043 as template and cloned as fusions with the Gal4 activation domain into pGADT7 (Clontech). In Fig. 4, the Cap-G fragment encoding amino acids 1-413 was analyzed. We scored activation of the HIS3 and ADE2 reporter genes. For control experiments, we used the plasmid combinations pGBKT7-p53/pGADT7-T-Ag (strong growth on all selective media), pGBKT7-SSE 1-247 (SSE-NT)/pGADT7-PIM (strong activation of ADE2; weak activation of HIS3), and pGBKT7-SSE 1-247/pGADT7-THR 1-933 (THR-NT) (activation of HIS3; weak activation of ADE2) (Jäger et al. 2001).
Assessment of Cap-G annotations
In release 3.2.0 of the Drosophila genome annotation database, five different annotations are proposed for Cap-G. In four of these annotations (“long” annotations), a group of closely spaced 5′-terminal exons is separated from the 3′-terminal exons by a >22 kb intron (Fig. 2i). In the fifth annotation (Cap-G-RB; “short” annotation), the same group of 5′-terminal exons is separated by a 4819 bp intron from a group of alternative 3′-terminal exons (Fig. 2i). Besides our experimental data, the following observation questions the significance of the long annotations. When compared with the DNA sequence of Drosophila pseudoobscura, a clear ortholog for the complete conceptual translation of the Cap-G-RB annotation can be identified encoded on a single contig (Contig 2230_Contig3351). In contrast, for the 42 amino acid sequence unique to the proposed C-terminus of the Cap-G-RD/Cap-G-RC annotations, no significant homologous sequence can be identified by TBLASTN searches in the entire D. pseudoobscura genome sequence. When a TBLASTN search with the proposed unique 40 C-terminal amino acids of the Cap-G-RA/Cap-G-RE annotations is performed, a clear match can be found (28 identical amino acids in a stretch of 29). This match, however, is not within the roughly 400 kb following the common Cap-G upstream exons, but rather on a different contig (contig3295_contig8282), making it unlikely that this region is co-transcribed with the upstream exons.
Our sequence analysis of SD10043, and also the published SD10043 sequence (Accession no. BT009931) revealed that, besides four non-synonymous polymorphisms, the second to last exon is 15 nt (encoding the sequence SNDSE) at its 3′ end longer than in the mRNA sequence predicted by the Cap-G-RB annotation. Taken together, we propose that Cap-G is most likely and completely represented by an annotation corresponding to SD10043 encoding a protein of 1351 amino acids.
Reverse transcribed polymerase chain reaction (RT-PCR)
Preparation of poly(A)+ RNA from different developmental stages of the Drosophila strain w1 and subsequent cDNA synthesis have been described previously (Jacobs et al. 2002). Primers were designed specifically to amplify the region between the fourth Cap-G exon common to all annotations and either the sixth exon of Cap-G-RB or the small exon common to all long annotations (Fig. 2i). The sequences of these primers as well as of the primers for amplification of cDNAs specific for the fzr2 and the RpL32 genes are available upon request.
Sequence analysis of the l(2)49Ff alleles
All three l(2)49Ff mutant chromosomes were balanced over a CyO, P[w+, Act5c-GFP] chromosome and homozygous mutant embryos from 14 to 17 h collections were identified by the lack of green fluorescent protein (GFP) fluorescence. Genomic DNA was prepared from these GFP-negative embryos and used as template in PCRs with primers designed to amplify the coding regions according to the Cap-G-RB annotation. The PCR products were sequenced directly.
Results
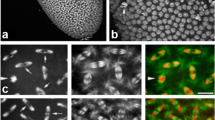
To identify factors that interact with the D. melanogaster centromere-specific histone H3 variant CID, we employed a genetic strategy. We used the GAL4/UAS system (Brand and Perrimon 1993) for ectopic expression of cid. Ubiquitous, low expression of UAS-cid using daughterless-GAL4 (da-GAL4) as driver complements the lethality associated with cid mutant individuals, thus demonstrating the functionality of the transgene (data not shown). However, overexpression of cid using eyeless-GAL4 (ey-GAL4), which directs expression of UAS transgenes early in eye development (Hazelett et al. 1998), results in a severe rough eye phenotype (Fig. 1b). Strong overexpression of cid in endoreduplicating cells of larval salivary glands using the driver F4-GAL4 (Weiss et al. 1998) had no obvious deleterious effect (data not shown). To investigate whether centromeric localization of overexpressed CID is required for generation of the rough eye phenotype, we constructed a CID variant in which helix II of its HFD was exchanged with the corresponding helix II of the HFD from histone H3 (CID[hx2H3]; Fig. 1f). An equivalent exchange in human CENP-A has been shown to result in the loss of centromeric targeting (Shelby et al. 1997). To assess, whether this exchange also results in mislocalization in Drosophila, the intracellular localization of this modified protein was analyzed (Fig. 1n–t). For detection of the transgene products, ten copies of the human c-myc epitope tag were fused to the C-termini of both CID and CID[hx2H3]. Expression of the UAS transgene in the embryonic epidermis was directed in stripes by use of the driver paired-GAL4 (Brand and Perrimon 1993). In these experiments, we used UASP transgenes, which result in lower levels of expression than the UAST transgenes, since high product levels were observed to obscure the detection of centromeric signals (data not shown). The lower expression level of myc-tagged cid from UASP transgenes still results in a rough eye phenotype (Fig. 1c). While CID-myc localizes to centromeres, as is evident by the co-localization of the anti-myc staining and anti-CID signals (Fig. 1l, m, arrowheads), the helix II exchange in CID abolished centromeric targeting (Fig. 1s, t). Importantly, overexpression of the cid[hx2H3]-myc transgene in the eye imaginal disc driven by ey-GAL4 also did not result in a rough eye phenotype (Fig. 1d). The same result was observed when a transgene without the 10 x myc tag was expressed using the stronger expressing vector pUAST (Fig. 1e). Quantitative immunoblotting experiments revealed equal expression of the UASP-cid-myc and UASP-cid[hx2H3]-myc transgenes, ruling out the possibility that differences in expression levels are accountable for differences in the eye phenotype (see Supplementary Material). Taken together, these results suggest that centromeric targeting is required for the generation of the rough eye phenotype.
The cid overexpression phenotype in the adult eye depends on the centromeric localization of the ectopically expressed protein. a–e Eyes of individuals with the genotypes a ey-GAL4/+ (ey), b ey-GAL4/+; UAST-cid III.9/+ (ey>T-cid), c ey-GAL4/+; UASP1-cid-myc III.5/+ (ey>P1-cid-myc), d ey-GAL4/+; UASP1-cid[hx2H3]-myc III.3/+ (ey>P1-cid[hx2H3]-myc), and e ey-GAL4/UAST-cid[hx2H3] II.2 (ey>T-cid[hx2H3]). f Schematic representation of the CID[hx2H3] construct. CID-derived sequences are illustrated in green and H3-derived sequences in orange. In the upper panel, the structural elements of the histone fold domain are indicated. g–i, n–p Embryos derived from crosses of prd-GAL4 females with UASP1-cid-myc males (g–i) or UASP1-cid[hx2H3]-myc males (n–p) were fixed at stage 14 and labeled with antibodies against the myc-epitope (i, p), against CID (h, o) and with a DNA stain (g, n). prd-GAL4 drives expression of UAS transgenes in stripes in the embryo. j–l, q–s Enlargements of the boxed areas in g–i, n–p, respectively, with the focus on the CNS. m, t Superimposed images of j–l and q–s, respectively. Blue, green, and red represent the labeling of DNA, CID and myc, respectively. Note the intense dotlike signals in l that superimpose with CID signals (m; arrowheads). Such dots are absent when cid[hx2H3]-myc is expressed (s, t)
As observed for cid, the phenotypes caused by ey-GAL4 driven overexpression are often quite variable, even when the eyes of the same individual are compared (data not shown; Tseng and Hariharan 2002). Overexpression of cid using GMR-GAL4 (Freeman 1996), which results in a later onset of UAS transgene expression than ey-GAL4, caused a milder, and more consistent rough eye phenotype, primarily in the posterior part of the eye (Fig. 2a). A genetic modifier screen was performed by crossing various deficiencies into a background resulting in GMR-GAL4 driven cid overexpression. Each of the deficiencies, which collectively remove a high proportion of the euchromatic genome of D. melanogaster, was scored for enhancement or suppression of the cid overexpression phenotype when present in a heterozygous state. Deficiencies that also modified the phenotype observed after the overexpression of the Drosophila genes pimples, or Cyclin D and cdk4, or the head gap gene orthodenticle (O. Leismann, J. Reischl, C. Meyer and C. Lehner, personal communication) were not investigated further. In that way deficiencies that most probably influence the phenotype in some non-specific way, were excluded from further analysis.
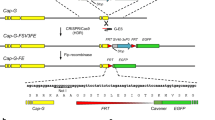
Identification of Cap-G alleles as enhancer of the cid overexpression phenotype. a–h Eyes of an individual ectopically expressing UAS-cid II.3A under control of the GMR-GAL4 driver (GMR>cid; a) and of individuals harboring in addition the deficiencies Df(2R)CX1 (b), Df(2R)vg56 (c), Df(2R)vgD (d), or the lethal alleles l(2)49Ff1 (e, f), l(2)49Ff3 (g), and l(2)49Ff6 (h). In f, one copy of the genomic Cap-G transgene gCap-G III.1 was also present. i Schematic representation of the 49C–50C region on the right arm of the second chromosome. Enhancing deficiencies are represented in green and the non-enhancing deficiency in red. The extent and location of the various deficiencies and of the Cap-G gene are indicated. The five different Cap-G annotations are illustrated in the middle part of the panel. Translated regions are represented by solid boxes and the 5′- and 3′- untranslated regions by open boxes. The insertion position of the P-element EP(2)2346 is indicated by a green arrow. Black arrows indicate the positions of the mutations identified in the l(2)49Ff alleles. The extent of the genomic Cap-G rescue fragment is denoted by a gray bar
Df(2R)CX1 was identified as a strongly enhancing deficiency (Fig. 2b). Df(2R)CX1 deletes the cytological region 49C–50C on the right arm of the second chromosome. Analysis of further deficiencies within this region allowed us to limit the location of the enhancer locus to approximately 170,000 bp (Fig. 2c, d, i). We next tested all available P-element insertions as well as alleles of lethal complementation groups within this region for enhancement of the cid overexpression phenotype. Three alleles of the l(2)49Ff complementation group did indeed enhance the cid overexpression phenotype (Fig. 2e, g, h).
In order to identify the gene mutated in the l(2)49Ff alleles, we crossed heterozygous l(2)49Ff mutant flies against strains carrying lethal P-element insertions in the cytological region 49F. We found that all three l(2)49Ff alleles were lethal over EP(2)2346, which is inserted 109 nucleotides upstream of the translational initiation codon of the gene Cap-G (CG17054) (Fig. 2i). Cap-G is orthologous to XCAP-G, a subunit of the condensin complex, which has been shown to possess chromosome condensation activity in vitro (Hirano et al. 1997). To analyze whether the l(2)49Ff alleles indeed represent alleles of Cap-G, we set out to perform rescue experiments. First, we scrutinized the Cap-G transcript annotations of the Drosophila genome project (Fig. 2i). The presence of Cap-G-RB transcripts was confirmed by RT-PCR experiments, while all other annotated transcripts (Cap-G-RA, Cap-G-RC, Cap-G-RD and Cap-G-RE, see Fig. 2i) could not be detected (data not shown). The Cap-G-RB transcripts were detected throughout development with maximal levels in early embryos and males (Fig. 3a). The higher expression of Cap-G-RB in males compared with females was confirmed using independent cDNA preparations (Fig. 3b). The identity of the male-specific cDNA preparation was corroborated by amplifying cDNA corresponding to fizzy-related 2 (fzr2), a gene known to be expressed exclusively in males (Fig. 3b; Jacobs et al. 2002).
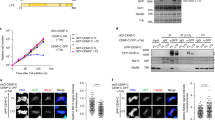
Expression of Cap-G-RB predominantly during embryogenesis and in adult males. a Reverse transcribed polymerase chain reaction (RT-PCR) experiment to monitor expression during embryonic stages, 0–2, 2–4, 4–8, and 8–16 h after egg deposition, during first (L1), second (L2), and third (L3) larval instar stages, during the pupal (P) stage and in females and males. The primers used were specific for a region spanning the large intron between exons four and five of Cap-G-RB, or for the RpL32 gene. For the control (−) no mRNA was added to the reverse transcription reaction. b In an independent preparation of cDNA from males and females, the identity of the male cDNA preparation was confirmed by amplification of the male-specific fzr2-cDNA
Based on the above results, we constructed transgenic lines harboring an 11.2 kb genomic fragment encompassing the region represented by Cap-G-RB (Fig. 2i). In a second approach, we expressed a Cap-G cDNA (SD10043), corresponding to the Cap-G-RB annotation, under UAS control using the da-GAL4 driver. In both cases, the transgene expression complemented the lethality associated with the l(2)49Ff mutations, albeit with lower than expected frequency (genomic transgene: 20.9% of expected; 1506 progeny scored; UAS-cDNA-transgene: 21.9% of expected, 2483 progeny scored). The genomic transgene (but not the UAS-cDNA transgene in combination with da-GAL4) resulted in a vast excess of rescued females over males (approximately 10:1). Therefore, the genomic transgene might lack regulatory elements required for efficient rescue of male flies. Nevertheless, we conclude that expression of Cap-G versions corresponding to the Cap-G-RB annotation is sufficient for complementing the lethality associated with Cap-G mutations.
To characterize the molecular nature of the l(2)49Ff alleles, we amplified the Cap-G coding region from l(2)49Ff homozygous embryos and sequenced the PCR products directly. Both l(2)49Ff1 and l(2)49Ff6 contain a nonsense mutation in Cap-G-RB introducing a premature stop codon at amino acid positions 343 and 77, respectively. The allele l(2)49Ff3 was found to contain a missense mutation changing the arginine at position 558 to a tryptophan (R558W). This mutation does not represent a polymorphism, because the respective nucleotide exchange is absent in the l(2)49Ff1 and l(2)49Ff6 alleles, which were created in the same mutagenesis experiment as l(2)49Ff3 (Lasko and Pardue 1988). This finding further demonstrates that the l(2)49Ff alleles represent Cap-G alleles.
If mutations in Cap-G enhance the cid overexpression phenotype, then one would expect that supplying a Cap-G+ transgene in this genetic background should suppress this enhancement. Indeed, one copy of the genomic Cap-G transgene reverts the enhancement of the phenotype caused by the presence of the l(2)49Ff1 or the l(2)49Ff6 alleles (Fig. 2f and data not shown).
Even though the condensin complex is thought to act along the entire chromosome, a specific function in the centromeric region cannot be ruled out. Alternatively, Cap-G may perform a centromere-specific function independently of its role in the condensin complex. To investigate whether CID and Cap-G interact directly, we performed yeast two-hybrid experiments. We observed an interaction between CID and a fragment of Cap-G comprising the N-terminal 413 amino acids (Cap-G-NT; Fig. 4). This interaction appeared to be stronger than interactions within the Drosophila separase complex (Fig. 4; PIM/SSE-NT and THR-NT/SSE-NT) (Jäger et al. 2001), as strong growth on both selective media was observed. The activation of the reporter genes in the two-hybrid assay is specific, as Cap-G-NT together with p53 or histone H3 does not trigger reporter gene activation. Furthermore, no activation of the reporter genes is observed, when CID is co-expressed in yeast with SV40 T-antigen (Fig. 4) or any of the other Cap-G fragments (data not shown). Interestingly, full length CID is required for the interaction with the Cap-G N-terminus, as both the isolated CID N-terminus (amino acids 1–125) as well as the isolated CID HFD (amino acids 126–225) fail to interact with the N-terminal Cap-G fragment (Fig. 4).
Interaction of CID and the N-terminus of Cap-G in the yeast two-hybrid system. a Pattern of the transformants plated onto the different selection media. The upper proteins/protein fragments (normal print) were fused to the Gal4 transcription activation domain and the lower proteins/protein fragments (italics) were fused to the Gal4 DNA-binding domain. Transformants were plated onto synthetic medium lacking leucine and tryptophan (b), selecting for the presence of both plasmids, onto medium lacking leucine, tryptophan, and adenine (c) or lacking leucine, tryptophan, and histidine, supplemented with 0.5 mM 3-aminotriazole (d) for assessing activation of the reporter genes
These results indicate that Cap-G, alone or in the context of the condensin complex, interacts with CID and may influence kinetochore structure and/or function.
The physical interaction between Cap-G and CID and the genetic interaction of the Cap-G alleles with cid may suggest kinetochore dysfunction and thus mitotic defects in Cap-G mutant individuals. The l(2)49Ff alleles have been described as embryonic lethals (Lasko and Pardue 1988). Therefore, we analyzed Cap-G mutant embryos by indirect immunofluorescence. We found that Cap-G mutant embryos fixed at a stage when epidermal cells are progressing through mitosis 15 displayed massive anaphase defects (Fig. 5j, l, p, r). Anaphase figures contained bridges and often a large part of the chromatin masses appeared to reside within the cleavage plane (arrowheads in Fig. 5j, l). No significant difference was observed between the phenotypes caused by the three Cap-G alleles in either homozygous or transheterozygous states (data not shown). Thus, chromosome segregation appears to be severely impaired in Cap-G mutant embryos. Sister chromatid separation in Drosophila requires the APC/C-dependent activation of the endoprotease separase (Jäger et al. 2001; Leismann et al. 2000). To investigate whether Cap-G mutant embryos can activate APC/C during mitosis 15, we analyzed the levels of Cyclin B, which is an APC/C substrate. Cyclin B levels dropped dramatically in cells progressing through the metaphase-to-anaphase transition in Cap-G mutant embryos, despite the failure to separate the sister chromatids efficiently (Fig. 5q, r). We conclude that APC/C is activated in Cap-G mutant cells in a timely manner and thus the defect in sister chromatid segregation is not due to a lack of APC/C mediated proteolysis. To analyze whether kinetochore function is affected, we labeled Cap-G mutant embryos with an antibody against CID. The CID signals were almost exclusively found at the poleward faces of the chromatin masses in anaphase figures, suggesting that kinetochores are functional and that dissolution of centromeric cohesion still occurs in mutant embryos (Fig. 5l).
Cytological phenotype of Cap-G mutants. Embryos derived from l(2)49Ff6/CyO, ftz-lacZ flies (a–c, g–i) or from a cross of l(2)49Ff6 /CyO, ftz-lacZ females with l(2)49Ff3/CyO, ftz-lacZ males (m–o) were fixed while progressing through epidermal mitosis 15 and stained with antibodies against CID (b, h), β-galactosidase (β-Gal; c, i, o), Cyclin B (CYCB; n), or a DNA stain (a, g, m). d, e, j, k, p, q Enlargements of the epidermis of embryos shown in a, b, g, h, m, n, respectively. f, l, r Merged images of d, e, j, k, and p, q, respectively. DNA is shown in red and CID (f, l) and CYCB (r) in green. Cap-G mutant embryos (g–i, m–o) were identified by the absence of the balancer-derived lacZ expression (i, o) while balanced siblings expressed lacZ under control of the ftz promoter (c). Open arrowheads in d, f indicate normal anaphase figures with well separated chromatin masses. Solid arrowheads in j, l denote anaphase bridges in Cap-G mutant embryos. Solid arrowheads in p–r denote prophase (p), metaphase (m), and anaphase (a) figures. Note that cyclin B in the Cap-G mutant is still high in the prophase cell, while it has largely disappeared in the metaphase cell and is absent in the anaphase cell
As Cap-G is a subunit of the condensin complex, defects primarily in mitotic chromosome condensation might have been expected in Cap-G mutant embryos. However, in Cap-G mutant embryos chromosome condensation does not appear to be substantially affected, as judged by the apparent normal morphology of metaphase plates (e.g., see Fig. 5r). This observation is consistent with the phenotype described for mutations in the genes for two other Drosophila condensin components, CAP-H/Barren and SMC4/Gluon (Bhat et al. 1996; Steffensen et al. 2001). Thus, the primary defect after loss of condensin function appears to be a difficulty in segregating the replicated chromatids rather than converting the chromatin into its compacted state during mitosis.
Discussion
Here, we provide evidence that the condensin subunit Cap-G interacts both genetically and physically with the constitutive kinetochore component CID of D. melanogaster. Our results demonstrate that the l(2)49Ff alleles identified as enhancers in our genetic experiments are indeed Cap-G alleles. The RT-PCR and rescue experiments furthermore suggest that the genomic organization of Drosophila Cap-G is best described by the short Cap-G-RB annotation.
Chromosome condensation involves a coupling of chromatin compaction and resolution of topological entanglements by topoisomerase II, thus resulting in individualization of chromosomes in mitosis (Hirano 2000). The term condensin was coined to emphasize the biochemical role of the five-subunit 13S protein complex in chromosome condensation in vitro (Hirano et al. 1997). However, studies of condensin function in Drosophila, C. elegans, and chicken indicate that the absence of condensin has only a limited effect on the longitudinal compaction of mitotic chromosomes in vivo. Rather, lack of condensin often results in difficulties in separating the chromosome arms during anaphase, probably due to residual topological links between sister chromatids that have not been resolved properly by topoisomerase II (Bhat et al. 1996; Coelho et al. 2003; Hagstrom et al. 2002; Hudson et al. 2003; Steffensen et al. 2001). The phenotype of Cap-G mutant embryos is very similar, thus expanding the published observations of segregation defects in mutants of condensin subunit genes.
While the Cap-G mutant phenotype is similar to the phenotypes observed due to the lack of SMC4/gluon and CAP-H/barren function (Bhat et al. 1996; Steffensen et al. 2001), mutant alleles of the latter two condensin subunit genes did not enhance the cid overexpression phenotype (H. Jäger and S. Heidmann, unpublished observation). Thus, Cap-G may be rate limiting in the condensin complex assembly and reduction of the gene dosage by half of the other condensin subunits may not reduce the levels of functional condensin complex significantly. Alternatively, Cap-G may perform a role independently of the other condensin subunits. Evidence is indeed accumulating that condensin subunits can assemble in different complexes performing functions different from chromatin condensation (for review see Hagstrom and Meyer 2003).
It has been recently found that besides the originally described condensin complex, a second condensin complex (condensin II) exists. In condensin II, the SMC2/SMC4 dimer associates with a different, but related, set of non-SMC subunits (Ono et al. 2003; Yeong et al. 2003). In this respect, it is interesting to note that the Drosophila genome harbors homologs for the non-SMC condensin II subunits hCAP-D3 and hCAP-H2, but not for hCAP-G2 (Ono et al. 2003). This suggests that Drosophila Cap-G may represent a common non-SMC component of both condensin complexes. Thus, Cap-G mutants may reflect the phenotype due to loss of both condensin I and condensin II function in Drosophila.
Our results indicate a link between the inner kinetochore structure and the chromosome condensation machinery. Evidence from C. elegans supports the notion that condensin might influence centromere organization. The C. elegans condensin components CAP-D2/HCP-6, SMC4, and SMC2/MIX-1 co-localize with CENP-A/HCP-3, the centromere-specific histone H3 variant of the worm (Hagstrom et al. 2002; Stear and Roth 2002). Moreover, depletion of SMC4 or SMC2/MIX-1 by RNAi prevents the restricted orientation of centromere proteins toward the spindle poles (Hagstrom et al. 2002). An important point to keep in mind is that C. elegans chromosomes are holocentric with their centromeres stretched along the entire poleward oriented faces of the metaphase chromosomes. Thus, any major disturbance of the rigid metaphase chromosome structure is expected to result in a distorted orientation of these centromeres. However, there is also evidence suggesting a link between condensin and centromere structure of monocentric chromosomes. Chromatin-immunoprecipitation experiments in S. pombe showed a preferential association of the CAP-H/Cnd2 and SMC4/Cut3 proteins with central centromeric sequences (Aono et al. 2002). Furthermore, condensin depletion in Xenopus extracts causes disorganized localization of the outer kinetochore protein CENP-E (Wignall et al. 2003). Finally, immunofluorescence analysis of the localization of the Drosophila condensin components SMC4/Gluon and CAP-H/Barren revealed an enrichment of these components on centromeres (Coelho et al. 2003; Steffensen et al. 2001). In our experiments, a major disturbance of kinetochore structure is not evident in Cap-G mutant embryos. CID is localized at the poleward faces of the chromatin in anaphase figures and in only a few cases are CID signals found in the cleavage plane. How can this obvious functional integrity of the centromeres in Cap-G mutant embryos be reconciled with the described interaction of CID and Cap-G? It is possible that Cap-G plays multiple roles. Its function in establishing a proper structure of mitotic chromosomes, probably in concert with the other condensin subunits and topoisomerase II, may be the first function that is affected when the maternal contribution runs low in Cap-G mutant embryos. Thus, the Cap-G levels in early Cap-G mutant embryos may still suffice to exert a hypothetical centromere-specific role and corresponding defects are not detectable at this stage.
In tissue culture cells it has been shown that overexpressed CID localizes to both centromeres and the euchromatin (Ahmad and Henikoff 2002). Even though overexpressed CID in the eye imaginal disc also localizes throughout the euchromatin (data not shown), we could clearly show a correlation of the ability of the overexpressed protein to localize to the centromere with the generation of the rough eye phenotype. However, we cannot rule out the possibility that the euchromatic localization of CID also contributes to the rough eye phenotype and that the enhancement seen in Cap-G mutants is due to an interaction of Cap-G and this euchromatic localized CID.
Our two-hybrid results show a direct interaction of the N-terminus of Cap-G with CID. The N-terminal regions of the frog and yeast Cap-G homologs were shown to contain HEAT (Huntingtin, Elongation factor 3, A subunit of protein phosphatase 2A, TOR lipid kinase) repeats (Neuwald and Hirano 2000). HEAT repeats can also be found with very high levels of significance in the N-terminal fragment (aa 1-413) of Drosophila Cap-G using fold recognition methods that have been used previously to obtain tertiary structure predictions for Drosophila THR and separases (S Heidmann, unpublished observation; Jäger et al. 2004). HEAT repeats have been shown to form a bent superhelical structure involved in protein–protein interactions (Andrade et al. 2001), consistent with our finding that the Cap-G N-terminus interacts with CID. This Cap-G/CID interaction is to our knowledge the first report of a direct interaction of a Cap-G homolog with another protein that is not part of the condensin complex. Interestingly, it has been shown that the human condensin subunit CAP-D2/CNAP1 can interact with histone H3 in vitro and in vivo (Ball et al. 2002). Thus, it is tempting to speculate that the condensin complex may be targeted to bulk chromatin by CAP-D2–H3 interactions and to centromeric chromatin by CAP-G–CID/CENP-A interactions. Future studies will reveal whether an interaction between CAP-G and CENP-A homologs is a general feature of eukaryotic chromosome biology, thus contributing to the unfolding complexity of condensin functions.
References
Ahmad K, Henikoff S (2002) Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A 99 (Suppl 4):16477–16484
Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struct Biol 134:117–131
Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M (2002) Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197–202
Ball AR Jr, Schmiesing JA, Zhou C, Gregson HC, Okada Y, Doi T, Yokomori K (2002) Identification of a chromosome-targeting domain in the human condensin subunit CNAP1/hCAP-D2/Eg7. Mol Cell Biol 22:5769–5781
Bhat MA, Philp AV, Glover DM, Bellen HJ (1996) Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell 87:1103–1114
Blower MD, Karpen GH (2001) The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol 3:730–739
Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415
Chuang PT, Albertson DG, Meyer BJ (1994) DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79:459–474
Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407–421
Coelho PA, Queiroz-Machado J, Sunkel CE (2003) Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J Cell Sci 116:4763–4776
Draviam VM, Xie S, Sorger PK (2004) Chromosome segregation and genomic stability. Curr Opin Genet Dev 14:120–125
Evan GI, Lewis GK, Ramsay G, Bishop JM (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol 5:3610–3616
Freeman M (1996) Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87:651–660
Fukagawa T (2004) Assembly of kinetochores in vertebrate cells. Exp Cell Res 296:21–27
Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ (2002) C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16:729–742
Hagstrom KA, Meyer BJ (2003) Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet 4:520–534
Hazelett DJ, Bourouis M, Walldorf U, Treisman JE (1998) decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125:3741–3751
Henikoff S, Ahmad K, Platero JS, van Steensel B (2000) Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci U S A 97:716–721
Hirano T (2000) Chromosome cohesion, condensation, and separation. Annu Rev Biochem 69:115–144
Hirano T, Kobayashi R, Hirano M (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511–521
Horn C, Wimmer EA (2000) A versatile vector set for animal transgenesis. Dev Genes Evol 210:630–637
Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A 97:1148–1153
Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC (2003) Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell 5:323–336
Jacobs H, Richter D, Venkatesh T, Lehner C (2002) Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Curr Biol 12:1435–1441
Jäger H, Herzig A, Lehner CF, Heidmann S (2001) Drosophila Separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev 15:2572–2584
Jäger H, Herzig B, Herzig A, Sticht H, Lehner CF, Heidmann S (2004) Structure predictions and interaction studies indicate homology of separase N-terminal regulatory domains and Drosophila THR. Cell Cycle 3:182–188
Knoblich JA, Lehner CF (1993) Synergistic action of Drosophila cyclin A and cyclin B during the G2-M transition. EMBO J 12:65–74
Lasko PF, Pardue ML (1988) Studies of the genetic organization of the vestigial microregion of Drosophila melanogaster. Genetics 120:495–502
Leismann O, Herzig A, Heidmann S, Lehner CF (2000) Degradation of Drosophila PIM regulates sister chromatid separation during mitosis. Genes Dev 14:2192–2205
Lupo R, Breiling A, Bianchi ME, Orlando V (2001) Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol Cell 7:127–136
McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19:519–539
Meluh PB, Koshland D (1995) Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell 6:793–807
Moore LL, Roth MB (2001) HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol 153:1199–1208
Neuwald AF, Hirano T (2000) HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res 10:1445–1452
Nishihashi A, Haraguchi T, Hiraoka Y, Ikemura T, Regnier V, Dodson H, Earnshaw WC, Fukagawa T (2002) CENP-I is essential for centromere function in vertebrate cells. Dev Cell 2:463–476
Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 153:1209–1226
Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T (2003) Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109–121
Rørth P (1998) Gal4 in the Drosophila female germline. Mech Dev 78:113–118
Shelby RD, Vafa O, Sullivan KF (1997) Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol 136:501–513
Stear JH, Roth MB (2002) Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev 16:1498–1508
Steffensen S, Coelho PA, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko SN, Bellen H, Heck MM, Sunkel CE (2001) A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr Biol 11:295–307
Sullivan BA, Blower MD, Karpen GH (2001) Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet 2:584–596
Swedlow JR, Hirano T (2003) The making of the mitotic chromosome: modern insights into classical questions. Mol Cell 11:557–569
Tseng AS, Hariharan IK (2002) An overexpression screen in Drosophila for genes that restrict growth or cell-cycle progression in the developing eye. Genetics 162:229–243
Van Hooser AA, Ouspenski, II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114:3529–3542
Weiss A, Herzig A, Jacobs H, Lehner CF (1998) Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr Biol 8:239–242
Westermann S, Cheeseman IM, Anderson S, Yates JR III, Drubin DG, Barnes G (2003) Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol 163:215–222
Wignall SM, Deehan R, Maresca TJ, Heald R (2003) The condensin complex is required for proper spindle assembly and chromosome segregation in Xenopus egg extracts. J Cell Biol 161:1041–1051
Wodarz A, Hinz U, Engelbert M, Knust E (1995) Expression of crumbs confers apical character on plasma-membrane domains of ectodermal epithelia of Drosophila. Cell 82:67–76
Yeong FM, Hombauer H, Wendt KS, Hirota T, Mudrak I, Mechtler K, Loregger T, Marchler-Bauer A, Tanaka K, Peters JM, Ogris E (2003) Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr Biol 13:2058–2064
Acknowledgements
We thank Katharina Neugebauer for technical support and C.-ting Wu, Steve DiNardo, and Volker Hartenstein for fly stocks. We are indebted to Thom Kaufman for providing the cid mutant lines prior to publication. We thank members of the laboratory for helpful discussions and Christian F. Lehner for generous support and critical reading of the manuscript. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (He 2354-1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Uhlmann
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Jäger, H., Rauch, M. & Heidmann, S. The Drosophila melanogaster condensin subunit Cap-G interacts with the centromere-specific histone H3 variant CID. Chromosoma 113, 350–361 (2005). https://doi.org/10.1007/s00412-004-0322-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-004-0322-4