Abstract
Purpose
Polycystic ovary syndrome is heterogeneity disease, and the association with DEEND1A gene has been discussed incompatibly for a long time. We conducted a meta-analysis to evaluate the rs10818854, rs2479106, and rs10986105 polymorphism in DENND1A gene with PCOS susceptibility.
Methods
Meta-analysis was performed for common allele versus rare allele using random effect model on published papers from January 1, 1980 to October 1, 2015. Subgroup analysis, sensitivity analysis and publication bias were also carried out ultimately. The combined odds ratio (OR) with 95 % confidence interval (95 % CI) was calculated to estimate the strength of the association.
Results
The results showed that rs10818854 (OR = 1.36, 95 % CI 1.12–1.61) and rs10986105 (OR = 1.39, 95 % CI 1.20–1.58) polymorphism increased the risk of PCOS probably. A significant association was also found between rs2479106 mutation and Asian PCOS patients but not Europeans (OR = 1.32, 95 % CI 1.25–1.39; OR = 1.01, 95 % CI 0.97–1.05, respectively).
Conclusions
In conclusion, the DENND1A gene variant is likely to have influence on PCOS risk. Further studies are warranted to assess these associations in greater detail, especially in different populations and different subtype of PCOS patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polycystic ovary syndrome (PCOS) is a complex endocrine and metabolic disease with a prevalence between 6 and 10 % based on the National Institute of Health (NIH) criteria and as high as 15 % when the broader Rotterdam criteria are applied [1]. Hyperandrogenism, oligo-ovulation or anovulation and polycystic ovarian morphology are the common features. Associated complications include obesity, type 2 diabetes, dyslipidemia and other cardiovascular disorders are noteworthy [2]. To date, the etiology of PCOS is poorly understood except the combined effect of environmental factor and genetic variants. It was a milestone event [3] for the first attempt of genome-wide association study (GWAS) of PCOS by Chen et al. [4] in Han Chinese cohort. Three independent susceptibility loci (2p16.3, 2p21, 9q33.3) were identified in this GWAS, which had remarkable association with PCOS. Recently, there is another large GWAS published among Europeans as well [5].
Among those, 9q33.3 mapped to the genomic areas of DENND1A gene, which encoded the connecdenn 1 proteins. The proteins contain N-terminal DENN domain, expressing differently in neoplastic and normal cells [6]. To our knowledge, they function as guanine nucleotide exchange factors (GEF) for Rab35, and play a role in clathrin-mediated endocytosis [7]. Three single-nucleotide polymorphisms (SNPs) in DENND1A gene, including rs10818854(A/G), rs2479106(A/G), and rs10986105(G/T), had been reported as susceptible loci, and all of them are in intron.
After the first publication of GWAS based on Han-Chinese PCOS patients, the association between DENND1A gene and PCOS was identified in subsequent replication studies covering different population around the world, but their results were inconsistent. Besides that, people across the world share a common genetic risk for PCOS, which is worth exploring. To figure out the contradictory issues, we performed this meta-analysis and systematic review to assess the DENND1A gene mutations on PCOS pathogenesis.
Materials and methods
Search strategy
We conducted a systematic search in the PubMed, Embase and CNKI (China National Knowledge Infrastructure) databases from January 1, 1980 to October 1, 2015. We used the terms “DENND1A”, “9q33.3”, “rs10818854”, “rs2479106”, “rs10986105” “polymorphism”, “polycystic ovary syndrome” and “Stein-Leventhal Syndrome” to identify all the literatures concerning DENND1A gene variants and PCOS. Mesh terms, EMTREE terms and free text words ([tiab] or/ti, ab.) were applied together for the entire search. The search was not restricted by languages.
Study inclusion
First, EndNote X7 (Thomson Reuter, Philadelphia, Pennsylvania) [8] was employed to remove the duplicates and triplicates gleaned from the searches in the above-mentioned databases. Next, two independent reviewers (Gao J and Xue JD) deleted the articles that had no relationship with our subjects according to the titles and abstracts. If the reviewers had conflicting results, the article was included and more information was used to determine its ultimate inclusion in the next iteration. Third, the two reviewers screened the full text of the remaining articles and used the following inclusion criteria: (1) sufficient allele data were presented to compute the odds ratios (ORs) and 95 % confidence intervals (CIs), (2) case–control study and the genotype distributions of control group conforming to Hardy–Weinberg equilibrium (HWE). The cardinal reasons for exclusion were: (1) animal experiments, (2) conference abstract, review, editorial etc. (3) not associated with gene polymorphism. If the reviewers’ reference lists were not in an agreement, another reviewer (Chen C) would be consulted to make a final decision for inclusion. Thus, we included a total of 10 articles for our meta-analysis.
Data extraction
According to the criteria described above, two authors (Gao J and Xue JD) extracted information based on the checklist for data collection from the Cochrane Handbook independently [9]. The following data were collected from all the eligible publications: first author, year of publication, country, ethnic of population, criteria for PCOS, total number of cases and controls and allele frequencies.
Quality assessment
The Newcastle Ottawa Scale (NOS) was used to estimate the quality of each study, as it has been used in genetic association studies [10]. NOS is one of the few scales previously recommended by a Cochrane working group [11, 12]. This measure assesses aspects of methodology in case–control studies, consisting of selection of cases, comparability of each study and ascertainment of exposure to risks. In all cases, disagreements among raters were resolved through discussion.
Statistics analysis
HWE in controls of all included studies was tested with exact test using an online HWE calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) and significant cutoff was set at 0.05. We applied the ORs along with 95 % CI to estimate the strength of association between DEEND1A and the risk of PCOS by the inverse-variance method [13]. A heterogeneity check was calculated based on Cochran’s Q statistic test [14], and the inconsistency index (I 2) < 25 %, p > 0.10 denoted that heterogeneity did not exist between the studies [9]. If there was no heterogeneity; the overall gene effect was evaluated with the fixed effects model, or else by the random-effects model. To evaluate the ethnicity and diagnostic criterion effect, subgroup analysis was performed by ethnic and criterion subgroup. To assess the influence of individual studies on the overall association, sensitivity analysis was also conducted by omitting one study at a time and re-analyzing the association for the remaining studies. Finally, the Egger’s weighted regression method [15] were carried out, p < 0.10 was considered as evidence of possible publication bias. All statistical analyses were performed using STATA 12.0 software (Stata Corporation, USA) and all p values are two-tailed.
Results
Description of studies
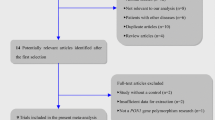
A total of 119 studies were acquired from the PubMed (N = 54), Embase (N = 59) and CNKI (N = 6) databases. The literature selection process used was illustrated in S1 Files. A total of 85 studies were included after removal of any duplicates. Next, 37 studies were excluded through screening of the titles and abstracts. The remaining 48 studies were full-text reviewed, and 38 studies were excluded according to the inclusion criteria as follows: 26 studies were reviews or conference abstracts, 10 studies were not related to our gene polymorphism, 2 studies had repeating data information. Finally, in the current study, 10 eligible case control studies [3, 5, 16–23] that satisfied the inclusion criteria were included in our meta-analysis and all of them were in HWE (Table 1). Allele frequencies, OR and 95 % CI of each SNP were displayed on Table 2.
Two diagnostic criteria for PCOS are applied: Rotterdamand NIH criteria. The former is defined by two out of three criteria: (1) menstrual irregularity (oligo-ovulation or anovulation), (2) hyperandrogenism (clinical or biochemical) and (3) polycystic ovarian morphology [24]. NIH criteria should satisfy a definition of women with hyperandrogenaemia and chronic anovulation [25]. Both Rotterdamand NIH criteria have to exclude specific disorders of the ovaries, adrenal orpituitary.
Methodological quality
Given the quality of the studies, the NOS scores were in high level in general. Table 3 presents the scores ranged from five to nine, with the average of 6.6.
Meta-analysis
Overall analysis and subgroup analysis
rs10818854
A total of six studies were involved in the rs10818854 polymorphism. Expect for three papers [3, 17, 19] suggesting an at-risk effect of rs1018854, the rest three papers [20–22] did not produce significant results. The overall analysis for evaluating the association of rs10818854 polymorphism with PCOS under allelic model showed a large heterogeneity between these six studies (I 2 = 76.5 %, P h = 0.002), and a significant random effects pooled OR (OR = 1.36, 95 % CI 1.12–1.61) (Table 4). Ethnicity and diagnostic criterion were not significantly responsible for the heterogeneity. However, subgroup analyses stratified by diagnostic criterion yielded different results; rs10818854 had strong impact on patients by the diagnosis of NIH criterion (OR = 1.69, 95 % CI 1.36–2.03; I 2 = 18.6 %, P h = 0.268). The application of Rotterdam criterion did not show the association between rs10818854 and PCOS statistically (OR = 1.23, 95 % CI 0.95–1.52, I 2 = 80.1 %, P h = 0.002) (Table 4).
rs2479106
With the whole 10 studies, we analyzed the effect of rs2479106 on the PCOS. Significant between-study heterogeneity was detected across studies for the A vs G model and thus was random-effects model. Overall, we found no significant association between rs2479106 polymorphism and PCOS in total analysis (OR = 1.11, 95 % CI 1.00–1.25, I 2 = 84.5 %, P h = 0.000, Table 4). Heterogeneity within each group disappeared when subgroup analyses stratified by Ethnicity were made. The results for were as follows: OR = 1.32, 95 % CI 1.25–1.39, I 2 = 0.0 %, P h = 0.824 for Asian; OR = 1.01, 95 % CI 0.97–1.05, I 2 = 0.0 %, P h = 0.754 for Europeans (Table 4). The results of diagnosis criteria subgroups also had the tendency to deny the effect of rs2479106 variants on the risk of PCOS (Table 4).
rs10986105
Moderate heterogeneity made the FEM available (I 2 = 50.0 %, P h = 0.091, Table 4). In this meta-analysis, we included a total of 5 case–control studies. The pooled result indicated that there was an obvious association between rs10986105 polymorphism and PCOS risk under allelic model (OR = 1.39, 95 % CI 1.20–1.58) with a low-to moderate heterogeneity level (I 2 = 50.0 %, P h = 0.091). We also tried to make subgroups classified by ethnicity and diagnosis criteria, but the results were poor because of the small sample size of included studies in each subgroup, showing in Table 4.
Sensitivity analysis
Owing to rs2479106 heterogeneity disappear after subgroup analysis, so ethnic background could be explained as the cause of heterogeneity. The heterogeneity in rs10818854 and rs10986105 studies was unacceptable, so we had to conduct the sensitivity analysis by excluding any single report at one time. The exclusion of any single report did not alter the significance of the final decision, suggesting that the outcomes were reliable and robust (Table S1).
Publication bias
Egger’s test was performed to evaluate publication bias of the literature on PCOS. The statistical results displayed no publication bias. (rs10818854: Egger’s test p = 0.510; rs2479106: Egger’s test p = 0.817; rs10986105: Egger’s test p = 0.875
Discussion
It has been demonstrated that genetic force could be more than 70 % pathogenesis of PCOS through twin studies [26]. Despite the numbers of candidate gene are up to one hundred, none of those has been validated. Since the DENND1A gene has been raised in 2010, there have been 10 independent studies to pursue the real association between gene variants and PCOS risk subsequently. The present meta-analysis reached the conclusion that rs10818854 and rs10986105 variants could be considered as meaningful SNPs significantly in Europeans and Asians, but we failed to find the source of its high heterogeneity. Furthermore, rs2479106 had impact on Asians but not Europeans, indicating that the susceptibility of PCOS varied from ethnicity, which totally denied the conclusions raised by Louwers et al. [21]. While a family-based analysis, containing 276 PCOS family trios used the transmission disequilibrium test (TDT) to detect rs10818854 and rs24709106 association to PCOS, they did not observe positive outcome [27]. Apart from this, endometrial adenocarcinoma (EA) as one of the long-term complications of PCOS, Wang et al. [28] performed a SNP comparison in Chinese individuals and acquired the DENND1A gene rs24709106 carrying risk allele to EA. It means that rs2479106 can serve as a gene marker of PCOS and EA. The mentioned SNPs above are all located in introns, and it is not conformed that they have responsibility of gene expression function. For the sake of characterize the coding protein sequence of DENND1A gene in PCOS cohort, one research came up with 8 SNPs in the coding region, in which a missense SNP (rs189947178, A/C) could potentially alter the structural conformation of the protein [29]. However, we need more replication studies and protein functional studies of this SNP in the further exploration.
Since the DENND1A gene locus served as a novel candidate gene in PCOS pathogenesis, the possible mechanism of promoting PCOS is urged to be recognized. There are two different transcripts of DENND1A gene due to the different division: DEEND1A.V1 and DEEND1A.V2. McAllister et al. [30]. found that DEEND1A.V2 was highly expressed in theca cells of PCOS patients, and they compelled the DEEND1A.V2 to express in normal theca cells, those would become PCOS-like manifestation with increasing expression of CYP17A1 and CYP11A1 gene as well as augmented androgen production. Notably, Studies have found that if DEEND1A.V2 gene was knocked down among PCOS patients, the theca cells would come to be normal phenotype [30]. All conveyed us the message that DENND1A gene might play a vital role in regulating the theca cell steroid genesis. In addition, they also discovered the significant elevated level of DENND1A.V2 RNA in PCOS patients’ urine. It provided the basis for a promising noninvasive PCOS diagnose, particularly in adolescent even preadolescent girls. Recently, the lentiviral vector carrying DENND1A gene has been constructed successfully [31]. They generated DENND1A recombinant virus by the means of lentivirus package, and the recombinant was employed to infect the primary theca-interstitial cells, which established fundamental foundation for further investigation of gene function in vivo and in vitro.
Our meta-analysis and systematic review still has some limitations and pities. To begin with, apart from Asian and European data, there were not relevant references based on other ethnic population, such as African. Secondly, three kinds of PCOS diagnostic criteria coexisted. The most common criteria is 2003 Rotterdam consensus criteria [24]: (1) oligo-ovulation or/and anovulation (OA); (2) clinical and/or biochemical hyperandrogenism (HA); (3) polycystic ovaries (PCO, by ultrasound). Two of them are required to confirm the diagnosis which yielded four PCOS subtypes: OA+HA+PCO;OA+HA;HA+PCO;OA+PCO. Nevertheless, all the patients were diagnosed as OA+HA+PCO type in Chen et al.’s study, meanwhile, the whole ones in Goodarizi et al.’s [17] and Welt et al’s studies [19] were identified as OA+HA type according to NIH criteria [25]. However, the patients in the reminding studies enrolled in our study ranged over all 4 subtypes. In our opinions, it will probably cause bias for the various phenotypes in genetic background. Therefore, it is advised that researchers should calculate the SNPs considering the subtypes discrepancy in the next step. For example, Cui et al. and Sun et al. [32, 33], they calculated the allele frequency distribution among different sub-phenotypes. Last but not least, our meta-analysis was based on unadjusted OR estimates because not all published studies presented adjusted ORs, or when they did, the ORs were not adjusted by the same potential confounders, such as age, BMI, ethnicity and exposures. Lacking of the information for the data analysis might cause serious confounding bias.
In conclusion, our current meta-analysis suggest that DENND1A gene may contribute to the pathogenic of PCOS, whereby, rs10818854 and rs10986105 have impact both in Asian and European, rs2479106 just affect Asians yet. Our meta-analysis provided an emphasis on the genetic risk profile. In the future, it is critical that larger and well-designed multi-centric studies based on Africans and other ethnics should be performed to re-evaluate the association, taking the gene–environment interaction into consideration if necessary.
References
Fauser BC, Tarlatzis BC, Rebar RW, Legro RS, Balen AH, Lobo R, Carmina H, Chang RJ, Yildiz BO, Laven JS, Boivin J, Petraglia F, Wijeyeratne CN, Norman RJ, Dunaif A, Franks S, Wild RA, Dumesic D, Barnhart K, Amsterdam ESHRE/ASRM-Sponsored 3rd PCOS Consensus Workshop Group (2012) Consensus on women’s health aspects of polycystic ovary syndrome (PCOS). Hum Reprod 27(1):14–24. doi:10.1093/humrep/der396 (Oxford, England)
Legro RS, Arslanian SA, Ehrmann DA, Hoeger KM, Murad MH, Pasquali R, Welt CK, Endocrine S (2013) Diagnosis and treatment of polycystic ovary syndrome: an endocrine society clinical practice guideline. J Clin Endocrinol Metab 98(12):4565–4592. doi:10.1210/jc.2013-2350
Strauss JF 3rd, McAllister JM, Urbanek M (2012) Persistence pays off for PCOS gene prospectors. J Clin Endocrinol Metab 97(7):2286–2288. doi:10.1210/jc.2012-2109
Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J, Liang X, Zhao X, Zhao J, Sun Y, Zhang B, Jiang H, Zhao D, Bian Y, Gao X, Geng L, Li Y, Zhu D, Sun X, Xu JE, Hao C, Ren CE, Zhang Y, Chen S, Zhang W, Yang A, Yan J, Li Y, Ma J, Zhao Y (2011) Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet 43(1):55–59. doi:10.1038/ng.732
Day FR, Hinds DA, Tung JY, Stolk L, Styrkarsdottir U, Saxena R, Bjonnes A, Broer L, Dunger DB, Halldorsson BV, Lawlor DA, Laval G, Mathieson I, McCardle WL, Louwers Y, Meun C, Ring S, Scott RA, Sulem P, Uitterlinden AG, Wareham NJ, Thorsteinsdottir U, Welt C, Stefansson K, Laven JS, Ong KK, Perry JR (2015) Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun 6:8464. doi:10.1038/ncomms9464
Marat AL, McPherson PS (2010) The connecdenn family, Rab35 guanine nucleotide exchange factors interfacing with the clathrin machinery. J Biol Chem 285(14):10627–10637. doi:10.1074/jbc.M109.050930
Allaire PD, Marat AL, Dall’Armi C, Di Paolo G, McPherson PS, Ritter B (2010) The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol Cell 37(3):370–382. doi:10.1016/j.molcel.2009.12.037
Eapen BR (2006) EndNote 7.0. Indian J Dermatol Venereol Leprol 72(2):165–166
Higgins JPT, Green S (2014) Cochrane handbook for systematic reviews of interventions version 5.1.0 (updated March 2011). The cochrane collaboration 2011. Available from http://www.cochrane-handbook.org. Accessed 26 Mar 2014
Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, Tugwell P (2004) The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm. Accessed 29 Apr 2004
Lundh A, Gotzsche PC (2008) Recommendations by Cochrane Review Groups for assessment of the risk of bias in studies. BMC Med Res Methodol 8:22. doi:10.1186/1471-2288-8-22
Bauer T, Bouman HJ, van Werkum JW, Ford NF, ten Berg JM, Taubert D (2011) Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. BMJ 343:d4588. doi:10.1136/bmj.d4588
Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J (2005) A method for meta-analysis of molecular association studies. Stat Med 24(9):1291–1306. doi:10.1002/sim.2010
Midgette AS, Wong JB, Beshansky JR, Porath A, Fleming C, Pauker SG (1994) Cost-effectiveness of streptokinase for acute myocardial infarction: a combined meta-analysis and decision analysis of the effects of infarct location and of likelihood of infarction. Med Decis Mak 14(2):108–117
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634. doi:10.1136/bmj.315.7109.629
Lerchbaum E, Trummer O, Giuliani A, Gruber HJ, Pieber TR, Obermayer-Pietsch B (2011) Susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21, and 9q33.3 in a cohort of Caucasian women. Horm Metab Res 43(11):743–747. doi:10.1055/s-0031-1286279
Goodarzi MO, Jones MR, Li X, Chua AK, Garcia OA, Chen YD, Krauss RM, Rotter JI, Ankener W, Legro RS, Azziz R, Strauss JF 3rd, Dunaif A, Urbanek M (2012) Replication of association of DENND1A and THADA variants with polycystic ovary syndrome in European cohorts. J Med Genet 49(2):90–95. doi:10.1136/jmedgenet-2011-100427
Eriksen MB, Brusgaard K, Andersen M, Tan Q, Altinok ML, Gaster M, Glintborg D (2012) Association of polycystic ovary syndrome susceptibility single nucleotide polymorphism rs2479106 and PCOS in Caucasian patients with PCOS or hirsutism as referral diagnosis. Eur J Obstet Gynecol Reprod Biol 163(1):39–42. doi:10.1016/j.ejogrb.2012.03.020
Welt CK, Styrkarsdottir U, Ehrmann DA, Thorleifsson G, Arason G, Gudmundsson JA, Ober C, Rosenfield RL, Saxena R, Thorsteinsdottir U, Crowley WF, Stefansson K (2012) Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab 97(7):E1342–E1347. doi:10.1210/jc.2011-3478
Shi Y, Zhao H, Shi Y, Cao Y, Yang D, Li Z, Zhang B, Liang X, Li T, Chen J, Shen J, Zhao J, You L, Gao X, Zhu D, Zhao X, Yan Y, Qin Y, Li W, Yan J, Wang Q, Zhao J, Geng L, Ma J, Zhao Y, He G, Zhang A, Zou S, Yang A, Liu J, Li W, Li B, Wan C, Qin Y, Shi J, Yang J, Jiang H, Xu JE, Qi X, Sun Y, Zhang Y, Hao C, Ju X, Zhao D, Ren CE, Li X, Zhang W, Zhang Y, Zhang J, Wu D, Zhang C, He L, Chen ZJ (2012) Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet 44(9):1020–1025. doi:10.1038/ng.2384
Louwers YV, Stolk L, Uitterlinden AG, Laven JS (2013) Cross-ethnic meta-analysis of genetic variants for polycystic ovary syndrome. J Clin Endocrinol Metab 98(12):E2006–E2012. doi:10.1210/jc.2013-2495
Gammoh E, Arekat MR, Saldhana FL, Madan S, Ebrahim BH, Almawi WY (2015) DENND1A gene variants in Bahraini Arab women with polycystic ovary syndrome. Gene 560(1):30–33. doi:10.1016/j.gene.2015.01.034
Ha L, Shi Y, Zhao J, Li T, Chen ZJ (2015) Association study between polycystic ovarian syndrome and the susceptibility genes polymorphisms in Hui Chinese Women. PLoS One 10(5):e0126505. doi:10.1371/journal.pone.0126505
Rotterdam EA-SPcwg (2004) Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod 19(1):41–47 (Oxford, England)
DA Zawadzki JK (1992) Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, Givens JR, Haseltine FP, Merriam GR (eds) polycystic ovary syndrome. Blackwell Scientific, Boston, pp 377–384
Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI (2006) Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab 91(6):2100–2104. doi:10.1210/jc.2005-1494
Zhao H, Xu X, Xing X, Wang J, He L, Shi Y, Shi Y, Zhao Y, Chen ZJ (2012) Family-based analysis of susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Hum Reprod 27(1):294–298. doi:10.1093/humrep/der379 (Oxford, England)
Wang Z, Li T, Zhang W, You L, Zhao Y, Xia M, Zhao H, Chen ZJ (2012) Variants in DENND1A and LHCGR are associated with endometrioid adenocarcinoma. Gynecol Oncol 127(2):403–405. doi:10.1016/j.ygyno.2012.08.007
Eriksen MB, Nielsen MF, Brusgaard K, Tan Q, Andersen MS, Glintborg D, Gaster M (2013) Genetic alterations within the DENND1A gene in patients with polycystic ovary syndrome (PCOS). PLoS One 8(9):e77186. doi:10.1371/journal.pone.0077186
McAllister JM, Modi B, Miller BA, Biegler J, Bruggeman R, Legro RS, Strauss JF 3rd (2014) Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc Natl Acad Sci USA 111(15):E1519–E1527. doi:10.1073/pnas.1400574111
Mou C (2014) Effect of testosterone on the in vitro mouse follicle development and construction of lentiviral vector carrying mouse Denndla gene. Dissertation, Shandong University
Cui L, Zhao H, Zhang B, Qu Z, Liu J, Liang X, Zhao X, Zhao J, Sun Y, Wang P, Li T, Shi Y, Chen ZJ (2013) Genotype-phenotype correlations of PCOS susceptibility SNPs identified by GWAS in a large cohort of Han Chinese women. Hum Reprod 28(2):538–544. doi:10.1093/humrep/des424 (Oxford, England)
Sun M, Sheng Y, Ma Z, Chen Z, Tang R (2014) Correlation analysis between polycystic ovary syndrome susceptibility genes and metabolic phenotypes. Zhonghua fu chan ke za zhi 49(6):441–445
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
J. Gao and J.-D. Xue contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, J., Xue, JD., Li, ZC. et al. The association of DENND1A gene polymorphisms and polycystic ovary syndrome risk: a systematic review and meta-analysis. Arch Gynecol Obstet 294, 1073–1080 (2016). https://doi.org/10.1007/s00404-016-4159-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-016-4159-x