Abstract
Background
We analysed cell-free DNA (cfDNA) in the plasma of patients with both malignant and benign breast lesions by real-time quantitative PCR to determine whether the finding may have diagnostic and prognostic implications.
Methods
Plasma samples were obtained from 33 patients with breast cancer, 32 patients with benign breast lesions and 50 healthy women as normal controls. Circulatory cfDNA was extracted from the plasma samples and quantified by real-time quantitative PCR for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene.
Results
The mean concentrations of cfDNA in the plasma samples from patients with breast cancer, patients with benign breast lesions and normal controls were 2,285, 1,368 and 1,489 genome equivalents (GE) per millilitre, respectively. The level of cfDNA in the breast cancer group was significantly higher than those in the benign lesion group and control group (P = 0.007 and 0.013, respectively). These findings were associated with malignant tumour size. The levels of the cfDNA were high in patients with lymph node involvement and distant metastasis.
Conclusions
Our results suggest that levels of cfDNA in the plasma are elevated in malignant breast cancer and correlated with tumour size. These findings could have diagnostic and prognostic value for malignant breast tumours.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common malignant disease in women and the second leading cause of cancer death in Western industrial countries. Early diagnosis of breast cancer and accurate identification of haematogenic metastases can improve the success of treatment [1–4]. Therefore, the detection of single tumour cell shedding in the blood in early stage cancer could help physicians choose the optimal therapy for patients. In addition, accurate quantification of metastatic tumour cells could help physicians adjust the level of treatment. Determination of occult metastases at later stages or after primary treatment of the disease could assist in monitoring and following the progress of the disease. Many hospitals and institutes have performed the detection of disseminated tumour cells in bone marrow of patients with breast cancer [5–8]. A number of studies suggested that the detection of disseminated single tumour cells in bone marrow in primary breast cancer could eventually replace lymph node dissection in patients with small tumours and no clinical signs of lymph node involvement [4, 7]. However, the risks of invasive bone marrow aspiration have made its routine implementation in screening, tracking and follow-up of patients with breast cancer problematic.
The discovery of cell-free DNA in plasma and serum samples provides another possibility to examine tumour derived genetic materials in the circulation and to detect haematogenic spread of tumour cell DNA [9]. Qualitative alterations in circulating DNA, such as microsatellite alterations [10], oncogene mutations [11], mitochondrial DNA, tumour-specific methylated DNA [12] and viral DNA [13], have been found in patients with different types of cancer. Quantitative alterations of circulating cfDNA have also been observed in several cancers, such as prostate cancer [14], lung cancer [15], pancreatic cancer [16], leukaemia and lymphoma [17]. High levels of circulating cfDNA were correlated with tumour metastasis, response to therapy and recurrence [15, 18].
Therefore, tumour-derived circulating nucleic acids in the plasma or serum of cancer patients were introduced as a tool for detection and surveillance of cancers [19].
In our study, we present a quantification of cell-free plasma DNA in patients with malignant and benign breast lesions. The concentration of these circulating molecules in the cohort of breast cancer was compared with those in normal controls and in the patients with benign lesions. We analysed the correlation between the levels of cell-free plasma DNA in patients with breast cancer and the traditional staging parameters and clinical predictive markers.
Materials and methods
Sample collection
In total, 115 peripheral blood samples were recruited in this study, which included samples from 32 patients with benign breast lesions and 33 patients with malignant breast cancer at the University Women’s Hospital in Basel, Switzerland. The remaining 50 samples from healthy female patients without disease were used as controls. All diagnoses were biopsy-confirmed. The samples had been collected before any invasive procedures had been performed or any therapy had been administered. The clinical characteristics of the study cohorts are shown in Table 1.
The study was approved by the institutional review board of the University of Basel. Written informed consent was obtained in all cases.
DNA extraction
About 10 ml of peripheral blood from each subject were sent to our laboratory after blood draw and processed immediately by centrifugation at 1,600g for 10 min. The plasma layers were transferred to new Eppendorf tubes and centrifuged again at maximum speed (16,000g) for 10 min. Cell-free plasma DNA was extracted from 400 μl plasma sample using the high pure PCR Template Preparation Kit (Roche Diagnostics, Germany) according to the manufacturer’s protocol. The DNA preparations were eluted in 100 μl elution buffer. The eluted DNA was stored at −20°C until further use.
Quantitative analysis of cell-free DNA in plasma samples
A total of 5 μl of DNA elution were used as template for the TaqMan real time PCR analysis. The amount of cell-free plasma DNA was quantified using the following primer pairs and VIC labelled TaqMan MGB-probe for GAPDH gene (GenBank Accession No. J04038):
-
Forward: 5′ CCCCACACACATGCACTTACG 3′
-
Reverse: 5′ CCTAGTCCCAGGGCTTTGATT 3′
-
Probe 5′ (MGB): GTG AAC GTG GAT GAA GTT GG (VIC) 3′
Our standard TaqMan PCR conditions have been described in our previous publications, involving 40 cycles of 15 s at 95°C and 1 min at 60°C [20]. The cell-free plasma DNA equivalents were calculated according to very reproducible standard dilution curves using a known concentration of human genomic DNA. The concentrations of cell-free plasma DNA were expressed as genome-equivalents (GE) per millilitre of plasma.
Statistical analysis
Data were analysed with the SPSS software (Statistical Software Package for Windows). The Mann-Whitney U-test was used to compare the levels of cell-free plasma DNA in different groups categorized by normal healthy individuals, benign tumour group and breast cancer. The Pearson test was used to analyse the relationships between levels of cell-free plasma DNA and other established prognostic factors in breast cancer.
Results
Levels of cell-free plasma DNA in the three study groups
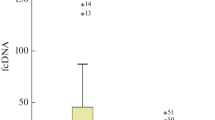
The results in Table 1 show that the mean concentrations of cell-free plasma DNA in the samples of healthy controls, patients with benign breast lesions and breast cancer were 1,489, 1,368 and 2,285 GE/ml, respectively. In breast cancer cases, the level was significantly elevated when compared to benign breast lesions and healthy controls (P = 0.013 and 0.007, respectively) (Fig. 1). No significant difference was found between the control group and the benign lesion group. In the study group of benign lesions, the patients were younger than those in the control group. We subdivided the benign lesion group into two different age cohorts, namely a younger group and an age-matched group to the controls. We compared the concentration of cell-free plasma DNA between the control group and age-matched benign lesion group, and control group and younger benign lesion group. No significant differences were found.
Box plot indicating cell-free plasma DNA levels in normal individuals and patients with benign lesion and breast cancer The medians are indicated by a line inside each box, the 25th and 75th percentiles by the box limits; the lower and upper error bars represent the 10th and 90th percentiles, respectively
Correlation between levels of cell-free plasma DNA and other established prognostic factors
In the cohort of breast cancer, associations between levels of cell-free plasma DNA and traditional clinical parameters, such as tumour size, lymph node involvement, extent of metastasis, histological grades, receptor status and HER-2/neu were analysed.
Elevated levels of cell-free plasma DNA were found to be associated with tumour size (Fig. 2, Table 2). The level of cell-free plasma DNA was significantly higher in the patients with breast cancer with a tumour size >2 cm than in those with a tumour size <2 cm (3,556 GE/ml vs. 1,470 GE/ml, P = 0.013) (Fig. 2).
In this study, lymph node involvement was present in 11 cases. The level of cell-free plasma DNA was overall higher in the nodal positive patients than in the nodal negative patients (Fig. 3), but no statistical significant difference was found between the two groups.
Box plot indicating cell-free plasma DNA levels in the circulation of the patients with or without lymph node involvement (nodal positive or nodal negative) and the patients with distant metastases. The medians are indicated by a line inside each box, the 25th and 75th percentiles by the box limits; the lower and upper error bars represent the 10th and 90th percentiles, respectively
Distant metastases were found in 2 of 33 patients. The concentrations of the cell-free plasma DNA in the two cases amounted to 4,416 and 5,294 GE/ml, respectively, which were two folds higher than the mean concentration of cell-free plasma DNA in the overall cohort of breast cancer cases (2,285 GE/ml) (Fig. 3). The cumulative percentages of the two cases in the cohort of breast cancer were 84 and 87%.
Oestrogen and progesterone are involved in breast cancer growth. Hormone receptors are measured to guide clinical hormonal therapy. In our study, both the oestrogen and progesterone receptor status in the tumour tissues of the patients with breast cancer were examined by immunohistochemistry. Positive oestrogen and progesterone receptors were found in 29 of the 33 cases. The receptor-positive tumour cells in tumour tissues ranged from 10 to 100%. The level of cell-free plasma DNA did not correlate with the percentage of oestrogen and progesterone receptors.
HER-2/neu (also called erbB-2) encodes a transmembrane tyrosine kinase receptor. The amplification of HER-2/neu can serve as a predictive factor in breast cancer. In our study, 7 of the 29 cases showed HER-2/neu amplification in the tumour tissues. The levels of cell-free plasma DNA in the seven cases were not higher than in the other 22 cases, which were negative for HER-2/neu.
The histological grade of breast cancer, based on the frequency of cell mitosis, tubule formation and nuclear pleomorphism serves as a predictive factor. The level of cell-free plasma DNA in the cohort of grade I cases was not significantly different from the level in the cohorts of grade II or III cases.
Discussion
In this study, we found that the level of cell-free plasma DNA is elevated in patients with breast cancer. From these results, several questions arise. First, what is the origin of these circulatory molecules and the mechanism behind their increase? Second, what roles do they play in patients with breast cancer? Third, can the high level of the plasma DNA in breast cancer be used as a marker for patients in screening, early diagnosis, identification of metastases and/or follow-up testing?
Our experiences with cell-free fetal DNA and fetal cells in the maternal circulation may help to answer the first question. Preeclampsia is a pregnancy related disorder and is caused by apoptotic–necrotic shedding of cell-free debris from placental ischaemia into the maternal circulation. We found that both cell-free fetal and maternal DNA was increased in the preeclampsia [20]. The increments of both the cell-free fetal and maternal DNA levels corresponded to the degree of disease severity [20]. In addition, cell-free fetal DNA increased prior to the onset of preeclampsia, but not cell-free maternal DNA [21]. The number of fetal cells in the maternal circulation was also elevated in preeclampsia, but did not correlate with amount of cell-free fetal DNA in maternal plasma in preeclampsia [22]. However, cell-free fetal DNA has been found to be associated with placental syncytiotrophoblast microparticles (STBM), which are released into maternal blood stream during pregnancy [23]. The results suggest that exaggerated shedding of placental apoptotic and/or necrotic materials might be the main source of the high level of the cell-free fetal DNA, and also that the exaggerated inflammatory response and inappropriate endothelial activation of preeclampsia might be the main source of the high concentration of the maternal DNA. Along similar lines, the high level of cell-free plasma DNA in breast cancer could result from apoptosis and/or necrosis of tumour tissues. Using a methylation-specific tumour gene as a marker, Jahr et al. [24] were able to discriminate tumour and nontumour derived DNA in plasma. Tumour-derived cfDNA amounted to 3% of total cf DNA.
Viruses are encapsulated nucleic acid (DNA or RNA) that can only replicate in living host cells. Some viruses can convert normal host cells into tumour cells by viral transformation. Regarding the question of what role cell-free tumour-derived DNA plays, whether it can play similar role as viruses in transforming normal cells into malignant cells remains unclear. Several studies have shown that tumour-derived DNA could be transferred from one cell to another via the phagocytosis of apoptotic bodies and is stable over time, suggesting that tumour-derived DNA plays a possible role in the development of metastasis, and that stem cells might be naturally transformed with dominant oncogenes shedding in the plasma as a result of dissemination [25–28]. Thijssen et al. [18] reported that cell-free serum DNA was correlated with colorectal liver metastases. Garcia-Olmo et al. [29] were able to detect cell-free tumour DNA earlier and more frequently than circulating tumour cells in plasma samples of metastatic animal models, but neither the detection of tumour DNA in the plasma nor the presence of circulating tumour cells were strongly correlated with the presence of metastases.
In reference to the third question posed earlier, we compared the high level of cell-free plasma DNA with other traditional staging parameters, such as tumour size, lymph node involvement, extent of metastasis and predictive markers such as histological grades, receptor status and HER-2/neu. Tumour size and extent of spread are the most important factors in predicting the prognosis of a woman with this disease. High level of cell-free plasma DNA elevated in tumour size was found. A pronounced increase of cell-free plasma DNA in patients with lymph node involvement and distant metastases was observed.
In conclusion, our data suggest that using a robust Taqman real-time PCR, the level of cell-free plasma DNA in patients with breast cancer can be easily quantified and compared with normal individuals and patients with benign lesions. This may have diagnostic and prognostic value for malignant breast tumours. In addition, plasma sampling is much easier and less painful than bone marrow aspiration. Quantification of cell-free plasma DNA in patients with breast cancer could be considered for use in detecting occult tumour shedding before, during and after treatment. However, before the transition of a research technique to clinical use can be achieved, large-scale studies are needed to clarify the roles of the molecules in metastatic disease and to verify the value of its application in monitoring disease and evaluating prognosis. We believe that our findings provide a good platform for such future studies.
References
Pantel K, Muller V, Auer M, Nusser N, Harbeck N, Braun S (2003) Detection and clinical implications of early systemic tumour cell dissemination in breast cancer. Clin Cancer Res 9(17):6326–6334
Diel IJ, Solomayer EF, Bastert G (2000) Bisphosphonates and the prevention of metastasis: first evidences from preclinical and clinical studies. Cancer 88:3080–3088
Diel IJ (2000) Antitumour effects of bisphosphonates: first evidence and possible mechanisms. Drugs 59(3):391–399
Diel IJ, Mundy GR (2000) Bisphosphonates in the adjuvant treatment of cancer: experimental evidence and first clinical results. International Bone and Cancer Study Group (IBCG). Br J Cancer 82(8):1381–1386
Diel IJ, Solomayer EF, Bastert G (2000) Micrometastatic cells in the bone marrow of patients with breast carcinoma]. Radiologe 40(8):681–687
Pantel K, Woelfle U (2005) Detection and molecular characterisation of disseminated tumour cells: implications for anti-cancer therapy. Biochim Biophys Acta 1756(1):53–64
Solomayer EF, Diel IJ, Meyberg GC, Gollan C, Bastert G (2000) Metastatic breast cancer: clinical course, prognosis and therapy related to the first site of metastasis. Breast Cancer Res Treat 59(3):271–278
Bauer KD, de la Torre-Bueno J, Diel IJ, Hawes D, Decker WJ, Priddy C, Bossy B, Ludmann S, Yamamoto K, Masih AS, Espinoza FP, Harrington DS (2000) Reliable and sensitive analysis of occult bone marrow metastases using automated cellular imaging. Clin Cancer Res 6(9):3552–3559
Anker P, Stroun M (2000) Circulating DNA in plasma or serum. Medicina (B Aires) 60:699–702
Nawroz-Danish H, Eisenberger CF, Yoo GH, Wu L, Koch W, Black C, Ensley JF, Wei WZ, Sidransky D (2004) Microsatellite analysis of serum DNA in patients with head and neck cancer. Int J Cancer 111(1):96–100
Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B (2005) Detection and quantification of mutations in the plasma of patients with colorectal tumours. Proc Natl Acad Sci USA 102(45):16368–16373
Wong TS, Kwong DL, Sham JS, Wei WI, Kwong YL, Yuen AP (2004) Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clin Cancer Res 10(7):2401–2406
Leung SF, Chan AT, Zee B, Ma B, Chan LY, Johnson PJ, Lo YM (2003) Pretherapy quantitative measurement of circulating Epstein–Barr virus DNA is predictive of posttherapy distant failure in patients with early-stage nasopharyngeal carcinoma of undifferentiated type. Cancer 98(2):288–291
Boddy JL, Gal S, Malone PR, Harris AL, Wainscoat JS (2005) Prospective study of quantitation of plasma DNA levels in the diagnosis of malignant versus benign prostate disease. Clin Cancer Res 11(4):1394–1399
Gautschi O, Bigosch C, Huegli B, Jermann M, Marx A, Chasse E, Ratschiller D, Weder W, Joerger M, Betticher DC, Stahel RA, Ziegler A (2004) Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol 22(20):4157–4164
Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD (1998) Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 17(1):89–97
Wu TL, Zhang D, Chia JH, Tsao KH, Sun CF, Wu JT (2002) Cell-free DNA: measurement in various carcinomas and establishment of normal reference range. Clin Chim Acta 321(1–2):77–87
Thijssen MA, Swinkels DW, Ruers TJ, de Kok JB (2002) Difference between free circulating plasma and serum DNA in patients with colorectal liver metastases. Anticancer Res 22(1A):421–425
Goebel G, Zitt M, Zitt M, Muller HM (2005) Circulating nucleic acids in plasma or serum (CNAPS) as prognostic and predictive markers in patients with solid neoplasias. Dis Markers 21(3):105–120
Zhong XY, Laivuori H, Livingston JC, Ylikorkala O, Sibai BM, Holzgreve W, Hahn S (2001) Elevation of both maternal and fetal extracellular circulating deoxyribonucleic acid concentrations in the plasma of pregnant women with preeclampsia. Am J Obstet Gynecol 184(3):414–419
Zhong XY, Holzgreve W, Hahn S (2002) The levels of circulatory cell free fetal DNA in maternal plasma are elevated prior to the onset of preeclampsia. Hypertens Pregnancy 21(1):77–83
Zhong XY, Holzgreve W, Hahn S (2002) Cell-free fetal DNA in the maternal circulation does not stem from the transplacental passage of fetal erythroblasts. Mol Hum Reprod 8(9):864–870
Gupta AK, Holzgreve W, Huppertz B, Malek A, Schneider H, Hahn S (2004) Detection of fetal DNA and RNA in placenta-derived syncytiotrophoblast microparticles generated in vitro. Clin Chem 50(11):2187–2190
Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R (2001) DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 61(4):1659–1665
Bergsmedh A, Szeles A, Spetz AL, Holmgren L (2002) Loss of the p21(Cip1/Waf1) cyclin kinase inhibitor results in propagation of horizontally transferred DNA. Cancer Res 62(2):575–579
Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, Holmgren L (2001) Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci USA 98(11):6407–6411
Garcia-Olmo D, Garcia-Olmo DC, Ontanon J, Martinez E (2000) Horizontal transfer of DNA and the “genometastasis hypothesis”. Blood 95(2):724–725
Garcia-Olmo DC, Ruiz-Piqueras R, Garcia-Olmo D (2004) Circulating nucleic acids in plasma and serum (CNAPS) and its relation to stem cells and cancer metastasis: state of the issue. Histol Histopathol 19(2):575–583
Garcia-Olmo DC, Gutierrez-Gonzalez L, Ruiz-Piqueras R, Picazo MG, Garcia-Olmo D (2005) Detection of circulating tumour cells and of tumour DNA in plasma during tumour progression in rats. Cancer Lett 217(1):115–123
Acknowledgments
We thank Dr. Dorothy Huang and Professor Dennis Lo for their helpful comments and reviewing the English. We thank Vivian Kiefer for her excellent assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhong, X.Y., Ladewig, A., Schmid, S. et al. Elevated level of cell-free plasma DNA is associated with breast cancer. Arch Gynecol Obstet 276, 327–331 (2007). https://doi.org/10.1007/s00404-007-0345-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-007-0345-1