Abstract
Keloids are characterized as an “over-exuberant” healing response resulting in a disproportionate extracellular matrix (ECM) accumulation and tissue fibrosis. In view of the integral role of inflammation and cytokines in the healing response, it is logical to assume that they may play a part in orchestrating the pathology of this “abnormal” healing process. Tumor necrosis factor-α (TNF-α) is a potent proinflammatory cytokine involved in activation of signaling events and transcriptional programs, such as NFκB. This study attempts to determine the difference in NFκB and its related genes expression and DNA binding activity between keloid and normal skin fibroblasts. Three keloid and normal skin tissues (NSk) and their derived fibroblasts were used to determine NFκB signaling pathway expression using specific cDNA microarrays, Western blot analysis and immunohistochemistry. Electrophoretic mobility gel shift assay (EMSA) was used to assess NFκB-binding activity, all assays were performed in the presence and absence of TNF-α. TNF-α up-regulated 15% of NFκB signal pathway related genes in keloid fibroblast compared to normal skin. At the protein level, keloid fibroblasts and tissues showed higher basal levels of TNF- receptor–associated factors—TRAF1, TRAF2—TNF-α, inhibitor of apoptosis (c-IAP-1), and NFκB, compared with NSk. Keloid fibroblasts showed a constitutive increase in NFκB-binding activity in comparison to NSk both with and without TNF-α treatment. NFκB and its targeted genes, especially the antiapoptotic genes, could play a role in keloid pathogenesis; targeting NFκB could help in developing therapeutic interventions for the treatment of keloid scarring.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Keloids represent an abnormal healing response to injuries; characterized by excess accumulation of several matrix constituents, including glycoproteins, fibronectin, and collagen. These lesions are often characterized by a marked cellular infiltration, predominantly fibroblastic, with deposition of a dense collagenous meshwork and a rich vascular supply [15, 28]. Compared to normal skin, keloid has several well-defined morphologic characteristics. Histologically, the center of the keloid is highly collagenous, avascular, and relatively acellular. In contrast, the junction between the keloid lesion and its adjacent clinically normal skin periphery harbors large populations of lymphocytes and fibroblasts interspersed within a dense capillary network [1, 10, 29]. This observation is consistent with previous studies demonstrating that in keloid tissue, mixed biological effects including proliferation, apoptosis, and necrosis exist simultaneously, but in a distinctly compartmentalized fashion [2]. Most keloid studies have focused on the gene expression of extracellular matrix (ECM) molecules, growth factors, cytokines, and proteinases that might alter cell proliferation and migration in the formation of keloid. Given the complex molecular mechanisms of wound repair, it is unlikely that expression of these specific genes alone can explain aberration in wound healing products [8]. Therefore, a broad evaluation of differences in gene expression in the formation of keloid is necessary.
Tumor necrosis factor-α (TNF-α) is a potent proinflammatory cytokine produced by many cell types, including macrophages, lymphocytes, keratinocytes, and fibroblasts in response to inflammation, injury, infection, and other environmental stresses. TNF-α exerts its varied biological activities through two distinct cell surface receptors, termed TNFR1 and TNFR2, which are expressed on most cell types [19, 27]. Binding of TNF-α to its receptors especially to TNFR1 induces receptor trimerization and recruitment of several signaling proteinases to its cytoplasmic domain, which in turn, activates downstream signaling pathways including caspases and two major transcription factors, AP-1 and NFκB [11, 16]. Through a complex network of signaling cascades, TNF-α can activate both pro- and anti-apoptotic mechanisms depending on cell context. Thus the biologic response to TNF-α stimulation in a particular cell type depends on the balance between these two opposing pathways, as well as NFκB activation [5, 13]. It is also known that TNF-α induces the expression of various antiapoptotic genes, such as A20, the Bcl-2 family, and cellular inhibitors of apoptosis-2 (c-IAP), which are all targets of the NFκB family of transcription factors [3].
NFκB heterodimers are found in virtually all cell types and sequestrated in the cytoplasm in an inactive form bound to its inhibitors termed IκBs. NFκB is primarily regulated by phophorylation of IκBs. Upon exposure to TNF-α and other agonists, the IκBs are phophorylated by specific IκB kinases (IKK), resulting in their ubiquitination and degradation and the subsequent translocation of the freed NFκB into the nucleus [5]. The transcription factor NFκB can either repress or activate transcription of numerous genes whose products are critical to inflammation and immune responses such as Th2 cytokines, chemokines, leukocyte adhesion molecules, MMPs, Cox-2, and inducible nitric oxide synthase (iNOS), as well as genes involved in apoptotic signals [18, 26]. Interestingly, the TNF-α promoter itself contains a NFκB binding site and is subject to positive autoregulation that is essential to the amplification of inflammatory responses [6]. The NFκB pathway is also a key mediator of genes involved in the control of cellular proliferation and apoptosis [22]. Antiapoptotic genes that are directly activated by NFκB include the cellular inhibitors of apoptosis (c-IAP1, c-IAP2, and xIAP) and the TNF-receptor–associated factors (TRAF1 and TRAF2) [31, 33].
We have previously demonstrated that there is a significant difference in apoptotic profiles between normal skin and keloid fibroblasts, and that keloid fibroblasts are more resistant to cell death than their normal skin counterparts [20, 21, 24]. Differential changes in apoptotic profiles in keloid fibroblasts were also reported by other investigators [9, 17].
To further investigate the apoptotic mechanisms, particularly the NFκB signal pathways involved in keloid pathogenesis, we first employed the NFκB signal pathway microarray to determine whether there is a difference in the expression profile of a broad spectrum of genes in keloid fibroblasts, and their normal skin counterparts in response to TNF-α stimulation. Next, we evaluated the constitutive as well as TNF-α–induced NFκB DNA-binding activity in normal skin (NSk) and keloid (Kel) fibroblasts. Our results demonstrated a constitutive elevation of several components of the NFκB signal pathway in keloid tissues and derived fibroblasts, suggesting that this signal pathway may play a role in keloid pathogenesis.
Materials and methods
Tissue and cell culture origins
The demographics of patients are summarized in Table 1. Keloid and their associated normal peripheral skin samples (n=3, respectively) were collected from patients (age range, 18–60 years) who had undergone keloid resection in the Dermatology Clinic of the Martin Luther King Hospital, Los Angeles, as part of therapeutic procedures and after appropriate patient informed consent was obtained. Three replicate cultures each of human NSk and Kel fibroblasts were cultured as previously described [21]. Matched normal skin was also obtained via an elliptical incision which allowed a 2–4-mm rim of normal skin peripheral border to allow tension-free primary closure. No significant comorbid conditions such as diabetes, or having received drugs known to influence the transcriptional response of cells, were identified. All keloid included in this study had characteristic symptoms as confirmed by pathologic examination. After excision, half of the tissues were formalin fixed and used for pathologic identification and for immunohistochemistry, and the rest were used for cell cultures, and RNA and protein isolation. Primary fibroblast cells were isolated from the central region of keloid tissues and normal skin and cultured in DMEM supplemented with 10% fetal bovine serum. All cultures were maintained at 37°C in 5% CO2, 20% O2. Cells used for experiments were between passages 2 and 8.
Materials and reagents
The following antibodies were used at different concentrations and after different incubation periods depending on the specific assays: mouse IgG1 antibody (negative control), TNF-α (R&D, Minneapolis, Minn., USA); anti-TRAF1, TRAF2, (Santa Cruz Biotechnology, Santa Cruz, Calif., USA); and NFκB (p65) (Pharmingen, San Diego, Calif., USA). Other agents are all analytical grade (Sigma Chemicals, St Louis, Mo., USA).
RNA isolation and NFκB signaling pathway gene expression array
Cultured fibroblasts of low passages (4–8th) from normal skin and keloid tissues were seeded in 100-mm culture dishes and allowed to reach 70–80% confluency. Primary cultures were exposed to serum-free deprivation for 24 h followed by incubation with 50 ng/ml of TNF-α (Sigma Chemical, St Louis, Mo., USA) for an additional 24 h. Cells were then washed twice with PBS and scraped to isolate total RNA, using a commercially available kit to extract cellular RNA (Genflute Total Mamalian RNA Miniprep Kit, Sigma). Integrity and purity of the isolated RNA was verified by electrophoresis on an agarose gel. Concentrations were determined using a spectrophotometer and the integrity of the RNA samples verified by an OD 260/280 nm check. All total cellular RNA samples were stored at −70°C.
To characterize gene expression associated with the NFκB signal pathway, prefabricated cDNA arrays were employed (GEArray Q series purchased from SuperArray, Bethesda, Md., USA). Each GEArray Q Series Human NFκB Signaling Pathway Gene Array contains 96 genes related to NFκB-mediated signal transduction. SuperArray’s TrueLabeling-RT Kit was employed for the cDNA probe synthesis. Labeling was performed by reverse transcription reaction of 5 μg of total RNA isolates to 30 μCi of [α−32P]-dCTP (10 μCi/μl; 3,000 Ci/mmol; Amersham Pharmacia Bio Tech). The labeled probe was hybridized overnight with the array membrane. The membrane was washed twice with 2X SSC/1% SDS buffer (10 min) and twice with 2X SSC/0.1% SDS buffer (10 min). The array membrane was exposed to Kodak Biomax X-ray film (Kodak, Rochester, N.Y., USA) for 24 h. The array image was scanned and converted into raw data using the ScanAlyze software provided by Michael Eisen of Stanford University. Subsequent data were then analyzed by GEArray Analyzer software provided by SuperArray. Numerous housekeeping cDNAs were used as positive control for RNA normalization. The signal from a blank portion of the cDNA array was taken as background, and each individual gene expression on the arrays was adjusted for background. Results were described as means and ratios of three separate RNA samples taken from keloid and normal skin fibroblasts following TNF-α treatment (Table 2).
Western immunoblot analysis
Keloid and normal skin tissue homogenates were prepared as described previously [34]. For whole cell lysates, TNF-α stimulated and unstimulated dermal fibroblasts were harvested, washed in cold phosphate-buffered saline (PBS), and incubated in two packed cell volumes of buffer A (10 mM HEPES, pH 8.0, 1.5 mM MgCl2, 10 mM Kcl, 0.5 mM DTT, 200 mM sucrose, 0.5 mM phenylmethane-sulfonyl fluoride [PMSF]; 10 μl each of leupeptin and aprotinin per ml, 0.5% Nonidet P-40) for 5 min at 4°C. The protein concentration of the different cell lysates and tissue homogenates were measured using the BCA protein assay kit (Pierce, Rockford, Ill., USA) and analyzed by a spectrophotometer. The cell lysates were washed with TSA buffer and denatured by boiling for 10 min in 2X SDS. Fifty micrograms of either cell lysates or tissue homogenates were run on a 10% acrylamide SDS gels and transferred onto Hybond nitrocellulose membranes (Amersham, Life Sciences, Arlington Heights, Ill., USA). The western blots were incubated overnight with antibodies specific against TRAF1, TRAF2, NFκBp65, and cIAP-1 (inhibitor of apoptosis-1) (Santa Cruz, Calif., USA) (1:200). The immunoblots were detected by chemiluminescent kit (SuperSignal Pico, Pierce). Film exposures were done on Kodak X-OMAT-AR autoradiographic film.
Immunohistochemistry
The presence of TNF-α in normal skin and keloid tissues was examined using immunohistochemistry. Tissues were fixed in 10% buffered formalin and processed with standard procedures for paraffin embedding. Sections (5 μm) were cut and loaded on frosted slides; following deparaffinization, pretreatment in 10 mM Tris-hydrochloride, pH 10, in 90°C for 10–20 min was performed. Sections were left until they are completely cooled to room temperature (RT), then were washed with Tris-hydrochloride buffer and incubated in 10% goat serum in bovine serum albumin (BSA) for 30 min. Sections were incubated overnight at 4°C with mouse monoclonal IgG (negative control) and anti- TNF-α (R&D, Minneapolis, Minn., USA) followed by incubation with antimouse secondary antibody (DAKO, Carpentaria, Calif., USA) (1:200) for 1 h at RT. Detection was performed using Streptavidine Biotene detection kit following the manufacturer’s protocol (Vector Laboratories, Burlingame, Calif., USA) for 30 min at RT. Each step was followed by a brief wash with sterile phosphate buffer solutions. The reactions were visualized by using 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma, St Louis, Mo., USA), followed by counterstaining with hematoxylin for optimal evaluation of positive reactions.
Preparation of nuclear extracts
Following treatment with TNF-α (50 ng/ml) for 6 h [30], cellular extracts were prepared. The crude nuclei released by lysis from the treated cells were collected by microcentrifugation (15 s), rinsed once in buffer A, and resuspended in 2/3 packed cell volume of buffer C (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 1.0 mM DTT, 0.5 mM PMSF, 10 μl each of leupeptin and aprotinin per milliliter). Nuclei were incubated on a rocking platform at 4°C for 30 min and clarified by centrifugation for 5 min. The resulting supernatants were diluted 1:1 with buffer D (20 mM HEPES, pH 7.9, 100 mM Kcl, 0.2 mM EDTA, 20% glycerol, 1.0 mM DTT, 0.5 mM PMSF, 10 μl of each of leupeptin and aprotinin per milliliter) [31]. Nuclear extracts were frozen on dry ice, aliquoted, and stored at −80°C.
Electrophoretic mobility gel shift assays (EMSA)
The nuclear extracts (5 μg) were preincubated for 20 min at 25°C with poly (dI-dC) (80 μg/ml) in a buffer containing 25 mM HEPES-KOH (pH 7.9), 40 mM NaCl, 1 mM EDTA, 5% glycerol, and 1 mM dithiothrietol (DTT). Following preincubation, radiolabeled double-stranded oligonucleotide (10,000 cpm) were added in each reaction and incubated for 20 min at 25°C. The sequence of the oligonucleotides that corresponds to the NFκB consensus DNA-binding site is forward, 5′-AGTTGAGGGGACTTTCCCAGGC-3′; reverse, 5′-GCCTGGGAAAGTCCCCTCAACT-3′ (Santa Cruz Biotechnology, Santa Cruz, Calif., USA) [31]. The probe was radiolabeled with [γ−32P] ATP by T4 polynucleotide kinase (ICN Biochemicals, Irvine, Calif., USA). DNA/nucleoprotein complex specificity was determined by coincubation of nuclear extracts with unlabeled homologous. For competition experiments, 100-fold excess of unlabeled oligonucleotides was included in the binding reaction. The reactions were immediately loaded onto 5% nondenaturing polyacrylamide gels via electrophoresis in 0.5×Tris/borate/EDTA buffer, and the electrophoresis was performed at 4°C for 1 h. Gels were dried and exposed to film for 16–24 h.
Statistical analysis
All experiments were performed at least three times and in triplicates. Data were analyzed using Student’s paired t-test.
Results
Analysis of gene expression associated with NFκB signaling pathway in keloid and normal skin fibroblasts
To identify expression profiles of genes involved in the NFκB signaling pathway, we used NFκB cDNA superarray to examine expression levels of 96 full-length genes in dermal fibroblasts following treatment with and without TNF-α. To this purpose, primary fibroblasts derived from three representative sets of keloid lesions and their corresponding control skin borders of three unrelated patients were established and cultured for total RNA extraction. Our results showed that the majority of gene-expression signals did not exceed the background hybridization level nor did they reach at least twofold differences between keloid and NSK fibroblasts. However, 15 genes (15%) were up-regulated and 5 genes (5.25%) were down-regulated in keloid fibroblasts relative to their normal skin counterparts in response to TNF-α treatment (Table 2). On the contrary, in the absence of TNF-α, no differences in the constitutive expression of target genes were observed above the twofold threshold in keloid fibroblasts as compared with normal skin (data not shown). As demonstrated in Table 2, genes of interest that exhibited a twofold increase include TNF-α TRAF1, TRAF2, NFκB, IAP-1 and IAP-2, and the inflammatory cytokines IL-1 α and β and IL-6. These results were in accordance with recent studies demonstrating that proinflammatory and antiapoptotic gene expression in normal dermal keratinocytes and mesencymal cells are induced by NFκB activation [12]. The associated greater increase in antiapoptotic genes in keloid fibroblasts suggests that keloid fibroblast resistance to cell death could be due to an increase in antiapoptotic genes regulated by the NFκB pathway.
Elevated expression of TNF-α in keloid tissues and fibroblasts
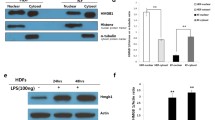
To confirm a constitutive elevation of TNF-α in keloid tissues and fibroblasts, Western blot analysis and immunohistochemistry were performed. Our results showed that keloid tissue homogenates contained a relatively higher level of TNF-α protein than normal skin (Fig. 1). The increased expression of TNF-α protein in keloid tissues was further confirmed by immunohistochemistry using a specific antibody against TNF-α, showing positively stained fibroblasts and mast cells (Fig. 2a, b), TNF-α is secreted in the ECM demonstrating the diffuse pattern of staining in the keloid ECM (Fig. 2a, b). Moreover, a similar pattern of elevated TNF-α level was also demonstrated by Western blot using cultured primary keloid fibroblasts derived directly from the above tissue samples (Fig. 1). The consistent findings of elevated TNF-α in both tissues and tissue-derived cells confirm that fundamental in vivo biological differences persist in vitro.
Western blot analysis of TNF-α protein expression in keloid tissues and their derived fibroblasts. Left hand panel homogenates were prepared from freshly biopsied keloid and normal skin tissues and Western blot analysis was performed to determine TNF-α protein levels; NSk normal skin, Kel keloid. Right hand panel whole cell lysates were isolated from primary cultured fibroblasts derived from the same sets of dermal tissues and Western blot was performed to determine TNF-α protein levels. Densitometric analysis was performed and the results were described as percentage relative to normal skin controls (100%). Data are expressed as the mean ± SEM and are representative of three independent experiments
Immunohistochemistry of paraffin-embedded tissue sections using an antibody specific against TNF-α protein. Strong positive signals are present in keloid tissues as compared with normal skin, positivity is also seen in the keloid extracellular matrix (ECM) as the antibody is against secretory TNF-α which is diffused in the keloid ECM. a Tissues at ×20 magnification. b Tissues at ×40 magnification. Arrows indicate mast cells and fibroblasts in keloid tissues, which are positively stained with TNF-α
Elevated expressions and activation of NFκB in keloid tissues and fibroblasts
Since keloid tissues and fibroblasts expressed a higher level of TNF-α, a well-known proinflammatory cytokine that exerts its biological effects by mediating the activation of NFκB signal pathways, we explored the expression of NFκB in keloid in vivo as well as in vitro. Results from Western blot analyses showed a consistent increase in the basal level of cytosolic NFκBp65/p50 isoforms in three separate keloid tissue homogenates as compared to their normal skin partners (Fig. 3a). Next, we investigated the translocation of NFκB to the nucleus of dermal fibroblasts, an essential process for its transcriptional activation. Nuclear extracts were prepared from both keloid and normal fibroblasts in the presence and absence of TNF-α (50 ng/ml) treatment for 30 min and Western blot analysis was performed. As shown in Fig. 3b, TNF-α treatment induced an increase in the nuclear level of NFκBp65/p50 isoforms in keloid fibroblasts compared to normal cells. However, a twofold to threefold elevated constitutive level of NFκB was observed in the nucleus of keloid fibroblasts, as compared to fibroblasts (Fig. 3b).
Western blot analysis of Rel-A/p65 in dermal tissues and nucleus of dermal fibroblasts. a Tissue lysates were extracted from three separate sets of freshly biopsied keloid tissues and their associated peripheral normal skin, and Western blot analysis was performed to determine the level of cytosolic Rel-A/p65 proteins. NSk normal skin, Kel keloid. b Nuclear extracts were isolated from primary cultured fibroblasts derived from the same sets of tissues with (+) or without TNF-α treatment (−) (50 ng/ml) for 30 min, and Western blot analysis was performed to determine the translocated nuclear Rel-A/p65 protein levels. Densitometric analysis was performed, and the results were described as intensity per pixel per area. Data are expressed as the mean ± SEM and are representative of three independent experiments
Determination of NFκB DNA-binding activity in dermal fibroblasts
We next examined the constitutive as well as TNF-α–induced DNA-binding activities of NFκB in dermal fibroblasts by using EMSA. Our results demonstrated an elevated basal level of NFκB activity in the nucleus of keloid fibroblasts as compared to normal skin (Fig. 4a, b). Although treatment with TNF-α led to an up-regulation of DNA-binding activities of NFκB in both normal and keloid fibroblasts, a greater enhancement was observed in keloid fibroblasts in response to TNF-α (Fig. 4a, b).
EMSA on DNA-binding activity of NFκB/p65 in keloid fibroblasts in response to TNF- α (50 ng/ml). a Nuclear extracts were prepared from keloid ( K) and normal skin (N) fibroblasts in the presence (+) or absence (−) of TNF-α (50 ng/ml) for 30 min. Then 5 μg of nuclear extract was submitted to EMSA for determination of the DNA-binding activity of NFκB/p65. Unlabeled NFκB/p65 and AP2 probes were used as nonspecific competitors. Nuclear extracts isolated from HeLa cells in response to TNF-α (50 ng/ml) served as a positive control. b Densitometric analysis of the bands and the results were described as percentage relative to the normal skin control without TNF-α treatment (100%). Data are expressed as the mean ± SEM and are representative of three independent experiments
Differential expression of NFκB-related proteins in keloid and normal fibroblasts
Results from our microarray assay have shown that treatment of keloid fibroblasts with TNF-α induced up-regulation of TRAF1, TRAF2, and cIAP-1 mRNA expressions (Table 2), all of which are important components associated with NFκB signaling pathways. To further confirm the effects of TNF-α on the expression of these NFκB-associated genes in keloid and normal fibroblasts, whole cell lysates were prepared from two sets of fibroblasts derived from both keloid tissues and their matched normal bordering skins, and Western blot analyses were performed. In contrast to results from microarray assays (Table 2), TNF-α did not modulate the expression of TRAF1 and TRAF2 at the protein level (Fig. 5), which could be due to posttranscriptional modification. However, our findings also showed that keloid fibroblasts consistently expressed an increased basal level of TRAF 1 and TRAF2 proteins as compared to their partners (Fig. 5). In addition, our results demonstrated a higher level of both basal and TNF-α–induced cIAP-1 protein expressions in keloid fibroblasts as compared with normal skin (Fig. 6).
Western blot analysis of TRAF-1 and TRAF-2 expressions in dermal fibroblasts in response to TNF-α treatment. Keloid and normal skin fibroblasts were exposed to TNF-α (50 ng/ml) for 24 h, and whole cell lysates were prepared for Western blot analysis on TRAF-1 (a) and TRAF-2 (b) protein levels. NSk normal skin, Kel keloid; with TNF-α treatment (−), without TNF-α treatment (+). Densitometric analyses were performed and the results were depicted as relative intensities (%) as compared with normal skin controls without TNF-α treatment (100%). Data are expressed as the mean ± SEM and are representative of three independent experiments
Western blot analysis of cIAP-1 protein expressions in dermal fibroblasts. Keloid and normal skin fibroblasts were exposed to TNF-α (50 ng/ml) for 24 h, and whole cell lysates were prepared for Western blot analysis on cIAP-1 protein levels. NSk normal skin, Kel keloid; with TNF-α treatment (−), without TNF-α treatment (+). Densitometric analyses were performed, and the results were depicted as relative intensities (%) as compared with normal skin controls without TNF-α treatment (100%). Data are expressed as the mean ± SEM and are representative of three independent experiments
Discussion
Previous studies in our laboratory have demonstrated a significant difference in apoptotic profiles between normal skin (NSk) and keloid fibroblasts, and that keloid fibroblasts are more resistant to cell death than their normal skin counterpart [20, 21, 24]. In view of the growing evidence on the role of NFκB in controlling apoptosis and inflammation, and the importance of both these factors during wound repair, we investigated the differential expressions of NFκB signal pathway related apoptotic genes in keloid tissues as well as their derived fibroblasts. We used the NFκB signal pathway cDNA microarray to profile changes in both the basal and TNF-α–induced gene expressions specifically associated with NFκB pathways, and found that approximately 15% of genes tested were up-regulated in keloid fibroblasts in comparison to NSk fibroblasts, among which most are proinflammatory cytokines including, IL-1α and β, TNF-α, IL-6, and antiapototic genes such as TRAF1, TRAF2, IAP-1, IAP-2, and xIAP (Table 2). Recently, Chen et al. 2003 [8] compared gene expression profiles between keloid and normal skin tissues using an 8,400-human-gene microarray and showed that approximately 4.79% of the genes exhibited altered expression, with 2.98% showing up-regulation and 1.81% showing down-regulation. Data presented in this study also showed that keloid tissues intrinsically or constitutively express an elevated level of NFκB pathway related proteins as compared with their normal skin partners. The elevated basal levels of these proteins in keloid tissues were recapitulated in their derived, in vitro cultured, keloid fibroblasts (Figs. 5 and 6). We also demonstrated that both keloid tissues (Fig. 3a) and keloid fibroblasts (Fig. 3b) showed constitutively higher levels of NFκB, as opposed to normal skin controls. The constitutive increase in the active NFκB isoforms in keloid fibroblasts was also confirmed by results from a DNA-binding assay using EMSA (Fig. 4a, b); similar constitutive NFκB activity has been found in several types of cancer such as breast, prostate, and colorectal cancers [18], which confirms the role of NFκB in the antiapoptotic regulatory pathway. Taken together, these findings suggest that activation of the NFκB pathway could play a role in keloid pathogenesis, partly by mechanisms involved in protecting keloid fibroblasts against apoptosis. Recently, the role of NFκB in skin biology has gained much interest, and studies have shown that the presence of NFκB in several dermal cells such as dermal keratinocytes and fibroblasts is essential in promoting inflammation and wound healing in response to trauma, findings which strengthen the role of NFκB in abnormal healing process and keloid formation [12, 14].
TNF-α, a homotrimer of 157 amino acid subunits, is a major mediator of apoptosis, inflammation, and immune responses [5]. Previous studies have established its involvement in the pathogenesis of a broad spectrum of human diseases, including sepsis, diabetes, osteoporosis, rheumatoid arthritis, inflammatory bowel disease, atherosclerosis, and a variety of cancers [33]. However, there is still a lack of convincing evidence to show whether TNF-α is involved in the pathogenesis of keloid, which is characterized by localized areas of prolonged inflammatory stage. In this study, our results showed that keloid tissues and their derived fibroblasts intrinsically express an elevated level of TNF-α mRNA (Table 2) and protein, as compared to their normal skin controls (Figs. 1 and 2), thus providing evidence that TNF-α may possibly play a role in the pathogenesis of keloid.
Binding to TNF-R1 results in the recruitment of various signal transducers such as TNF-receptor–associated death domain (TRADD), Fas-associated death domain (FADD), TRAF1, and TRAF2, and subsequently triggers a series of intracellular events leading to the activation of at least three distinct pathways [7]. For instance, caspase-8 is recruited by FADD to TNFR-1 complex, where it becomes activated and initiates a protease cascade that leads to apoptosis. TRAF2 recruits and activates cellular inhibitor of apoptosis protein-1 (cIAP-1) and cIAP-2, two important antiapoptotic proteins. Studies have shown that TRAF2 is also essential to the activation of extracellular signal-related kinase kinase 1 (MEKK1) or apoptosis-stimulated kinase 1 (ASK1), thereby activating a cascade of kinases leading to the activation of c-Jun NH2-terminal kinase (JNK), and increasing its transcriptional activity in response to various stresses [7]. The third arm of the TNF signaling network is the activation of the NFκB signal pathway. Activation of NFκB induced by TNF depends on the phosphorylation and degradation of IκB proteins, which retain NFκB within the cytoplasm of unstimulated cells [13]. Studies have shown that TRAF2 also plays an essential role in the activation of NFκB because overexpression of TRAF2 is sufficient to activate the signaling pathways leading to NFκB activation in the absence of extracellular stimuli [25]. Therefore, the ultimate biological activities of TNF-α rely on the existence and balance of extensive cross-talks between apoptosis, NFκB and JNK signaling pathways that emanate from TNF-R1. For example, absence of NFκB activity increases cellular susceptibility to TNF-α–induced apoptosis, whereas an enhancement of NFκB activation protects against apoptosis. Similarly, cells lacking NFκB exhibited a stronger and more prolonged activation of JNKs in response to TNF-α, and the products of several target genes of NFκB suppress the activation of JNK induced by TNF [7].
To date, the detailed mechanism by which NFκB protects cells against apoptosis is not well understood, but it is suggested that NFκB probably functions to regulate the expressions of genes whose protein products are involved in the regulation of apoptosis [32]. The most common antiapoptotic genes that are directly activated by NFκB include the cellular inhibitors of apoptosis (c-IAP1, c-IAP2, and IXAP), the TNF-receptor–associated factors (TRAF1 and TRAF2). In this study, our results showed a consistent increase in the basal levels of c-IAP-1, and TRAF1 and TRAF2 protein levels in keloid fibroblasts as compared to normal fibroblasts, further supporting the notion that activation of NFκB pathway in keloid may contribute, at least in part, to the prolonged and persistent inflammation as well as the disruption of the fibroblast apoptotic pathway that ultimately leads to keloid formation.
In conclusion, data presented here demonstrated that keloid tissues and fibroblasts expressed a constitutive elevation of TNF-α as well as several important components in the apoptotic-related down-stream signaling networks, including TRAF1, TRAF2, c-IAP1, and NFκB as compared to their associated normal skin partners. These findings have provided a partial explanation of the resistance of keloid fibroblasts to cell death as previously reported [20, 21, 24], and also underscore the concept of fibroblastic heterogeneity in keloid: keloid-type fibroblastic substrains, if they exist, might exhibit an intrinsically greater capacity for producing matrix, an altered responsiveness to biological response modifiers known to be present in healing wounds (such as TNF-α and TGF-β1), or an inability to respond to known death signaling genes. Moreover, the pattern of up-regulation of NFκB and TNF-α observed in keloid tissues was also apparent in primary cultured keloid fibroblasts, confirming that fundamental biological differences found in vivo also persist in vitro. Altogether, our present study has demonstrated the existence of an intrinsic activation of NFκB signal pathway in keloid tissues and their derived fibroblasts, thus contributing to an aberrant cellular proliferation and apoptosis in keloid pathogenesis. Therefore, inhibition of the NFκB signal pathway could lead to the development of potential therapeutic interventions for the treatment of keloid scarring.
References
Alster TS, Tanzi EL (2003) Hypertrophic scars and keloids: etiology and management. Am J Clin Dermatol 4:235–243
Appleton I, Brown NJ, Willoughby DA (1996) Short communication: apoptosis, necrosis, and proliferation. Possible implications in the etiology of keloids. Am J Pathol 149:1441–1446
Baldwin AS (2001) The transcription factor NF-κB and human disease. J Clin Invest 107(1):3–6
Baldwin AS, Mayo MW (2000) The transcription factor NF-κB: control of oncogenesis and cancer therapy resistance. Biochim Biophys Acta 1470:55–62
Baud V, Karin M (2001) Signal transduction by tumour necrosis factor and its relatives. Trends Cell Bio 11(9):372–377
Beg AA, Baltimore D (1996) An essential role for NFκB in preventing TNF-α induced cell death. Science 274:782–784
Chen GQ, Goeddel DV (2002) TNF-R1 signaling: a beautiful pathway. Science 296:1634–1635
Chen W, Fu X, Sun X, Sun T, Zhao Z, Sheng Z (2003) Analysis of differentially expressed genes in keloids and normal skin with cDNA microarray. J Surg Res 113:208–216
Choudon T, Sugihara T, Igawa HH, Funyama E, Furukawa H (2000) Keloid derived fibroblasts are refractory to Fas-mediated apoptosis, and neutralization of autocrine TGF-β 1 can abrogate this resistance. Am J Pathol 157:1661–1669
Ehrlich HP, Desmuliere A, Drgeiman RF, Cohen IK, Compton CC, Garner WL, Kapanci Y, Gabbiani G (1994) Morphological and immunochemical differences between keloid and hypertrophic scars. Am J Pathol 145:105
Granville DJ, Carthy CM, Hunt DC, McManus BM (1998) Apoptosis: molecular aspects of cell death and disease. Biol Dis 78:893–913
Hinata K, Gervin AM, Zhang YJ, Khavari PA (2003) Divergent gene regulation and growth effects by NF-κB in epithelial and mesenchymal cells of human skin. Oncogenes 22:1955–1964
Karin M, Lin A (2002) NF-κB at the crossroads of life and death. Nat Immunol 3(3):221–227
Kaufman CK, Fuchs E (2000) It’s got you covered: NF-κB in the epidermis. J Cell Biol 149(5):999–1004
Kischer CW, Shetlar MR, Chavpil M (1982) Hypertrophic scars and keloids: a review and new concept concerning their origin. Scan Elect Micros 4:1699–1713
Kyriakis J (2001) Life-or-death decisions. Nature 414:265–266
Ladin DA, Hou Z, Patel D et al (1998) p53 and apoptosis alterations in keloids and keloid fibroblasts. Wound Repair Regen 1(1):28–37
Lin A, Karin M (2003) NF-κB in cancer: a marked target. Semin Cancer Biol 13:107–114
Loetschcer H, Pan Y-CE, Lahm H-W, Gentz R, Brockhaus M, Tabuchi H, Lesslauer W (1990) Molecular cloning and expression of the human 55 kDa tumor necrosis factor receptor. Cell 61:351–359
Messadi DV, Le A, Berg S, Bertolami CN, Jewett A (1998) Role of apoptosis in keloid formation. Int J Oral Biol 23:31–36
Messadi DV, Jewett A, Le A, Berg S, Zhuang W, Bertolami CN (1999) Apoptosis associated genes and abnormal scar formation. Wound Repair Regen 7:511–517
Mohan RR, Mohan RR, Kim W, Wilson SE (2000) Modulation of TNF-α-induced apoptosis in corneal fibroblasts by transcription factor NF-κB. Invest Ophthalmol Vis Sci 41(6):1327–1335
Rothe M, Wong SC, Henzel WJ, Goeddel DV (1994) A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell 78:681–692
Sayah D, Soo C, Shaw W, Watson J, Messadi DV, Longaker MT, Zhang X, Ting K (1999) Downregulation of apoptosis-related genes in keloid tissues. J Surg Res 87:209–216
Song HY, Regnier CH, Kirschning CJ, Goeddel DV, Rothe M (1997) Tumor-necrosis factor-mediated kinase cascades: bifurcation of nuclear factor-κB and c-Jun N-terminal kinase (JNK/SAPK) pathways at TNF-receptor-associated factor-2. Proc Natl Acad Sci U S A 94:9792–9796
Tak PP, Firestein GS (2001) NF-κB: a key role in inflammatory diseases. J Clin Invest 107:7–11
Tartaglia LA, Goeddel DV (1992) Two TNF receptors. Immunol Today 13:151–153
Tredget EE, Nedelec B, Scott PG, Ghahary A (1997) Hypertrophic scars, keloids, and contractures: the cellular and molecular basis for therapy. Surg Clin North Am 77:701–707
Tuan TL, Nichter S (1998) The molecular basis of keloid and hypertrophic scar formation. Mol Med Today 4:19–24
Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM (1996) Suppression of TNF-α-induced apoptosis by NFκB. Science 274:787–789
Wang CY, Mayo MW, Korneluck RG, Goeddel DV, Baldwin AS Jr (1998) NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Sci Mag 281:1680–1683
Wang CY, Mayo MW, Baldwin AS Jr (2001) TNF- and cancer therapy induced apoptosis: potentiation by inhibition of NF-κB. Sci Mag 274(5288):784
Yamamoto Y, Gaynor RB (2001) Therapeutic potential of inhibition of the NF-κB pathway in treatment of inflammation and cancer. J Clin Investi 107(2):135–141
Zhang Q, Wu Y, Ann DK, Messadi DV, Tuan TL, Kelly PA, Bertolami CN, Le AD (2003) Mechanisms of hypoxic regulation of plasminogen activator inhibitor-1 gene expression in keloid fibroblasts. J Invest Dermatol 121:1005–1012 DOI:10.1046/j.1523-1747.2003.12564.x
Acknowledgments
This work was supported in part by grants from California Cancer Program (D. V. M.) and by the National Institutes of Health Research Grant, 1S11 AR47359 (A. Le).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Messadi, D.V., Doung, H.S., Zhang, Q. et al. Activation of NFκB signal pathways in keloid fibroblasts. Arch Dermatol Res 296, 125–133 (2004). https://doi.org/10.1007/s00403-004-0487-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00403-004-0487-y