Abstract
The TCF4 gene encodes for the basic helix–loop–helix transcription factor 4 (TCF4), which plays an important role in the development of the central nervous system (CNS). Haploinsufficiency of TCF4 was found to cause Pitt-Hopkins syndrome (PTHS), a severe neurodevelopmental disorder. Recently, the screening of a large cohort of medulloblastoma (MB), a highly aggressive embryonal brain tumor, revealed almost 20% of adult patients with MB of the Sonic hedgehog (SHH) subtype carrying somatic TCF4 mutations. Interestingly, many of these mutations have previously been detected as germline mutations in patients with PTHS. We show here that overexpression of wild-type TCF4 in vitro significantly suppresses cell proliferation in MB cells, whereas mutant TCF4 proteins do not to the same extent. Furthermore, RNA sequencing revealed significant upregulation of multiple well-known tumor suppressors upon expression of wild-type TCF4. In vivo, a prenatal knockout of Tcf4 in mice caused a significant increase in apoptosis accompanied by a decreased proliferation and failed migration of cerebellar granule neuron precursor cells (CGNP), which are thought to be the cells of origin for SHH MB. In contrast, postnatal in vitro and in vivo knockouts of Tcf4 with and without an additional constitutive activation of the SHH pathway led to significantly increased proliferation of CGNP or MB cells. Finally, publicly available data from human MB show that relatively low expression levels of TCF4 significantly correlate with a worse clinical outcome. These results not only point to time-specific roles of Tcf4 during cerebellar development but also suggest a functional linkage between TCF4 mutations and the formation of SHH MB, proposing that TCF4 acts as a tumor suppressor during postnatal stages of cerebellar development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription factor 4 (TCF4, also known as SEF2, ITF2, E2-2, ME2, and others) is a basic helix–loop–helix (bHLH) transcription factor that plays a crucial role in the differentiation and specification of various cell types and organs including the central nervous system (CNS) [26]. Mutations in TCF4 are the underlying cause of Pitt-Hopkins syndrome (PTHS) [83], and very recently, TCF4 was found to be frequently mutated in sporadic Sonic Hedgehog-associated medulloblastoma (SHH MB) [43, 59].
Eighteen TCF4 isoforms have been identified to date [71], all of whom contain a bHLH domain necessary for dimerization (homo- or heterodimer) and DNA binding [72]. Depending on the dimerization partner, TCF4 functions as a transcriptional activator or suppressor [24, 73]. Known binding partners include NeuroD2, ID2, and MATH1 (ATOH1) [25, 71]. TCF4 mRNA is ubiquitously expressed [71], but especially high levels can be detected throughout pre- and postnatal stages of CNS development in humans and mice [16, 74]. The relevance of TCF4 for neurodevelopment is further illustrated by the fact that mice homozygous for a Tcf4 knockout barely survive the first postnatal week [24].
Still, the particular role of TCF4 in neurodevelopment remains largely enigmatic. Recent animal studies propose that TCF4 is important for the plasticity and the development of hippocampal and cortical neurons [40], as well as TCF4 being part of the process of language production, comprehension, and recall [41]. Furthermore, Tcf4 is involved in dendrite development and outgrowth and synapse formation [15, 45]. Consequentially, TCF4 has been associated with different neurodevelopmental disorders [11, 32, 62], and TCF4 was found to interact with various genes linked to intellectual disability, the autism-spectrum disorder, and schizophrenia [56]. Most importantly, germline mutations in TCF4 were identified as the cause of Pitt-Hopkins syndrome (OMIM #610954) [3, 83].
PTHS is an autosomal-dominant, neurodevelopmental disorder characterized by facial dysmorphism, stereotypic movements, intellectual disability, seizures, and hyperventilation [63]. Very recently, the ‘First International Consensus Statement’ on diagnosis and management of PTHS has been published [82]. The prevalence of PTHS is thought to be 1/225.000–1/300.000 [82]. However, no reliable prevalence figures on PTHS are available. PTHS is seen in a 1:1 ratio in males and females and no preference for ethnicity has been observed [51]. Previous publications suspected PTHS patients to have an increased risk of death [32].Yet, causes of death remain questionable, with a few reported due to breathing abnormalities [63, 83], and current guidelines report typical life expectancy [82]. The mutational spectrum found in PTHS encompasses chromosomal deletions, partial gene deletions, frame-shift, splice site, missense, and nonsense mutations [63].
Although it was previously suggested that PTHS patients are at increased risk of developing malignancies, only three cases have been described so far [16, 82], including one case of a PTHS patient with MB (Blanluet et al. 2019). With respect to its role in tumor growth and progression, opposing functions for TCF4 have been proposed. While some studies support the idea that TCF4 exhibits oncogenic potential [5, 42, 57], more recent reports describe TCF4 as a tumor suppressor [10, 30, 34]. Interestingly, two recent studies on SHH MB showed that TCF4 is one of the most frequently mutated genes in adult SHH MB (8/53 > 16 years, 15% in Kool et al. 2014 and 13/48 > 16 years, 27% in Northcott et al. 2017) [43, 59].
MB is a tumor of the posterior fossa and the most common malignant CNS tumor in children [61], also accounting for 1% of all adult CNS tumors [28]. MB are a heterogeneous class of tumors and are subdivided into four distinct groups according to histological and genetic criteria, namely SHH, WNT, Group 3, and Group 4 MB [60]. While SHH and WNT MB are named after the deregulation of the respective signaling pathway, Group 3 and Group 4 MB cannot be traced down to a single pathway being the underlying cause of tumor development [47, 60]. SHH MB arise from cerebellar granule neuron precursor cells (CGNP) of the external granule cell layer (EGL) [70]. Intriguingly, TCF4 is known to be expressed in the CGNP of the EGL and interacts with the bHLH transcription factor MATH1 (ATOH1), which plays a crucial role in the formation of MB [24, 25]. The investigation of TCF4 during the development of SHH MB thus appears a promising approach to better understand and tackle this specific subset of CNS tumors.
To this end, we isolated and cultured CGNPs and investigated their proliferation behavior with and without a Tcf4 knockout. We used different transgenic mouse models to study the effects of a pre- as well as a postnatal Tcf4 knockdown on the development of the mouse brain and examined the proliferation of CGNPs in vivo. To examine the role of Tcf4 in the context of tumor development in vivo, we used a previously established mouse model for SHH MB [70] and introduced a postnatal knockout of Tcf4. While the prenatal knockdown of Tcf4 led to a significant decrease in proliferation of CGNPs, postnatal in vitro and in vivo knockdown of Tcf4 led to increased proliferation rates in CGNPs. This effect was also prominent in the context of a constitutively activated SHH pathway. Additionally, RNA sequencing (RNA-seq) of human SHH MB cells overexpressing TCF4 confirmed the upregulation of various genes involved in suppressing proliferation and cell-cycle control. Hence, our results suggest that TCF4 acts as a tumor suppressor in SHH MB following a postnatal knockout.
Materials and methods
Mice
hGFAP-cre (JAX #004600) [81], Math1-creERT2 (JAX #007684) [49] and SmoM2-YFPfl/fl (JAX #005130) [50] mice were obtained from The Jackson Laboratory (Bar Harbour, ME, USA). Tcf4 fl/fl mice have previously been generated and described [8]. hGFAP-cre::Tcf4 fl/fl, hGFAP-cre::Tcf4 fl/flSmoM2-YFPfl/+, Math1-creERT2::Tcf4 fl/fl, and Math1-creERT2::Tcf4 fl/flSmoM2-YFPfll+ mice were generated by crossing Tcf4 fl/fl with the respective mouse strains. Genotyping of genomic DNA from mouse ear biopsies or tail tips was performed by PCR. Mice were kept on a 12 h dark/light cycle; water and food was available ad libitum. Animals of both sexes were used for the experiments. All experimental procedures were approved by the Government of Upper Bavaria, Germany and Hamburg, Germany.
Mouse weights
Whole body weights of mice were measured every second day. Brain and cerebellar weights were determined separately on postnatal days 7, 14, and 21.
Mice treatments
To induce Cre activity, pups were injected intraperitoneally with 1 mg of tamoxifen (Sigma) dissolved in corn oil (Sigma) on postnatal day 5. For the in vivo analysis of proliferation, mice were pulse-labeled with BrdU (5-bromo-2-deoxyuridine; Sigma) at a concentration of 25 µg BrdU/g body weight 2 h before dissection.
Analysis of human tumors, survival analysis and differential methylation analysis
Analysis of TCF4 in DNA extracted from SHH MB from 15 adult patients was performed by Sanger sequencing of all coding exons (NM_003199) as described previously [84] and by MLPA in samples from 9 patients using the Kit P075 by MRC-Holland according to the manufacturer’s instructions. Analysis of mRNA and survival data was performed with the statistical programming language R [17]. Raw gene expression profiles published by Cavalli et al. [13] were obtained from Gene Expression Omnibus (GEO) [18] and preprocessed as described previously [9]. A total of 612 MB including 172 SHH samples with available gene expression and survival data were retained for the survival analysis and 215 SHH MB were used for the correlation analysis. Optimal cutoffs for survival analysis were determined using Cutoff Finder [12] with a minimum of ten samples per group. Kaplan–Meier analysis was performed with the R-package survival [77] and significance of difference in survival between groups was assessed with the logrank test. Proportional Hazards modeling as implemented by the R-function coxph was used for multivariate survival analysis, including TCF4 expression (as binary variable according to the optimal cutoff), age groups (0–3 years, 4–15 years, ≥ 16 years), metastatic status as well as TP53 mutational status. For validation of the cutoffs, an independent cohort of 396 cases with available survival and molecular subgroup information was obtained from Affymetrix U133P2 expression profiles of previously reported series through GEO accession numbers GSE10327 [44], GSE12992 [22], GSE37418 [68], GSE49243 [43], and published in Northcott et al. [59]. All data were MAS5.0 normalized and the datasets were combined using the genomics analysis and visualization platform R2 (http://r2.amc.nl). Probes corresponding to the same gene were averaged. To allow for the use of the same cutoffs, the expression of TCF4 was scaled to have the identical mean and standard deviation as in the initially used cohort published by Cavalli et al., 2017 [13].
For the differential methylation analysis, methylation profiles from 5 TCF4-mutated SHH MB were obtained together with 10 wild-type controls from GEO (GSE49243 [43] and GSE49576 [43]). CpG sites with an FDR < 5% were considered differentially expressed. Enrichment analysis of differential CpG sites was performed with the R-package methylGSA [66] using the function methylRRA with default parameters.
Cell culture
HEK293T (ATCC, Cat#CRL-3216) and DAOY (ATCC, Cat#HTB-186) cells were cultivated in DMEM (Dulbecco’s Modified Eagle Medium, PAN-Biotech) supplemented with 10% FCS (Invitrogen), 1% glutamax (Invitrogen), 1% HEPES (Sigma) and 1% Penicillin/Streptomycin 100x (Invitrogen) at 37 °C, 5% CO2. HEK293T cells were originally derived from a female donor [29]. DAOY cells were isolated from a desmoplastic cerebellar medulloblastoma of a 4-year-old Caucasian male [38]. DNA sequencing of DAOY cells revealed no mutations in TCF4, PTCH1, SUFU, or SMO. However, DAOY cells were previously reported to carry a TP53 mutation (COSMIC, https://cancer.sanger.ac.uk/cosmic). Cells were seeded in T-75 cell culture flasks and split 1:10 every 3–4 days.
CGNP cultures
For CGNP cultures, Tcf4fl/fl mice aged 5–8 days were used. Preparation of CGNP cultures was performed as described previously [55]. Production of retroviral particles used to induce the Tcf4 knockout was also described before [55]. Following incubation with viral supernatant, cells were grown in SHH-supplemented medium for another 24 h and pulsed with BrdU (Sigma, final concentration 25 µg BrdU/ml) 2 h prior to fixation with 4% paraformaldehyde (PFA).
Transfection
DAOY cells were seeded at a confluency of 40% the day before transfection. The transfection mix (50 µl OptiMEM (Gibco), 1.5 µg respective plasmid DNA, and 1.5 µl TransIT-2020 transfection reagent (Mirus)) was added to the cells. Two days post transfection, the DAOY cells were pulsed with BrdU (final concentration 25 µg BrdU/ml) 2 h prior fixation with 4% PFA.
Immunofluorescence
After fixating the cell cultures with 4% PFA, they were briefly washed with PBS. Subsequently, cells were treated with 4 N HCl and 0.1 M sodium borate for 10 min each. After blocking with 10% NGS in 0.3% Triton X-100 in PBS (PBS-T), cells were incubated at 4 °C over night with the following antibodies diluted in blocking solution: rabbit anti-GFP 1:200 (Santa Cruz Biotechnology, Cat#sc-8334, RRID:AB_641123), and mouse anti-BrdU 1:500 (MoBU-1, Thermo Fisher Scientific, Cat#B35128, RRID:AB_2536432), or rabbit anti-TCF4 1:500 (Sigma-Aldrich, Cat#HPA025958) and mouse anti-GFP 1:200 (Thermo Fisher Scientific, Cat#A11120, RRID:AB_221568). The next day, the cells were incubated with the following secondary antibodies diluted 1:500 in blocking buffer for 1 h at room temperature: anti-mouse Alexa488 (Invitrogen, Cat#A11029), anti-rabbit Alexa488 (Invitrogen, Cat#A11034), anti-rabbit Alexa546 (Invitrogen, Cat#A11035) or anti-mouse Alexa546 (Invitrogen, Cat#A11003). Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole, Roth, 1:1000 from a 1 mg/ml stock solution).
Immunohistochemistry
All stains on paraffin-embedded sections were performed on a Ventana System using standard protocols optimized for each antibody. Used antibodies were: mouse anti-Pax6 (Developmental Study Hybridoma Bank, PRID: AB_528427), rabbit anti-Casp3 (Asp175) (Cell Signaling Technology, Cat#9664, RRID: AB_2070042), rabbit anti-pHH3 (Ser10) (Cell Signaling Technology, Cat#9701, RRID: AB_2536432) and mouse anti-BrdU (MoBU-1, Thermo Fisher Scientific, Cat#B35128, RRID:AB_2536432). Nuclei were counterstained with hematoxylin.
Image quantifications
For all quantification of cellular markers in the EGL, ML, and IGL, pictures from lobules IV and V of the cerebellum were used, unless indicated otherwise. For embedded tumor tissue, CGNP and DAOY cell cultures, randomly picked areas were analyzed. Fractions of cell counts were determined by counting the total number of cells per field of vision/layer (identified by staining of hematoxylin or DAPI) and the number of cells that stained positive for the respective marker (chromogenic or fluorescent signal). For all other counts, i.e. cells/mm2, cells/mm and cells/section, only the absolute number of cells that were positive for the marker were included in the analysis. For all cell counts ImageJ software was used.
Cloning and site-directed-mutagenesis
A pcDNA3 plasmid containing the human TCF4 sequence (isoform ITF-2B−) was received from the group of Dr. Frank Kolligs (Helios Klinikum Berlin). For further use, the TCF4 sequence was cut out and inserted into the MSCV-IRES-GFP backbone (MSCV-IRES-GFP was a gift from Dr. Tannishtha Reya, Addgene plasmid #20672; http://n2t.net/addgene:20672; RRID: Addgene_20672). TCF4 mutants were generated through site-directed mutagenesis using overlap extension polymerase chain reaction [33]. The TCF4 mutants were generated in the pcDNA3 plasmid, extracted using restriction enzyme cloning and cloned into the MSCV-IRES-GFP backbone. Primer sets used for site-directed mutagenesis are available upon request. Successful cloning was verified by sequencing and control restriction digest.
In silico analysis of TCF4 variants
The TCF4 variants were analyzed with various in silico methods. First, ExAc (http://exac.broadinstitute.org/) and gnomAD (http://gnomad.broadinstitute.org/) databases were searched to test, whether the variant was present in healthy controls and obtains possible allele frequencies. To determine, if the TCF4 mutations had previously been found in monogenic disorders, ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/) were screened. Further, the data available on Catalogue of Somatic Mutations in Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic) were used to identify other tumors with the same TCF4 mutations. Finally, the Variant Effect Predictor on the Ensemble website (https://www.ensembl.org/Homo_sapiens/Tools/VEP) was used to obtain further information on the variants [54], such as SIFT and PolyPhen scores, and for the prediction of the variant effects. The inferred effects were annotated on the Criteria suggested by the American College of Medical Genetics and Genomics. Variants were given as ‘pathogenic’, if the mutation was seen in PTHS previously, absent from controls and the in silico analysis showed a strong impact on the protein function, as ‘likely pathogenic’ if the mutation was absent from controls and the in silico analysis showed a strong impact on the protein function, and as ‘variant of unknown significance (VUS)’, if the mutation was absent from healthy controls, but no clear impact on the function of the mutant protein could be inferred.
RNA sequencing
48 h after transfecting, DAOY cells transfected with either MSCV-TCF4WT-IRES-GFP, MSCV-TCF4V613F-IRES-GFP, or MSCV-IRES-GFP were sorted by FACS and RNA was isolated using the NucleoSpin RNA Kit (Macherey–Nagel) following the manufacturer’s protocol. After isolation of total RNA, the RNA integrity was analyzed with the RNA 6000 Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). From total RNA, mRNA was extracted using the NEBNext Poly(A) mRNA Magnetic Isolation module (New England Biolabs) and RNA-Seq libraries were generated using the NEXTFLEX Rapid Directional qRNA-Seq Kit (Bioo Scientific) as per the manufacturer's recommendations. Concentrations of all samples were measured with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and fragment lengths distribution of the final libraries was analyzed with the DNA High Sensitivity Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). All samples were normalized to 2 nM and pooled equimolar. The library pool was sequenced on the NextSeq 500 (Illumina) with 1 × 75 bp, with 13–20 mio reads per sample.
Raw Reads have been demultiplexed and trimmed with Illuminas bcl2fastq v2.18, and QC was done by FastQC [4]. Trimmed reads have been mapped to the human reference genome GRCh38 with STAR v2.5.3a [17] and counted per gene on thy fly via the ‘–quantmode GeneCounts’ parameter. Counts are based on the annotation Ensembl Release 95. Based on these counts, differentially expressed genes have been estimated with DESeq2 v1.18.1 [48]. A gene was called differentially expressed when FDR < 0.1 and log2FoldChange ≥ ± 1.
Statistical analyses
Statistical Analyses were performed using the Prism 5.02 and Prism 7.01 software (Graph Pad). All data are presented as mean ± SD. The level of significance for all statistical analysis was set to 5% (p ≤ 0.05), with *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001. Means of two groups were compared using two-tailed, unpaired t test (in case of normal distribution) or Mann–Whitney-U test, unless indicated otherwise. Chi-squared test was used to compare cell counts between groups. Kaplan–Meier plots were drawn to analyse the survival of patients; a logrank test was performed to test for significance of results. For all analysis, at least three samples (n = 3) were used.
Results
Mutated TCF4 proteins lose their function to suppress proliferation in a human SHH MB cell line
As previously described, TCF4 belongs to the most frequently mutated genes in SHH MB samples from adult patients [43]. Mutations in this gene encompass five different types [i.e. frame-shift (fs), deletion (del), nonsense (X), missense as well as splice site], affecting different domains of the protein. Figure 1a, Table 1, and Suppl. Table 1 give an overview on TCF4 mutations in SHH MB from previously published cohorts [43, 59] and from two additional tumors identified by us. To determine to what extent the functionality of TCF4 was hampered by these mutations and whether they could be accountable for any alterations in growth or progression of MB, we designed five of the identified mutations using site-directed mutagenesis. These mutations were cloned into an MSCV-IRES-GFP backbone and subsequently transfected into DAOY cells that had been derived from a desmoplastic human MB [38]. We then compared their proliferation rates with DAOY cells transfected with a vector coding for the wild-type (WT) TCF4. As further controls, we used untransfected DAOY cells as well as DAOY cells transfected with the empty MSCV-IRES-GFP vector to exclude any effects on the proliferation of the vector itself.
Overview of TCF4 mutations found in human medulloblastoma and their impact on DAOY cell proliferation. a Positions of TCF4 mutations identified in human MB [43, 59] shown on a scheme of TCF4 (Isoform TCF4-B−). Asterisks mark mutations also identified in the germline of patients with Pitt-Hopkins syndrome [51, 52, 79, 84] (in part, also documented at ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/). The G414fs mutation in bold has been detected in the patient described by Blanluet et al. (2019). Note that the found splice site mutations as well as the deletion resulting in a fusion protein (see Table 1) are not depicted in this figure. b TCF4 expression is not detectable in DAOY cells. c–i Transfection of DAOY cells with MSCV-IRES-GFP plasmids containing either no TCF4 (c), TCF4 WT (d) or TCF4 mutants shown in a (e–i). j–q DAOY cells untransfected (j) or transfected with MSCV-IRES-GFP plasmids containing either no TCF4 (k), TCF4 WT (l) or the TCF4 mutants shown in a (m–q) stained with antibodies against BrdU (red), GFP (green) and DAPI (blue). r Analysis of proliferation of transfected DAOY cells. Plotted were the fraction of BrdU+ cells from transduced cells (GFP+) and the baseline proliferation rate, respectively. Analysis was done comparing cell counts using Chi-squared tests. Each transfection was carried out three times (n = 3). Error bars show mean + SD. ****p ≤ 0.0001, **p ≤ 0.01, n.s. p > 0.05. AD1/2 transcription activation domain, NLS nuclear localization signal, bHLH basic helix-loop-helix domain. Scale bar in b corresponds to 25 µm for all images
Using antibodies against TCF4 we hardly detected any expression of the protein in untransfected DAOY cells or in DAOY cells transfected with the empty vector (Fig. 1b, c, respectively). However, TCF4 expression was clearly detectable, when WT TCF4 was introduced (Fig. 1d), as well as the TCF4 mutations 216del (Fig. 1g), 451del (Fig. 1h) and V613F (Fig. 1i). In line with this, RNA-sequencing did not reveal any expression of TCF4 mRNA in DAOY cells, but massive expression of TCF4 mRNA when transfected with respective vectors (data not shown). As expected, the used TCF4 antibody was not able to detect its epitope (AA sequence 468–525) when the two different truncating nonsense mutations R157X and R174X were introduced into the cells (Fig. 1e, f, respectively), although successful transcription of the vector was assessed due to the positive staining for GFP (green staining).
As summarized in Fig. 1r, analysis of the proliferation rates determined by a positive staining for 5-bromo-2-deoxyuridine (BrdU) showed that an introduction of the WT TCF4 significantly decreased the proliferation of DAOY cells (14 ± 3%, Fig. 1l) compared to untransfected cells (49 ± 2%, Fig. 1j). Mutants R157X (Fig. 1m) and R174X (Fig. 1n), which did not generate a functioning TCF4 variant detectable by the used antibody, showed no alteration in the proliferation rates compared to the control situations. Despite expressing a detectable TCF4, the other three introduced mutant TCF4 versions, namely 216del (Fig. 1o), 451del (Fig. 1p) and V613F (Fig. 1q) showed the following results: while the latter mutation showed no alteration in proliferation rates, 216del and 451del suppressed proliferation of DAOY cells only partially. However, compared to the WT TCF4, proliferation was still significantly elevated (Fig. 1r), supporting our hypothesis that mutant TCF4 variants have lost their proliferation suppressing function to some extent.
To better understand the lowered proliferation rates of TCF4 WT expressing DAOY cells, we performed RNA-seq. Comparing the transcriptome of DAOY cells expressing WT TCF4 with the transcriptome of DAOY cells expressing either a TCF4 mutant (V613F) or no TCF4 (empty vector), revealed a striking upregulation of various genes (Fig. 2a, left and central panel, respectively, see also Suppl. Table 2). In contrast, the transcriptome of DAOY cells expressing the mutant TCF4 largely corresponds to the one of DAOY cells transfected with an empty vector (Fig. 2a, right panel, see also Suppl. Table 2). Bioinformatic analyses confirmed these observations as we found 81 and 111 genes to be significantly upregulated in TCF4 WT expressing DAOY cells in comparison to those expressing the TCF4 mutant and empty vector controls, respectively (Fig. 2b and Suppl. Table 2). Interestingly, among those upregulated genes were several genes that have previously been described to be involved in either cell-cycle control or to be tumor suppressors themselves, e.g. CDKN1C [1, 39, 53], RASAL1 [46], CCND2 [20], RASSF6 [2, 31], and BMP4 [21, 80]. In contrast, only 10 genes were upregulated in TCF4 mutant expressing cells compared to empty vector controls (Fig. 2b and Suppl. Table 2). These findings underline our suspicion that TCF4 might act as a tumor suppressor.
TCF4 expression in DAOY cells leads to distinct changes of DAOY transcriptome. a Transfected DAOY cells expressing TCF4 WT show massive changes in their mRNA expression pattern when compared to DAOY cells expressing a non-functional TCF4 mutant (V613F) (left panel) or to DAOY cells transfected with an empty vector (center panel). In contrast, DAOY cells expressing TCF4 mutant (V613F) hardly differ from empty vector controls (right panel). b Bioinformatic analyses of RNA-sequencing data revealed 81 and 111 significantly differentially expressed genes in DAOY cells expressing TCF4 WT compared to DAOY cells expressing TCF4 mutant (V613F) and empty vector controls, respectively (for whole gene list see Suppl. Table 2). In contrast, when comparing DAOY cells expressing TCF4 mutant (V613F) and empty vector controls, only 9 genes are significantly expressed differentially (n = 3)
Prenatal loss of Tcf4 leads to reduced migration of CGNP and severe cerebellar hypoplasia in vivo
To investigate the influence of Tcf4 in cerebellar development, we established a new mouse model, knocking out Tcf4 in a large subset of neuronal cells. Thus, we bred mice expressing the Cre recombinase under the hGFAP promoter [81] with Tcf4 fl/fl mice [8] to generate hGFAP-cre::Tcf4 fl/fl mice. In comparison to other published mouse models with Tcf4 loss [40, 41, 76] our mice were viable following a homozygous knockout of Tcf4, giving us the opportunity to study a full loss of the gene in the CNS to elucidate the role of Tcf4 in early cerebellar development.
In a first step, we compared hGFAP-cre::Tcf4 fl/fl with hGFAP-cre mice phenotypically and noticed that mutant mice were lighter in weight (Fig. 3a). Weighing of whole brains and cerebella up to postnatal day 21 (P21) revealed that both are significantly lighter in the mutant mice (Fig. 3b, c, respectively). When we analyzed and compared brains of early postnatal and adult mice histologically, we observed a prominent change in cerebellar architecture, including a severe hypoplasia (Fig. 3d–g). To unravel the underlying mechanisms of the observed architectural changes, we performed several proliferation and apoptosis assays. First, we pulsed mice at P7 with BrdU 2 h before decapitation and stained sagittal sections for BrdU to compare proliferation rates of CGNPs in the EGL. Surprisingly, we detected a significantly lower fraction of BrdU+ cells in the EGL of mutant mice in comparison to the controls (Fig. 3h–j). This result strongly argued against a tumor suppressive role of Tcf4 in the cerebellum when knocked out during early embryonic stages. Staining the brains for Caspase 3 (CASP3) at P7 revealed no differences in the apoptosis rates (Fig. 3k–m). Since, under physiological conditions, CGNPs of the EGL migrate in an inwards directed manner to form the internal granular layer (IGL) [58], we wanted to determine whether the CGNPs of our mutant mice showed any deficits in their migratory behavior. We, therefore, pulsed the mice with BrdU again at P7, but this time killed them one week later at P14. As depicted in Fig. 3n and o, the mutant mice showed a striking number of BrdU+ cells in the molecular layer (ML) of the cerebellum. Quantification confirmed our observations, as mutant mice had a significantly elevated number of BrdU+ cells per mm ML as compared to control animals (Fig. 3p). In the last step, we wanted to further confirm the observed migrational deficit of CGNPs in hGFAP-cre::Tcf4 fl/fl mice at P21. As the migration of granule cells to the IGL should be completed by P20, we stained sagittal brain slices of 21-day-old mice for the granule cell marker paired box protein 6 (PAX6) [19]. In line with the previous results, we observed a significantly higher number of PAX6+ cells in ML of the mutant mice than in the control mice (Fig. 3q–s). In addition, the mutant mice showed a slightly but not significantly elevated number of cleaved CASP3+ cells at this stage of development in lobules VI/V, displaying higher apoptosis rates (Fig. 3t–v). However, number of CASP3+ cells was significantly higher in Lobules XI/X (data not shown). These results suggest that the migration of granule cells is either slowed down or not completed when Tcf4 is lost at an early stage of development, leading to severe architectural malformations of the cerebellum.
Prenatal deletion of Tcf4 leads to cerebellar hypoplasia and migration deficits. a–c Body weight (a), brain weight (b), and cerebellar weight (c) are significantly lower in Tcf4-deficient mice than in controls. Groups were compared using t tests for values at P21 only. For whole body weights, a one-tailed t test was used as patients with PTHS are known to be smaller and exhibit growth retardation. d–g Severe hypoplasia is observed in hGFAP-cre::Tcf4fl/fl mice. h–m Proliferation as measured by BrdU incorporation is significantly lower in mutant cerebellar granule cell precursors at postnatal day 7 (h, i, and j) while apoptosis appears unaffected (k–m). n–v Migration of granule cells into the internal granule cell layer is severely affected by a loss of Tcf4 as visualized by BrdU pulse chase experiments (n–p) and detection of significantly more PAX6+ granule in the molecular layer of 21-day-old mutant mice (q–s). Also, apoptosis was slightly higher in Tcf4-deficient animals (t–v). Analysis was done comparing cell counts using Chi-squared tests (n = 3). Error bars show mean ± SD. ****p ≤ 0.0001, **p ≤ 0.01, *p ≤ 0.05, n.s. p > 0.05. Scale bar in d corresponds to 250 µm in d and e, scale bar in f corresponds to 500 µm in f and g, scale bar in h corresponds to 25 µm in h, i, k, l, n, and o and scale bar in q corresponds to 50 µm in q, r, t, and u
Postnatal loss of Tcf4 leads to elevated proliferation rates of CGNPs in vitro and in vivo
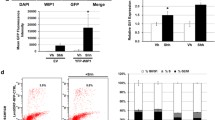
Due to the fact that most TCF4 mutations in SHH MB were detected in tumors from adult patients, we wanted to elucidate the effect of a postnatal Tcf4 knockdown on the proliferation of CGNPs, which are the cells of origin of MB [70]. First, we knocked out Tcf4 in CGNPs in vitro. To this end, we isolated and cultured CGNPs of Tcf4 fl/fl mice at postnatal days 5–8, when CGNP proliferation peaks [69] and subsequently transduced them either with a plasmid coding for a Cre recombinase, and therefore, disrupting the Tcf4 gene (MSCV-Cre-IRES-GFP) or an empty control plasmid (MSCV-IRES-GFP). Cells were pulsed with BrdU, fixed and then stained for BrdU and GFP to identify transduced and proliferating cells. As depicted in Fig. 4a–i, cells transduced with the MSCV-Cre-IRES-GFP plasmid, visualized through a positive GFP signal, showed a significantly higher fraction of BrdU+ cells than the cells transduced with the control plasmid. This result suggests that Tcf4 suppresses the proliferation of CGNPs at this stage of cerebellar development.
Postnatal knockout of Tcf4 increases proliferation of CGNP cells. a–c Granule cell precursors from Tcf4 fl/fl mice after transduction with IRES-GFP stained for DAPI (blue, a), GFP (green, b), and BrdU (red, c). d Merged image of (a–c). e–g Granule cell precursors of Tcf4 fl/fl mice after transduction with Cre-IRES-GFP stained for DAPI (blue, e), GFP (green, f), and BrdU (red, g). h Merged image of (e–g). i Analysis of proliferation of granule cell precursors after knockout of Tcf4 showed a significant increase in proliferation in mutant mice. j–p Proliferation of mutant granule cell precursors as measured by BrdU incorporation is significantly higher at P7 (m) and P15 (o) compared to controls (j and l, respectively, and p). No difference of proliferation was detectable at P12 (k and n). q–w In comparison to CGNPs with constitutive SHH pathway activation through SmoM2 (q–s), CGNPs with an additional loss of Tcf4 display significantly accelerated proliferation as measured by BrdU incorporation (t–v, also w). Cell counts were compared using a Chi-squared test (n = 5 in vitro, n = 3 in vivo). Error bars show mean + SD. ****p ≤ 0.0001, **p ≤ 0.01, *p ≤ 0.05, n.s. p > 0.05. Scale bar in a, j, and q corresponds to 25 µm in a–h, j–o, and q–v, respectively
To test whether this holds true for the situation in vivo, we made use of another mouse model. This time, mice expressed the fusion protein of the Cre recombinase and the estrogen receptor (creERT2) conditionally under the Math1 promoter [49]. Thus, Tcf4 disruption took place only upon administration of tamoxifen, a selective estrogen-receptor modulator, in cells expressing Math1, meaning predominantly CGNPs of the rhombic lip [49]. We injected tamoxifen at postnatal day 5 and killed the mice 2 h after pulsing them with BrdU on postnatal days 7, 12, or 15. Following the staining of the brain slices for BrdU, we analyzed the proliferation rates of the CGNPs in the EGL. When comparing the EGL of P7 and P15 control mice (Fig. 4j, l, respectively) with homozygous knockout mice of the same age (Fig. 4m, o, respectively) we detected a significantly higher fraction of BrdU+ cells in the mutant mice indicating higher proliferation rates (Fig. 4p). This difference was not detectable at P12 (Fig. 4k, n, and p). Still, these in vitro and in vivo results indicate that TCF4 may have a significant impact on regulating the proliferation of CGNPs at a critical time of the cerebellar development.
Reduced expression of TCF4 in murine and human SHH medulloblastoma increases proliferation and is associated with reduced survival
Eventually, since our approach arose from the finding that TCF4 mutations are in most MB cases occurring together with mutations within the SHH pathway, we started to investigate this particular scenario in vivo. For this purpose, we used the established SmoM2-YFPfl/fl mouse model [50] and bred them with Math1-creERT2::Tcf4 fl/fl mice to generate Math1-creERT2::Tcf4 fl/flSmoM2-YFPfl/+ mice. In these mice, upon injection of tamoxifen, Tcf4 is lost in the CGNP, while, simultaneously, the SHH pathway is constitutively activated due to an oncogenic mutation in the Smoothened (Smo) receptor (SmoM2). To identify, if the loss of Tcf4 has an impact on the tumor cell proliferation rates and the development of a tumor in this setting, we used Math1-creERT2::SmoM2-YFPfl/+ mice as controls. For investigation of the proliferation rates, we used the same approach as described above. As depicted in Fig. 4q–w, we detected a significantly elevated fraction of BrdU+ cells in the EGL of Math1-creERT2::Tcf4 fl/flSmoM2-YFPfl/+ mice on postnatal days 7, 12, and 15 (Fig. 4t–v, respectively) in comparison to their age-matched controls (Fig. 4q–s, respectively). Of note, the difference between the proliferation rates of Tcf4-deficient and Tcf4 wild-type cells was strongly decreasing between postnatal days 12 and 15 (Fig. 4w). We, therefore, conclude that a loss of Tcf4 has not only an impact on the proliferation of CGNPs in healthy developing mice, but also in a tumorigenic setting mimicking the very beginnings within SHH MB development.
After having observed a significant increase in proliferation in murine tumor cells with a Tcf4 deletion, we finally wondered, whether lower expression levels of TCF4 mRNA in human MB samples would be associated with a worse prognosis. Using publicly available data from 612 MB patients [13], we did not find significant differences in the patients’ overall survival when dichotomizing the cohort using the median expression. However, after calculation for the most meaningful cutoff with one group consisting of at least ten patients, we found significant differences regarding overall survival. The 185 patients with lower expression of TCF4 survived significantly shorter than the remaining 427 patients with higher expression of TCF4 (p = 0.0019, Fig. 5a). Since this effect was very likely to be confounded by the fact that Group 3 MB with a well-known bad prognosis show the lowest levels of TCF4 mRNA among MB subgroups (Fig. 5b), we also looked at SHH MB separately, being the subgroup carrying TCF4 mutations. By again using the most meaningful cutoff, this subgroup of MB still included a minority of 19 patients with low expression of TCF4 and a significantly worse overall survival than the remaining 153 patients with higher TCF4 expression (Fig. 5c, p = 0.00085). As multiple cutoff testing might yield to an overestimation of the significance, we used an independent cohort of 396 MB from other previously reported series to validate the identified cutoffs. The expression of TCF4 was scaled to the same mean and standard deviation as in the initially used cohort and the same cutoffs were applied for the survival analysis without any further cutoff testing. This yielded very similar results and corroborates the relevance of the identified cutoffs (Supp. Figure 1). In multivariate survival analyses of the initial cohort of 612 samples, TP53 mutations and metastatic disease were the most significant adverse prognostic factors (p = 0.0057 and p = 0.0023, respectively) followed by low TCF4 expression levels (p = 0.014). Age groups had no significant influence on overall survival (Fig. 5d). In addition, correlation analyses in human MB samples revealed that in infant SHH MB there were significantly more genes correlating with TCF4 expression (1722/20659) than in children or adult SHH MB cases (507/20659 and 489/20659, respectively) (p < 2.2e−16). We next wanted to know, if these TCF4-correlating genes were also upregulated in the mRNA from DAOY cells overexpressing TCF4. A significant overlap was found for the infant SHH MB (p = 1.62e−7), but not for the other age groups. Lastly, we analyzed publicly available methylation data from five human SHH MB samples with TCF4 mutation and compared them to 10 SHH MB samples with wild-type TCF4. We detected 189 differentially expressed CpG sites and enrichment analysis subsequently revealed 52 enriched categories that were related to transcription factor activity and brain development (Suppl. Figure 2 and Suppl. Table 3). These results suggest that TCF4 signalling is properly functioning in infant MB and might be impaired in older patients, the tumors of whom harbour TCF4 mutations and significantly more frequently display low TCF4 expression (p = 0.00094).
Low expression of TCF4 is associated with poor survival in SHH MB patients. a–c In MB patients the association between low TCF4 expression and poor survival (a) is likely to be confounded by low TCF4 expression in Group 3 MB (b), which are known to have a bad prognosis. Still, separate analysis of patients with SHH MB, where TCF4 mutations are actually detected, revealed the same poor survival of patients with low TCF4 expression (c). d Multivariate survival analysis revealed TP53 mutations, metastatic disease and low TCF4 expression levels were the most significant adverse prognostic factors (p = 0.0057, p = 0.0023, and p = 0.014, respectively). Age groups had no significant influence on overall survival
Discussion
As opposed to the situation in children, MB plays only a minor part among the adult CNS tumors [28], but research on the underlying mechanisms of MB development and classification has flourished the last years emphasizing the heterogeneity of the different MB subgroups [13]. Personalized therapy is desirable since patients still suffer from the common side-effects of classical chemotherapy. Recent studies by Kool et al. in 2014 and Northcott et al. in 2017 revealed that among adult patients with SHH MB, the most prevalent subtype in adult MB, 15% (8/53) and 27% (13/48) showed a mutation in the TCF4 gene coding for the Transcription factor 4, respectively [43, 59]. Germline mutations in TCF4 are the cause of Pitt-Hopkins syndrome (PTHS), a severe neurodevelopmental disorder [83], and, interestingly enough, 7 out of 16 mutations observed in MB samples have previously been identified in the germline of patients with PTHS (Fig. 1a, Table 1, and Suppl. Table 1). One might, therefore, speculate that these mutations indeed predispose to SHH MB and a respective surveillance might be appropriate. In line with this, Blanluet et al. describe a new patient with PTHS, who developed a SHH MB at the age of 27 (Blanluet et al. 2019). However, except for this case, there is no evidence so far that patients with Pitt Hopkins have an increased risk for tumor development, and Waszak et al. did not identify any TCF4 germline mutations in approximately 50 adult patients with SHH MB [78]. Our findings are in accordance with observations that there are several recurring genes and pathways implicated in both neurodevelopmental disorders (germline de novo mutations) and different forms of cancer (somatic mutations) [36]. However, individuals with neurodevelopmental disorders carrying a pathogenic variant in any of these genes do not necessarily have an increased tumor risk. In fact, timing, genetic background and cellular context might have an important contribution on the outcome of de novo variants in these genes and this is supported by the opposing roles of Tcf4 that we describe during mouse cerebellar development.
We analyzed mRNA expression data of human MB and discovered that high levels of TCF4 mRNA were favorable for the patients’ outcome. A study from 2015 on colorectal carcinoma patients demonstrated the same correlation of high TCF4 expression and a better clinical outcome, underlining the potential role of TCF4 as a tumor suppressor [10]. Already in 2009, Herbst et al. proposed that TCF4 shows tumor suppressive function through inducing cell cycle arrest in colorectal cancer cells [35]. We were able to further confirm this hypothesis by introducing different TCF4 mutations into a human SHH MB cell line (DAOY). Whereas introducing WT TCF4 into DAOY cells led to a significantly decreased number of proliferating cells, the introduced mutant TCF4 versions seemed to have lost their suppressive function. Accordingly, when we performed RNA-seq on DAOY cells overexpressing wild-type TCF4 we detected several genes involved in tumor suppression and cell-cycle control to be upregulated, which was not the case in DAOY cells overexpressing a TCF4 mutant. Certainly, these in vitro findings must be interpreted with caution. Firstly, DAOY cells are originally from a MB of a young boy [38], while the TCF4 mutations are almost exclusively found in adult SHH MB. As none of the currently available SHH MB cell lines is derived from an adult patient [37], the often used DAOY cell line still seemed like the best approach for initial studies. Secondly, as other groups even suggest that TCF4 shows oncogenic potential [5, 57], further in vivo studies on the role of TCF4 in brain and especially tumor development were inevitable.
In a first step, we established the hGFAP-cre::Tcf4 fl/fl transgenic mouse, in which TCF4 is homozygously knocked-out at around embryonic day 13.5, as soon as the hGFAP promoter becomes active [81]. Of note, a homozygous knockout of Tcf4 in mice is not identical to the situation in human patients with PTHS, who usually carry heterozygous TCF4 mutations. Indeed, our in vitro results demonstrate that heterozygous mutations are sufficient to cause functional impairments. However, at least for the development of the CNS and the formation of MB in mice, such impairments could only be modeled by a homozygous knockout. In contrast, a heterozygous knockout remained without any obvious phenotype, possibly because of a lack of the dominant negative effect that is conveyed by specific heterozygous mutations in human patients with Pitt-Hopkins syndrome or MB. Measurements of whole mouse brain and cerebellar weights on different time points after birth and histological analysis of mutant mice brains revealed that a homozygous loss of Tcf4 not only led to a severe cerebellar hypoplasia and significantly lower number of proliferating granule cells in the EGL, but also to a severe migrational deficit of those cells from the EGL to the IGL in the following days of development. During normal development of the cerebellum granule cells migrate into the inner regions of the cerebellar cortex along the radial fibers of the Bergmann glia [7, 19]. The used hGFAP promoter is expressed in the neural progenitors that generate a variety of cerebellar cell types including granule neuron precursors, interneurons or radial glia [70, 75]. It is, therefore, feasible to suggest that not only the granule cells, but also the glia is affected by the loss of Tcf4 and contributes to the observed phenotype. This would explain why some of the architectural alterations were not present in the Math1-creERT2::Tcf4 fl/fl mice and also why Math1-cre::Tcf4 fl/fl mice show a much milder phenotype (data not shown).
As the detected TCF4 mutations in SHH MB are somatic, late occurring mutations, we then analyzed the postnatal knockout of Tcf4 and were able to show elevated levels of proliferation in CGNPs both in vitro and in vivo, also in the context of a constitutively activated SHH pathway as found in SHH MB. This is in contrast to the data obtained with a prenatal Tcf4 loss. However, it is known that depending on its dimerization partner, TCF4 can display transcriptionally activating as well as suppressing functions [24, 73]. It is, therefore, possible that the function of TCF4 is time-sensitive and TCF4 exhibits opposing effects, as dimerization partners may not only change in a spatial but also in a temporal manner. Similar findings of opposing and time-sensitive effects of mutations in CREBBP (CREB binding protein), also in the context of SHH MB, have been published recently [55]. Likewise, members of the Bone morphogenic protein (BMP) family, which have been shown to interact with Tcf4 [14, 27], and are upregulated by TCF4 in our in vitro experiments, may promote tumor cell survival in some circumstances [23], but have also been shown to antagonize SHH-mediated proliferation of CGNPs and to induce their differentiation [67, 80].
In summary, the data presented in this manuscript suggest that TCF4 plays a subtle, but significant role in the differentiating process of CGNPs and acts as a tumor suppressor postnatally. Still, further research is needed to elucidate how the opposing functions of Tcf4 and the interaction with other proteins can be explained and to fully understand the role of TCF4 in the context of MB development.
References
Algar EM, Muscat A, Dagar V, Rickert C, Chow CW, Biegel JA et al (2009) Imprinted CDKN1C is a tumor suppressor in rhabdoid tumor and activated by restoration of SMARCB1 and histone deacetylase inhibitors. PLoS One 4:e4482. https://doi.org/10.1371/journal.pone.0004482
Allen NP, Donninger H, Vos MD, Eckfeld K, Hesson L, Gordon L et al (2007) RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene 26:6203–6211. https://doi.org/10.1038/sj.onc.1210440
Amiel J, Rio M, de Pontual L, Redon R, Malan V, Boddaert N et al (2007) Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet 80:988–993. https://doi.org/10.1086/515582
Andrews S (2014) FastQC a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Appaiah H, Bhat-Nakshatri P, Mehta R, Thorat M, Badve S, Nakshatri H (2010) ITF2 is a target of CXCR4 in MDA-MB-231 breast cancer cells and is associated with reduced survival in estrogen receptor-negative breast cancer. Cancer Biol Ther 10:600–614
Blanluet M, Masliah-Planchon J, Giurgea I, Bielle F, Girard E, Andrianteranagna M et al (2019) SHH medulloblastoma in a young adult with a TCF4 germline pathogenic variation. Acta Neuropathol. https://doi.org/10.1007/s00401-019-01983-4
Benard M, Lebon A, Komuro H, Vaudry D, Galas L (2015) Ex vivo imaging of postnatal cerebellar granule cell migration using confocal macroscopy. J Vis Exp. https://doi.org/10.3791/52810
Bergqvist I, Eriksson M, Saarikettu J, Eriksson B, Corneliussen B, Grundstrom T et al (2000) The basic helix-loop-helix transcription factor E2-2 is involved in T lymphocyte development. Eur J Immunol 30:2857–2863. https://doi.org/10.1002/1521-4141(200010)30:10%3c2857:Aid-immu2857%3e3.0.Co;2-g
Bockmayr M, Mohme M, Klauschen F, Winkler B, Budczies J, Rutkowski S et al (2018) Subgroup-specific immune and stromal microenvironment in medulloblastoma. Oncoimmunology 7:e1462430. https://doi.org/10.1080/2162402x.2018.1462430
Brandl L, Horst D, de Toni E, Kirchner T, Herbst A, Kolligs FT (2015) ITF-2B protein levels are correlated with favorable prognosis in patients with colorectal carcinomas. Am J Cancer Res 5:2241–2248
Brzozka MM, Radyushkin K, Wichert SP, Ehrenreich H, Rossner MJ (2010) Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol Psychiatry 68:33–40. https://doi.org/10.1016/j.biopsych.2010.03.015
Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S et al (2012) Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One 7:e51862. https://doi.org/10.1371/journal.pone.0051862
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B et al (2017) Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31:737.e736–754.e736. https://doi.org/10.1016/j.ccell.2017.05.005
Chen T, Wu Q, Zhang Y, Lu T, Yue W, Zhang D (2016) Tcf4 controls neuronal migration of the cerebral cortex through regulation of Bmp7. Front Mol Neurosci 9:94. https://doi.org/10.3389/fnmol.2016.00094
D’Rozario M, Zhang T, Waddell EA, Zhang Y, Sahin C, Sharoni M et al (2016) Type I bHLH proteins daughterless and Tcf4 restrict neurite branching and synapse formation by repressing neurexin in postmitotic neurons. Cell Rep 15:386–397. https://doi.org/10.1016/j.celrep.2016.03.034
de Pontual L, Mathieu Y, Golzio C, Rio M, Malan V, Boddaert N et al (2009) Mutational, functional, and expression studies of the TCF4 gene in Pitt–Hopkins syndrome. Hum Mutat 30:669–676. https://doi.org/10.1002/humu.20935
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210
Edmondson JC, Hatten ME (1987) Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J Neurosci 7:1928–1934
Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B et al (2001) Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res 61:2782–2787
Fang WT, Fan CC, Li SM, Jang TH, Lin HP, Shih NY et al (2014) Downregulation of a putative tumor suppressor BMP4 by SOX2 promotes growth of lung squamous cell carcinoma. Int J Cancer 135:809–819. https://doi.org/10.1002/ijc.28734
Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S et al (2009) Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol 218:86–94. https://doi.org/10.1002/path.2514
Fiaschetti G, Castelletti D, Zoller S, Schramm A, Schroeder C, Nagaishi M et al (2011) Bone morphogenetic protein-7 is a MYC target with prosurvival functions in childhood medulloblastoma. Oncogene 30:2823–2835. https://doi.org/10.1038/onc.2011.10
Flora A, Garcia JJ, Thaller C, Zoghbi HY (2007) The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc Natl Acad Sci USA 104:15382–15387. https://doi.org/10.1073/pnas.0707456104
Flora A, Klisch TJ, Schuster G, Zoghbi HY (2009) Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science 326:1424–1427. https://doi.org/10.1126/science.1181453
Forrest M, Chapman RM, Doyle AM, Tinsley CL, Waite A, Blake DJ (2012) Functional analysis of TCF4 missense mutations that cause Pitt-Hopkins syndrome. Hum Mutat 33:1676–1686. https://doi.org/10.1002/humu.22160
Forrest MP, Waite AJ, Martin-Rendon E, Blake DJ (2013) Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One 8:e73169. https://doi.org/10.1371/journal.pone.0073169
Giordana MT, Schiffer P, Lanotte M, Girardi P, Chio A (1999) Epidemiology of adult medulloblastoma. Int J Cancer 80:689–692
Graham FL, Smiley J, Russell WC, Nairn R (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36:59–74. https://doi.org/10.1099/0022-1317-36-1-59
Grill JI, Herbst A, Brandl L, Kong L, Schneider MR, Kirchner T et al (2015) Inactivation of Itf2 promotes intestinal tumorigenesis in Apc(Min/+) mice. Biochem Biophys Res Commun 461:249–253. https://doi.org/10.1016/j.bbrc.2015.04.009
Harvey K, Tapon N (2007) The Salvador–Warts–Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer 7:182–191. https://doi.org/10.1038/nrc2070
Hasi M, Soileau B, Sebold C, Hill A, Hale DE, O’Donnell L et al (2011) The role of the TCF4 gene in the phenotype of individuals with 18q segmental deletions. Hum Genet 130:777–787. https://doi.org/10.1007/s00439-011-1020-y
Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. https://doi.org/10.1038/nprot.2007.132
Herbst A, Bommer GT, Kriegl L, Jung A, Behrens A, Csanadi E et al (2009) ITF-2 is disrupted via allelic loss of chromosome 18q21, and ITF-2B expression is lost at the adenoma-carcinoma transition. Gastroenterology 137:639–648. https://doi.org/10.1053/j.gastro.2009.04.049
Herbst A, Helferich S, Behrens A, Goke B, Kolligs FT (2009) The transcription factor ITF-2A induces cell cycle arrest via p21(Cip1). Biochem Biophys Res Commun 387:736–740. https://doi.org/10.1016/j.bbrc.2009.07.102
Hoischen A, Krumm N, Eichler EE (2014) Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci 17:764–772. https://doi.org/10.1038/nn.3703
Ivanov DP, Coyle B, Walker DA, Grabowska AM (2016) In vitro models of medulloblastoma: choosing the right tool for the job. J Biotechnol 236:10–25. https://doi.org/10.1016/j.jbiotec.2016.07.028
Jacobsen PF, Jenkyn DJ, Papadimitriou JM (1985) Establishment of a human medulloblastoma cell line and its heterotransplantation into nude mice. J Neuropathol Exp Neurol 44:472–485. https://doi.org/10.1097/00005072-198509000-00003
Jia H, Cong Q, Chua JF, Liu H, Xia X, Zhang X et al (2015) p57Kip2 is an unrecognized DNA damage response effector molecule that functions in tumor suppression and chemoresistance. Oncogene 34:3568–3581. https://doi.org/10.1038/onc.2014.287
Jung M, Haberle BM, Tschaikowsky T, Wittmann MT, Balta EA, Stadler VC et al (2018) Analysis of the expression pattern of the schizophrenia-risk and intellectual disability gene TCF4 in the developing and adult brain suggests a role in development and plasticity of cortical and hippocampal neurons. Mol Autism 9:20. https://doi.org/10.1186/s13229-018-0200-1
Kennedy AJ, Rahn EJ, Paulukaitis BS, Savell KE, Kordasiewicz HB, Wang J et al (2016) Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Rep 16:2666–2685. https://doi.org/10.1016/j.celrep.2016.08.004
Kolligs FT, Nieman MT, Winer I, Hu G, Van Mater D, Feng Y et al (2002) ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell 1:145–155
Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V et al (2014) Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25:393–405. https://doi.org/10.1016/j.ccr.2014.02.004
Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P et al (2008) Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One 3:e3088. https://doi.org/10.1371/journal.pone.0003088
Li H, Zhu Y, Morozov YM, Chen X, Page SC, Rannals MD et al (2019) Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol Psychiatry. https://doi.org/10.1038/s41380-019-0353-0
Liu D, Yang C, Bojdani E, Murugan AK, Xing M (2013) Identification of RASAL1 as a major tumor suppressor gene in thyroid cancer. J Natl Cancer Inst 105:1617–1627. https://doi.org/10.1093/jnci/djt249
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al (2016) The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Machold R, Fishell G (2005) Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48:17–24. https://doi.org/10.1016/j.neuron.2005.08.028
Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D et al (2006) A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res 66:10171–10178. https://doi.org/10.1158/0008-5472.Can-06-0657
Marangi G, Ricciardi S, Orteschi D, Lattante S, Murdolo M, Dallapiccola B et al (2011) The Pitt–Hopkins syndrome: report of 16 new patients and clinical diagnostic criteria. Am J Med Genet A 155A:1536–1545. https://doi.org/10.1002/ajmg.a.34070
Marangi G, Ricciardi S, Orteschi D, Tenconi R, Monica MD, Scarano G et al (2012) Proposal of a clinical score for the molecular test for Pitt–Hopkins syndrome. Am J Med Genet A 158a:1604–1611. https://doi.org/10.1002/ajmg.a.35419
Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A et al (1995) p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev 9:650–662
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A et al (2016) The ensembl variant effect predictor. Genome Biol 17:122. https://doi.org/10.1186/s13059-016-0974-4
Merk DJ, Ohli J, Merk ND, Thatikonda V, Morrissy S, Schoof M et al (2018) Opposing effects of CREBBP mutations govern the phenotype of rubinstein–Taybi syndrome and adult SHH medulloblastoma. Dev Cell 44:709.e706–724.e706. https://doi.org/10.1016/j.devcel.2018.02.012
Moen MJ, Adams HH, Brandsma JH, Dekkers DH, Akinci U, Karkampouna S et al (2017) An interaction network of mental disorder proteins in neural stem cells. Transl Psychiatry 7:e1082. https://doi.org/10.1038/tp.2017.52
Mologni L, Dekhil H, Ceccon M, Purgante S, Lan C, Cleris L et al (2010) Colorectal tumors are effectively eradicated by combined inhibition of {beta}-catenin, KRAS, and the oncogenic transcription factor ITF2. Cancer Res 70:7253–7263. https://doi.org/10.1158/0008-5472.CAN-10-1108
Muller F, O’Rahilly R (1990) The human brain at stages 21–23, with particular reference to the cerebral cortical plate and to the development of the cerebellum. Anat Embryol (Berl) 182:375–400
Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T et al (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547:311–317. https://doi.org/10.1038/nature22973
Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ et al (2012) Medulloblastomics: the end of the beginning. Nat Rev Cancer 12:818–834. https://doi.org/10.1038/nrc3410
Northcott PA, Robinson GW, Kratz CP, Mabbott DJ, Pomeroy SL, Clifford SC et al (2019) Medulloblastoma. Nat Rev Dis Primers 5:11. https://doi.org/10.1038/s41572-019-0063-6
O’Donnell L, Soileau B, Heard P, Carter E, Sebold C, Gelfond J et al (2010) Genetic determinants of autism in individuals with deletions of 18q. Hum Genet 128:155–164. https://doi.org/10.1007/s00439-010-0839-y
Peippo M, Ignatius J (2011) Pitt-Hopkins syndrome. Mol Syndromol. https://doi.org/10.1159/000335287
Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J et al (2012) Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. https://doi.org/10.1038/nature11329
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-proje ct.org/
Ren X, Kuan PF (2018) methylGSA: a Bioconductor package and Shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty892
Rios I, Alvarez-Rodriguez R, Marti E, Pons S (2004) Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development 131:3159–3168. https://doi.org/10.1242/dev.01188
Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L et al (2012) Novel mutations target distinct subgroups of medulloblastoma. Nature 488:43–48. https://doi.org/10.1038/nature11213
Roussel MF, Hatten ME (2011) Cerebellum development and medulloblastoma. Curr Top Dev Biol 94:235–282. https://doi.org/10.1016/b978-0-12-380916-2.00008-5
Schüller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG et al (2008) Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14:123–134. https://doi.org/10.1016/j.ccr.2008.07.005
Sepp M, Kannike K, Eesmaa A, Urb M, Timmusk T (2011) Functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5′ exon usage and splicing. PLoS One 6:e22138. https://doi.org/10.1371/journal.pone.0022138
Sepp M, Pruunsild P, Timmusk T (2012) Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum Mol Genet 21:2873–2888. https://doi.org/10.1093/hmg/dds112
Skerjanc IS, Truong J, Filion P, McBurney MW (1996) A splice variant of the ITF-2 transcript encodes a transcription factor that inhibits MyoD activity. J Biol Chem 271:3555–3561
Soosaar A, Chiaramello A, Zuber MX, Neuman T (1994) Expression of basic-helix-loop-helix transcription factor ME2 during brain development and in the regions of neuronal plasticity in the adult brain. Brain Res Mol Brain Res 25:176–180
Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C et al (2008) Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol 317:246–259. https://doi.org/10.1016/j.ydbio.2008.02.026
Thaxton C, Kloth AD, Clark EP, Moy SS, Chitwood RA, Philpot BD (2018) Common pathophysiology in multiple mouse models of Pitt–Hopkins SYNDROME. J Neurosci 38:918–936. https://doi.org/10.1523/JNEUROSCI.1305-17.2017
Therneau T (2015) A package for survival analysis in S. Version 2.38. https://CRAN.R-project.org/package=survival
Waszak SM, Northcott PA, Buchhalter I, Robinson GW, Sutter C, Groebner S et al (2018) Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol 19:785–798. https://doi.org/10.1016/s1470-2045(18)30242-0
Whalen S, Heron D, Gaillon T, Moldovan O, Rossi M, Devillard F et al (2012) Novel comprehensive diagnostic strategy in Pitt-Hopkins syndrome: clinical score and further delineation of the TCF4 mutational spectrum. Hum Mutat 33:64–72. https://doi.org/10.1002/humu.21639
Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF (2008) Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev 22:722–727. https://doi.org/10.1101/gad.1636408
Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A (2001) hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31:85–94
Zollino M, Zweier C, Van Balkom I, Sweetser DA, Alaimo J, Bijlsma EK et al (2019) Diagnosis and management in Pitt-Hopkins syndrome: first international consensus statement. Clin Genet. https://doi.org/10.1111/cge.13506
Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, Clayton-Smith J et al (2007) Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome). Am J Hum Genet 80:994–1001. https://doi.org/10.1086/515583
Zweier C, Sticht H, Bijlsma EK, Clayton-Smith J, Boonen SE, Fryer A et al (2008) Further delineation of Pitt-Hopkins syndrome: phenotypic and genotypic description of 16 novel patients. J Med Genet 45:738–744. https://doi.org/10.1136/jmg.2008.060129
Acknowledgements
We thank Margarethe Gregersen, Michael Schmidt, and Anne Reichstein for excellent technical support. This study was supported by the German Cancer Aid, the Wilhelm Sander-Stiftung, the Kind-Philipp-Stiftung, the Burkhard Meyer Stiftung, and the Fördergemeinschaft Kinderkrebs-Zentrum Hamburg.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hellwig, M., Lauffer, M.C., Bockmayr, M. et al. TCF4 (E2-2) harbors tumor suppressive functions in SHH medulloblastoma. Acta Neuropathol 137, 657–673 (2019). https://doi.org/10.1007/s00401-019-01982-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-01982-5