Abstract
Brain accumulation and aggregation of amyloid-β (Aβ) peptides is a critical step in the pathogenesis of Alzheimer’s disease (AD). Full-length Aβ peptides (mainly Aβ1–40 and Aβ1–42) are produced through sequential proteolytic cleavage of the amyloid precursor protein (APP) by β- and γ-secretases. However, studies of autopsy brain samples from AD patients have demonstrated that a large fraction of insoluble Aβ peptides are truncated at the N-terminus, with Aβ4–x peptides being particularly abundant. Aβ4–x peptides are highly aggregation prone, but their origin and any proteases involved in their generation are unknown. We have identified a recognition site for the secreted metalloprotease ADAMTS4 (a disintegrin and metalloproteinase with thrombospondin motifs 4) in the Aβ peptide sequence, which facilitates Aβ4–x peptide generation. Inducible overexpression of ADAMTS4 in HEK293 cells resulted in the secretion of Aβ4–40 but unchanged levels of Aβ1–x peptides. In the 5xFAD mouse model of amyloidosis, Aβ4–x peptides were present not only in amyloid plaque cores and vessel walls, but also in white matter structures co-localized with axonal APP. In the ADAMTS4−/− knockout background, Aβ4–40 levels were reduced confirming a pivotal role of ADAMTS4 in vivo. Surprisingly, in the adult murine brain, ADAMTS4 was exclusively expressed in oligodendrocytes. Cultured oligodendrocytes secreted a variety of Aβ species, but Aβ4–40 peptides were absent in cultures derived from ADAMTS4−/− mice indicating that the enzyme was essential for Aβ4–x production in this cell type. These findings establish an enzymatic mechanism for the generation of Aβ4–x peptides. They further identify oligodendrocytes as a source of these highly amyloidogenic Aβ peptides.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disorder and is responsible for an estimated 60–80% of all dementia cases [1]. According to the amyloid hypothesis, the disease process starts with the accumulation of amyloid-β (Aβ) peptides in the brain, which form neurotoxic oligomers and insoluble amyloid plaques and trigger detrimental downstream events including Tau aggregation and chronic inflammation [67]. This chain of events is strongly supported by the genetic and biochemical analyses of rare, familial forms of AD with early onset, in which modest overproduction of Aβ peptides in the brain is sufficient to cause the disease with complete penetrance [4]. Full-length Aβ peptides (mainly Aβ1–40 and Aβ1–42) are generated through sequential processing of the amyloid precursor protein (APP) by the proteases β- and γ-secretase [14]. In this process, the cleavage of APP by the β-secretase BACE1 is the rate-limiting step for the production of Aβ peptides with an aspartate residue (Asp) at position 1 [78]. However, biochemical studies have revealed that a substantial fraction of Aβ peptides extracted from autopsy brain samples of AD patients did not start with an Asp but were truncated at the N-terminus. Already in their seminal study in 1985, Masters et al. had demonstrated by N-terminal sequencing analysis that only around 10% of the formic acid extractable Aβ peptides began with an Asp while > 60% started with a phenylalanine residue (Phe) at position 4 [48]. While Aβ peptides truncated at various N-terminal residues have been detected in brain tissue and cerebrospinal fluid samples of AD patients [6, 40], subsequent studies have largely confirmed that Aβ4–42 and peptides with a pyroglutamate modification at position 3 (AβpE3–42) are particularly abundant [45, 50, 51, 58, 59, 68]. Recently, two studies reported novel Aβ4–x-specific antibodies and the consistent finding that in the brains of AD patients and mouse models Aβ4–x peptides were largely confined to amyloid plaque cores, which are composed of the most insoluble, fibrillar Aβ peptides in β-sheet conformation [11, 79]. In accordance, biophysical studies have demonstrated that Aβ4–x peptides rapidly assembled into soluble oligomers and fibrillar, high-molecular-weight aggregates [7, 11, 56]. Intriguingly, a comparison of the aggregation properties of Aβ1–40 and Aβ4–40 showed that the latter formed trimers and thioflavin-T-binding aggregates almost instantly [11]. Thus, truncation of the first three amino acids might therefore convert Aβ1–40 peptides, which are highly abundant in AD brains but have only limited tendency to aggregate, into a more pathogenic species. In vitro, Aβ4–40 and Aβ4–42 peptides were toxic to primary neurons with effect sizes comparable to Aβ1–42 [7, 56]. Finally, expression of Aβ4–42 in transgenic mice resulted in age-dependent spatial memory deficits with a severe 50% neuron loss in the hippocampus at 12 months of age [7].
The biophysical properties, distribution, and toxicity of Aβ4–x peptides point towards an important role for this Aβ species in the pathogenesis of AD. However, while these N-truncated peptides were detected more than 30 years ago, any proteases involved in their generation have remained elusive. In this study, we show that cleavage of APP within the Aβ peptide sequence by the secreted metalloprotease ADAMTS4 (a disintegrin and metalloproteinase with thrombospondin motifs 4) resulted in the release of Aβ4–x peptides both in vitro and in vivo. ADAMTS4 belongs to a family of Zn2+-metalloproteases with a major role in extracellular matrix (ECM) remodeling, e.g., during morphogenesis and angiogenesis [3, 38, 73, 74]. One of its substrates, the hyaluronan-binding proteoglycan aggrecan, is abundantly present in the articular cartilage, and ADAMTS4 has been linked to osteo- and rheumatoid arthritis [60, 71, 75]. We further demonstrate in the present study that Aβ4–x production was not dependent on prior cleavage of APP by BACE1, and that purified ADAMTS4 was able to convert Aβ1–40 into Aβ4–40 peptides, indicating that Aβ4–x peptides might be generated both within the cell and in the extracellular space. Immunohistochemistry in reporter mice demonstrated that in the adult mouse brain ADAMTS4 expression was restricted to oligodendrocytes. In agreement, white matter structures in aged 5xFAD mice showed abundant immunoreactivity for Aβ4–x peptides, which was absent in 5xFAD/ADAMTS4−/− mice. In summary, we report an enzymatic mechanism for the generation of Aβ4–x peptides and the unexpected finding that oligodendrocytes are a source of these highly amyloidogenic peptides.
Materials and methods
Antibodies
The following primary antibodies were used in this study: anti-APP antibodies: CT-15 (rabbit pAb raised against the C-terminal 15 amino acids of human APP, 1:3500 immunoblotting, 1:500 immunocytochemistry) [77]; 22C11 (mouse mAb raised against residues 66–81 of human APP, 1:1000, kindly provided by Dr. Stefan Kins, University of Kaiserslautern, Germany) [66]; IG7/5A3 (mouse mAbs recognizing non-overlapping epitopes of APP between residues 380–665, 1:1000) [77]; 6A1 (mouse mAb recognizing the C-terminal neoepitope generated by β-secretase cleavage of Swedish APP, 1:50, IBL Cat. No. 10321); anti-APPs-β (rabbit pAb recognizing the C-terminal neoepitope generated by β-secretase cleavage of wild-type APP, 1:50, IBL Cat. No. 18957); anti-APP (rabbit pAb raised against residues 756–770 of rat APP, 1:500, Synaptic Systems Cat. No. 127003). Anti-Aβ antibodies: IC16 (mouse mAb that preferentially detects Aβ peptides starting with Asp in position 1) [2]; 029-2 (guinea pig pAb that exclusively detects Aβ4–x with Phe in position 4, 1:500 immunohistochemistry) [79]; BAP-24 (Aβ40 C-terminus-specific mouse mAb) [24]; BAP-29 (Aβ38 C-terminus-specific mouse mAb) [24]; BAP-15 (Aβ42 C-terminus-specific mouse mAb) [8, 24]; 80C2 (mouse mAb against Aβ residues 1–5, 1:500, Synaptic Systems Cat. No. 218231); 4G8 (mouse mAb against Aβ residues 17–24, BioLegend Cat. No. 800701); D3E10 (rabbit mAb specific for the C-terminus of Aβx–42 peptides, 1:500, Cell Signaling Cat. No. 12843). Other antibodies: anti-V5 Tag (mouse mAb, 1:500 immunoblotting, 1:250 immunocytochemistry, ThermoFisher Scientific Cat. No. R960-25); anti-CNPase (mouse mAb, 1:1000; Sigma-Aldrich Cat. No. C5922); anti-β-gal (rabbit pAb, 1:500, Millipore Cat. No. AB986); anti-GFAP (rabbit pAb, 1:500; DAKO Cat. No. Z0334); anti-Iba1 (guinea pig pAb, 1:300; Synaptic Systems Cat. No. 234004); anti-MAP2 (guinea pig pAb, 1:500, Synaptic Systems Cat. No. 188004); anti-Actin (rabbit pAb, 1:2000, Sigma-Aldrich Cat. No. A2066); anti-calnexin (rat mAb, 1:250, BioLegend Cat. No. 699401); anti-TGN46 (sheep pAb, 1:100, Bio-Rad Cat. No. AHP500G); anti-Rab11 (goat pAb, 1:100, Santa Cruz Biotechnology).
Generation of cell lines with doxycycline-inducible ADAMTS4 expression
Cell lines with inducible ADAMTS4 expression based on HEK293 cells were generated using a lentiviral tetracycline-controlled expression system. The expression system was a gift by Dr. Eric Campeau and consisted of the lentiviral expression vectors pLenti CMVtight Hygro DEST (Addgene plasmid#26433) and pLenti CMV rtTA3 Blast (Addgene Plasmid #26429) encoding the reverse tetracycline-controlled transactivator 3 (rtTA3). Full-length human ADAMTS4 with a C-terminal V5-tag was cloned into pLenti CMVtight Hygro DEST by Gateway cloning (ThermoFisher Scientific). Viral particles were produced in 293FT cells using a third-generation lentivirus packaging system [20]. To generate double-stable cell lines expressing both rtTA3 and tetracycline-inducible ADAMTS4, either HEK293 cells with endogenous APP expression, or HEK293 cells with stable overexpression of the human APP695 isoform, or HEK293 cells with stable overexpression of the human APP695 isoform containing the “Swedish” double-mutation (the APP-overexpressing cell lines were a gift of Dr. Edward Koo, UC San Diego, USA) were first infected with lentiviral particles expressing rtTA3 and a stable mass culture was selected with blasticidin. Subsequently, rtTA3 expressing cells were infected with ADAMTS4 lentiviral particles and stable clones were selected with hygromycin. Double-stable cell lines were maintained in antibiotic selection medium containing DMEM with 10% FCS (v/v), 1% sodium pyruvate (v/v), 100 U/ml penicillin/streptomycin, blasticidin, and hygromycin (all media components from ThermoFisher Scientific).
Immunoblotting
Cultured cells were lysed in Nonidet P40 buffer (50 mM Tris–HCl pH 7.8, 150 mM NaCl, 1% NP40 (v/v), 0.02% NaN3 (w/v), with cOmplete protease inhibitor cocktail (Roche Diagnostics)) and total protein concentrations were determined with a BCA Protein Assay Kit (ThermoFisher Scientific). Equal amounts of protein were separated by 7% or 12% Bis–Tris SDS-PAGE and transferred onto PVDF membranes (Millipore) by electroblotting. The membranes were blocked with 3% gelatin from cold water fish skin (w/v) in PBS for 1 h at RT, and then incubated overnight at 4 °C with the primary antibody diluted in TBST (25 mM Tris, 137 mM NaCl, 2.7 mM KCl, 0.1% Tween-20 (v/v), pH 7.4). Subsequently, a secondary antibody labeled with a near-infrared fluorescent dye (IRDye 800CW goat-anti-mouse IgG or anti-rabbit IgG, LI-COR Biosciences) diluted in TBST was added and incubated for 1 h at RT. Fluorescence signals were detected with the Odyssey CLx Imaging System and quantified using the Image Studio Software 2.1 (LI-COR Biosciences).
Aβ enzyme-linked immunosorbent assays
Aβ levels in brain extracts and conditioned tissue culture supernatants were determined using specific sandwich enzyme-linked immunosorbent assays (ELISA) as described [29, 79]. Standard curves were generated with synthetic Aβ peptides (JPT). To detect full-length Aβ1–38, Aβ1–40, and Aβ1–42 peptides, monoclonal antibody IC16 was used as a capture antibody and combined with Aβ C-terminus-specific detection antibodies. To detect N-truncated Aβ4–40 peptides, the Aβ40 C-terminus-specific antibody BAP-24 was used as a capture antibody and combined with the Aβ4–x-specific antibody 029-2 as a detection antibody. 96-well high-binding microtiter plates (Greiner Bio-One) were incubated overnight at 4 °C with the capture antibodies in PBS, pH 7.2. After excess capture antibody was removed, freshly diluted brain samples, tissue culture media or Aβ peptide standards (in PBS, 0.05% Tween-20 (v/v), 1% BSA (w/v)) were added. Then, the detection antibodies labeled with horseradish peroxidase using the Pierce EZ-Link Plus Activated Peroxidase kit (ThermoFisher Scientific) and diluted in PBS, 0.05% Tween-20 (v/v), 1% BSA (w/v) were added to each well and incubated overnight at 4 °C. Plates were washed three times with PBS containing 0.05% Tween-20 (v/v) and once with PBS. Subsequently, 50 μl of trimethylbenzidine ELISA peroxidase substrate (ThermoFisher Scientific) was added and incubated for 1–5 min at RT in the dark. The reaction was terminated by adding 50 μl of 2 M H2SO4, and the absorbance was recorded using a Paradigm microplate reader (Beckman Coulter) at 450 nm.
Immunoprecipitation/mass spectrometry analysis of secreted Aβ peptides
Mass spectrometry analysis of Aβ peptides was performed as previously described [18, 21]. Conditioned cell culture media were collected, cleared by centrifugation, and incubated overnight at 4° C with antibody 4G8 (1 μg/ml) and 30 μl of protein G (Sigma-Aldrich) to precipitate Aβ. The beads were washed 3 times with washing buffer (50 mM HEPES, 150 mM NaCl, 0.1% n-octyl-β-d-glycopyranoside (w/v); pH 7.4), and immunoprecipitated proteins were eluted with trifluoroacetic acid (1%):acetonitrile:H2O (1:20:20). The elution was then mixed at a volume ratio of 1:1 with saturated α-cyano-4-hydroxycinnamic acid for the analysis by MALDI mass spectrometry in reflectron mode.
In vitro digest of synthetic Aβ1–40/Aβ1–42 peptides with purified ADAMTS4
To investigate whether Aβ1–40 and Aβ1–42 peptides could serve as substrates for ADAMTS4, two different in vitro digest assays with purified ADAMTS4 were performed. In the first assay, mature, catalytically active ADAMTS4 was immunoprecipitated from culture supernatants of HEK293 cells with DOX-inducible expression of V5-tagged ADAMTS4 (HEK–TetOn–ADAMTS4 cells). Anti-V5 tag antibodies were coupled to magnetic M-280 Dynabeads (ThermoFisher Scientific) according to the manufacturer’s instructions. 25 μl of antibody coupled beads were incubated with 1 ml of conditioned cell culture supernatant for 2 h at RT. The beads were immobilized with a magnet, washed twice with PBS containing 0.1% BSA (w/v), and then incubated with 350 μl of DMEM containing 15 ng of synthetic Aβ1–40 peptides (JPT Peptide Technologies) for 24 h at 37 °C. The beads were removed and the supernatant was stored at -80 °C prior to ELISA analysis of Aβ4–40 levels. In the second assay, a recombinant, catalytically active fragment of human ADAMTS4 was used (Phe213-Cys685, R&D Systems). 1 µg of the recombinant ADAMTS4 was pre-incubated for 20 min at 37 °C in assay buffer (50 mM HEPES, 50 mM NaCl, 1 mM CaCl2, 0.05% Brij-35, pH 7.5) in the presence or absence of 10 µM of the broad-spectrum metalloprotease inhibitor batimastat (BB-94, Sigma-Aldrich). The reaction was started by the addition of 25 ng synthetic Aβ1–42 peptides (JPT Peptide Technologies) in a total volume of 300 µl assay buffer. The reaction mixture was incubated for 24 h at 37 °C, and the resulting Aβ peptide species were analyzed by immunoprecipitation followed by MALDI mass spectrometry.
Animals
The generation of 5xFAD mice (Tg6799) has been described [54]. 5xFAD mice overexpress the human APP695 isoform carrying the Swedish, Florida and London mutations, and human presenilin-1 carrying the M146L and L286V mutations, both under the control of the murine Thy1-promoter. 5xFAD mice have been backcrossed for more than 10 generations to C57Bl/6J wild-type mice from the Jackson Laboratory (Bar Harbor, ME, USA) and were maintained on a C57BL/6J genetic background as a heterozygous transgenic line. ADAMTS4+/− mice were obtained from the Jackson Laboratory (Strain #005770), and maintained on a C57BL/6J background. Detailed genotyping information is available at https://www.jax.org/strain/005770. ADAMTS4+/− mice were crossed with 5xFAD mice to finally obtain heterozygous 5xFAD mice on a homozygous ADAMTS4 KO background (5xFAD/ADAMTS4−/−). All animals were housed in a 12 h/12 h light–dark cycle with food and water ad libitum.
Brain tissue preparation
Mice were killed via CO2 anesthetization followed by cervical dislocation. For biochemical studies, frozen brain hemispheres were weighed and sequentially extracted. First, brains were homogenized in 700 μl of TBS (120 mM NaCl, 50 mM Tris, pH 8.0, with cOmplete protease inhibitor cocktail) per 100 mg of tissue using a Dounce homogenizer (800 rpm). The resulting solution was centrifuged at 17,000 xg for 20 min at 4 °C. The pellet was dissolved in 800 μl of 2% SDS (w/v) in ddH2O and sonicated, followed by centrifugation at 17,000 xg for 20 min at 4 °C. The supernatant containing SDS-soluble proteins was incubated while rotating with 250 U of benzonase (Sigma-Aldrich) for 10 min at RT. Both TBS and SDS brain fractions were stored at − 80 °C before use for Western blotting or ELISA analysis. For immunohistochemistry, brains were dissected, immersion-fixed in 4% phosphate-buffered formalin at 4 °C, dehydrated in a series of alcohols and embedded in paraffin.
Immunohisto- and immunocytochemistry
Immunohistochemistry was performed on 4 µm sagittal brain paraffin sections. After treatment with 0.3% H2O2 (v/v) in PBS to block endogenous peroxidases, antigen retrieval was achieved by boiling sections in 0.01 M citrate buffer, pH 6.0, followed by 3 min incubation in 88% formic acid. Non-specific binding sites were blocked by treatment with skim milk and fetal calf serum in PBS. Sections were incubated with primary antibodies overnight at RT. For conventional immunostaining, biotinylated secondary anti-guinea pig (1:200; Dianova) or anti-mouse (DAKO) antibodies were used, and staining was visualized using the ABC method with a Vectastain Elite kit (Vector Laboratories) and diaminobenzidine (DAB) as a chromogen. Counterstaining was carried out with hematoxylin. For immunofluorescence labeling, corresponding fluorescent secondary antibodies (1:750; Invitrogen or ThermoFisher Scientific) in combination with 4′,6-diamidin-2-phenylindol (DAPI) were used. Images of DAB-stained sections were taken using a BX-51 microscope (Olympus) with a Moticam Pro 282 camera (Motic) and analyzed using ImageJ 1.47b software (http://imagej.nih.gov/ij). Fluorescent images were recorded using a Nikon TiE microscope (Nikon) and analyzed with NIS Elements imaging software (Nikon). For immunocytochemistry, HEK–APP695sw/TetOn–ADAMTS4 cells were seeded on poly-l-lysine (Sigma-Aldrich) coated glass coverslips, and ADAMTS4 expression was induced with DOX for 24 h. Cells were fixed with 4% PFA (w/v) in ddH20 for 10 min, permeabilized with 1% saponin (w/v) in TBS for 20 min, and blocked with 5% BSA (w/v) in TBS for 1 h, all at RT. To reduce cross reactions, instead of applying all primary and all secondary antibodies at the same time, primary antibodies were applied one after another followed by their respective fluorophore-tagged secondary antibodies. Each antibody was applied in 4% BSA (w/v) in TBS for 1 h. The following secondary antibodies were used: Alexa Fluor 488 goat-anti-rat (1:1000, ThermoFisher Scientific Cat. No. A11006); Alexa Fluor 546 goat-anti-rabbit (1:1000, ThermoFisher Scientific Cat. No. A11071); Alexa Fluor 647 goat-anti-mouse (1:500, ThermoFisher Scientific Cat. No. A21235); Alexa Fluor 488 goat-anti-rabbit (1:1000, ThermoFisher Scientific Cat. No. A11070); Alexa Fluor 594 goat-anti-mouse (1:500, ThermoFisher Scientific Cat. No. A11005); Cy3 donkey-anti-sheep (1:500, Jackson ImmunoResearch Cat. No. 713-165-147); Cy3 donkey-anti-rabbit (1:500, Jackson ImmunoResearch Cat. No. 711-165-152); Cy5 donkey-anti-goat (1:200, Jackson ImmunoResearch Cat. No. 705-175-147); Cy2 donkey-anti-mouse (1:100, Jackson ImmunoResearch Cat. No. 715-225-151). Coverslips were mounted on glass slides with ProLong Gold Antifade Mountant with DAPI (ThermoFisher Scientific), cured for 24 h at RT in the dark, and analyzed with a LSM 710 confocal laser-scanning microscope (Zeiss).
Culture and differentiation of murine oligodendrocyte precursor cells (OPCs)
OPCs were prepared from whole brains of newborn (P1–P3) ADAMTS4−/− KO or C57Bl/6J wild-type mice. Meninges were removed, brains were disintegrated with a scalpel, and pooled from animals with the same genotype. Brain tissue was further dissociated with the Neural Tissue Dissociation Kit, and A2B5+ glial-restricted progenitors were isolated by labeling with anti-A2B5 microbeads and magnetic enrichment using MS columns and a MiniMACS Separator according to the manufacturer’s instructions (Miltenyi Biotec). 1.5 × 106 progenitor cells were seeded on poly-l-lysine coated 10 cm dishes and cultured in proliferation medium (DMEM/F12 without glutamine, 1× B-27 supplement without vitamin A, 1× GlutaMAX, 100 U/ml penicillin/streptomycin, 20 ng/ml each of mouse FGF-b and mouse PDGF-AA, all media components from ThermoFisher Scientific, growth factors from ImmunoTools). Confluency was kept below 50% and cells were passaged approximately every 4 days after dissociation of the cell monolayer with accutase (Sigma-Aldrich). For differentiation of OPCs, cells were switched to differentiation medium (same media components, except 1× B-27 with vitamin A, and without growth factors but instead supplemented with 40 ng/ml each of L-thyroxine (T4) and triiodo-L-thyronine (T3), Sigma-Aldrich) for 1–5 days.
Adenovirus construction and infection of OPCs
Adenoviral vector construction was performed as previously described [46] with the following modifications: a cDNA encoding human APP695sw was cloned into the pAdTrack-CMV shuttle vector (kindly provided by Dr. Bert Vogelstein, Johns Hopkins University, Addgene Plasmid #16405) using NotI/HindIII restriction sites. This shuttle vector incorporates a GFP marker and was transformed into AdEasier-1 cells (derivative of BJ5183 cells containing the adenoviral backbone plasmid pAdEasy-1, Addgene bacterial strain #16399), which were made chemically competent by standard methods, to obtain recombinant adenovirus plasmids by homologous recombination. The recombinant adenovirus plasmids were packaged in HEK293 cells to obtain a low titer adenovirus preparation, which was further amplified and purified by a commercial vendor (GeneCust). To determine the functional titer of the adenovirus preparation, Hela cells were seeded in 6-well plates at a density of 70,000 cells per well. Serial dilutions of the adenovirus stock were prepared in 1 ml DMEM growth medium and used to infect the cells for 3 h. After 24 h, the percent of fluorescent-positive cells was determined using a Neubauer cell counting chamber. To calculate the transduction units (TU) per ml, the following formula was used: (total number of cells transduced x percent fluorescent cells x dilution factor)/transduction volume in ml. The assay was repeated three times and the average functional titer of the adenovirus stock was determined to be 5x108 TU/ml. For adenoviral transduction, 4x106 OPCs were seeded on poly-l-lysine coated 10 cm culture dishes in proliferation medium. The following day, cells were infected with 0.5 TU of adenovirus per cell for 4 h. 24 h after the infection, 4 ml of fresh proliferation medium were added and conditioned for another 24 h prior to ELISA and mass spectrometry analysis of Aβ levels.
Quantitative PCR
For mRNA expression analysis, total RNA was isolated from primary oligodendrocyte cultures at different time points after initiation of differentiation using the ReliaPrep RNA Cell Miniprep System (Promega) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg RNA by reverse transcription in a final volume of 20 μl 1 x M-MLV reverse transcriptase reaction buffer (Promega) containing 1.25 μM random hexamers (ThermoFisher Scientific), 0.5 mM dNTPs (ThermoFisher Scientific) and 200 units M-MLV (H–) reverse transcriptase (Promega). The mixture was incubated at 25 °C for 10 min, at 40 °C for 50 min, and at 70 °C for 15 min. For quantitative PCR analysis, the Platinum qPCR Super Mix (ThermoFisher Scientific) based on the fluorescent nucleic acid dye SYBR Green was used. 10 μl of the SYBR Green Mix, 500 nM of each primer, and 0.5 µl ROX reference dye were mixed and adjusted with RNase-free water to a final volume of 17 µl. 3 μl of the diluted cDNA (1:5 in RNase-free water) were added and the reaction mixture was transferred into a fast optical 96-well reaction plate (Applied Biosystems). Quantitative PCR was performed in a StepOnePlus Real-Time PCR System (Applied Biosystems) using the following program: 95 °C for 10 min followed by 40 cycles of repeated denaturation and hybridization/elongation at 95 °C for 15 s and 60 °C for 1 min. The relative gene expression levels were calculated with the StepOnePlus Software 2.0 using the 2−ΔΔCT-method. GAPDH served as the housekeeping gene, and the expression levels were normalized to the values of non-differentiated oligodendrocytes at day 0. Primer sequences are available upon request.
Statistical analysis
Analyses were performed using Prism 5 (GraphPad Software), and data are represented as mean ± SD. Comparisons between two groups were performed by unpaired t test, three or more groups were analyzed by one-way ANOVA followed by the Tukey’s post hoc test.
Results
Proteolytic cleavage of APP by ADAMTS4 generates N-truncated Aβ4–x peptides
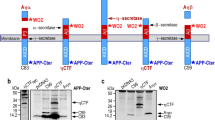
By inspecting the N-terminal region of the Aβ peptide sequence within APP, we noticed that it contains two putative cleavage sites for the secreted zinc-metalloprotease ADAMTS4. Previously, through the screening of peptide libraries, a 7-amino acid recognition motif for ADAMTS4 was defined as E-(AFVLMY)-X(0,1)- (RK)-X(2,3)-(ST)-(VYIFWMLA) with Glu at the P1 position [27]. This motif is partially conserved in the Aβ peptide sequence starting with the Glu residues 3 and 11 (Fig. 1a). Therefore, sequential cleavage of APP by ADAMTS4 and γ-secretase could potentially result in N-truncated Aβ peptides, e.g., Aβ4–40 and Aβ12–40. To test this hypothesis in a cellular system that allows careful adjustment of ADAMTS4 expression, we generated HEK293 cells with stable co-expression of Swedish mutant human APP695 and doxycycline (DOX)-inducible V5-tagged human ADAMTS4 (HEK-APP695sw/TetOn-ADAMTS4 cells). DOX-induced single-cell clones displayed near equal expression of the immature pro-form and the furin-processed, catalytically active form of ADAMTS4 in cell lysates and in culture media [76], with slight background expression in non-induced cells (Fig. 1b). To examine whether ADAMTS4 overexpression would affect the secretion of full-length Aβ peptides with an intact N-terminus (Aβ1–x), Aβ levels in conditioned culture supernatants were measured with a sandwich ELISA system that combined monoclonal antibody IC16, which preferentially detects Aβ peptides starting with Asp at position 1, with Aβ C-terminus-specific antibodies [2, 24]. In agreement with the predicted ADAMTS4 cleavage sites, measurements of Aβ1–40, Aβ1–42, and Aβ1–38 levels did not show differences between DOX-induced ADAMTS4 expressing and non-induced control cells for three independent cell clones (Fig. 1c). Next, the secreted Aβ peptides were analyzed by MALDI-TOF mass spectrometry, which allowed detection of all Aβ peptides including N- and C-truncated species for which no specific antibodies are available. Mass spectra from culture supernatants of DOX-induced cells showed clear peaks for Aβ4–40 and Aβ12–40, providing qualitative support that ADAMTS4 can generate Aβ4–x peptides and that the second, less-preserved recognition site starting with Glu11 is also cleaved in this ADAMTS4 overexpression system leading to the secretion of Aβ12–40 peptides (Fig. 1d). Due to the differential ionization properties of similar peptides, the quantitative analysis of MALDI-TOF measurements is limited and the peak heights in the mass spectra do not correspond to the abundance of individual peptides [9]. To obtain quantitative data and to confirm these results with an independent method, culture supernatants were analyzed with a sandwich ELISA that combined an Aβ40 C-terminus-specific antibody with the polyclonal antibody 029-2, which exclusively detects Aβ4–x peptides with Phe at position 4, as described [79]. Measurements revealed around 2 ng/ml of Aβ4–40 peptides in culture supernatants after induction of ADAMTS4 expression, while non-induced control cells showed no background signal (Fig. 1e). In sum, these data indicated that ADAMTS4 is able to cleave APP at the predicted sites, resulting in the secretion of Aβ4–40 and Aβ12–40 peptides.
ADAMTS4 generates N-truncated Aβ4–x peptides. a The N-terminal part of the Aβ peptide sequence is shown with the β-secretase cleavage site prior to Asp1 and the α-secretase cleavage site after Lys16. A recognition site for the metalloprotease ADAMTS4 starts with Glu3 and conforms entirely to the recognition motif as defined by Hills et al. with the exception of Gly9 [27]. A second putative recognition site starting with Glu11 is less preserved. b ADAMTS4 protein levels in HEK-APP695sw/TetOn-ADAMTS4 cells. ADAMTS4 expression was induced with DOX (100 ng/ml) for 48 h and cell lysates and culture supernatants were analyzed by Western blotting with anti-V5 antibody. Three single-cell clones displayed near equal expression of both the pro-form (immature) and the catalytically active form (mature) of ADAMTS4. Mature ADAMTS4 migrated below the 100 kDa molecular weight marker (M) consistent with previous results [76]. c Measurement of Aβ1–x peptide levels in conditioned culture supernatants of HEK-APP695sw/TetOn-ADAMTS4 cells. Aβ1–40, Aβ1–42, and Aβ1–38 levels were unchanged after induction of ADAMTS4 expression as compared to non-induced control cells. Three independent biological experiments for each cell clone (n = 3) with two technical replicates per experimental condition were performed. d Mass spectrometry analysis of Aβ peptides in conditioned culture supernatants of HEK–APP695sw/TetOn–ADAMTS4 cells. ADAMTS4 expression was induced with DOX and supernatants were immunoprecipitated with the monoclonal anti-Aβ antibody 4G8 and analyzed by MALDI-TOF mass spectrometry. Spectra of non-induced control cells (-DOX) showed peaks for the common Aβ species such as Aβ1–40, Aβ1–42, and Aβ1–38. Only after induction of ADAMTS4 expression (+DOX), Aβ4–40 and Aβ12–40 peptides were clearly detected while other Aβ species remained unchanged. Nearly identical spectra were acquired from three single-cell clones, and representative spectra are shown. The measured monoisotopic masses and the expected masses, and the primary amino acid sequences of the detected peptides are shown in Supplementary Table 4. e Robust levels of Aβ4–40 peptides were detected in culture supernatants of HEK–APP695sw/TetOn–ADAMTS4 cells after induction of ADAMTS4 expression with no background in non-induced control cells. Three independent biological experiments for each cell clone (n = 3) with two technical replicates per experimental condition were performed
Prior cleavage of APP by BACE1 is not required for the generation of N-truncated Aβ4–x peptides, and recombinant Aβ1–x can serve as a substrate for ADAMTS4
The experiments shown above did not clarify whether ADAMTS4 by itself could cleave APP and generate Aβ4–x peptides within the cell, or whether ADAMTS4 might only remove the first three amino acids from Aβ1–x peptides with an intact N-terminus, presumably in the extracellular space. To address this question, two sets of experiments were performed. First, we examined whether Aβ4–x peptides could still be generated by ADAMTS4 in the presence of a potent β-secretase inhibitor. Treating HEK–APP695sw/TetOn–ADAMTS4 cells with BACE inhibitor IV abolished Aβ1–40 peptides in culture supernatants with or without ADAMTS4 expression (Fig. 2a). However, analyzing the same culture supernatants with the Aβ4–40 specific ELISA showed that these N-truncated peptides were not reduced, indicating that Aβ4–x production was not dependent on BACE1 activity (Fig. 2a). Second, to explore whether ADAMTS4 would be able to convert Aβ1–x into Aβ4–x peptides, we established HEK293 cells with DOX-inducible expression of V5-tagged ADAMTS4 (HEK-TetOn-ADAMTS4), and immunoprecipitated ADAMTS4 from conditioned media with anti-V5 antibody. The immunoprecipitated mature, catalytically active form of ADAMTS4 (Fig. 2b) was then incubated with recombinant Aβ1–40 peptides in fresh culture medium for 24 h, and the generation of Aβ4–40 peptides was detected by ELISA. An approximately fivefold increase in Aβ4–40 levels was observed when ADAMTS4 was immunoprecipitated from DOX-induced cells as compared to non-induced control cells (Fig. 2b). To further prove that ADAMTS4 is the responsible enzyme activity and to demonstrate that recombinant Aβ1–42 could also serve as a substrate, we performed a cell-free in vitro assay followed by mass spectrometry analysis (Supplementary Fig. S1). After the co-incubation of recombinant ADAMTS4 with Aβ1–42 peptides, peaks corresponding to both Aβ4–42 and Aβ12–42 were observed in the mass spectra. Importantly, these peaks were not seen when enzyme and substrate were incubated in the presence of the broad-spectrum metalloprotease inhibitor batimastat (BB-94; 10 μM) (Supplementary Fig. S1). Taken together, these experiments demonstrated that ADAMTS4 was able to cleave APP and to generate Aβ4–x peptides within the cell without prior cleavage of APP by β-secretase. In addition, they showed that ADAMTS4 can convert Aβ1–x into Aβ4–x peptides, providing evidence that ADAMTS4 might truncate Aβ1–x peptides at the N-terminus in the extracellular space.
ADAMTS4 generates Aβ4–x peptides without prior cleavage of APP by BACE1, recognizes recombinant Aβ1–40 as substrate, and affects APPs levels. a HEK-APP695sw/TetOn-ADAMTS4 cells were pre-treated with BACE1 inhibitor IV (1 μM) for 24 h. Subsequently, medium was changed and ADAMTS4 expression was induced with DOX for 24 h in the presence of the BACE1 inhibitor. Medium was changed again and conditioned for another 24 h in the presence of DOX and the BACE1 inhibitor. Finally, conditioned media were analyzed with Aβ1–40 or Aβ4–40-specific ELISA assays. In control experiments, cells were not treated with the BACE1 inhibitor, and/or induction of ADAMTS4 expression was omitted. Consistent with the experiments shown in Fig. 1c, Aβ1–40 levels were not affected by the induction of ADAMTS4 expression. In contrast, Aβ1–40 peptides were completely abolished by treatment with the BACE1 inhibitor. Aβ4–40 peptides were only detected after induction of ADAMTS4 expression but not in non-induced control cells. Concomitant inhibition of BACE1 did not reduce Aβ4–40 levels, indicating that ADAMTS4 was able to cleave APP and to generate Aβ4–x peptides within the cell without prior cleavage of APP by BACE1. Three independent biological experiments for each cell clone (n = 3) with two technical replicates per experimental condition were performed. b ADAMTS4 was immunoprecipitated from conditioned culture supernatants of DOX-induced (48 h) or non-induced HEK–TetOn–ADAMTS4 cells with anti-V5 antibody. These cells express endogenous APP and secrete only minor amounts of Aβ undetectable with our ELISA assays. The immunoprecipitated material was analyzed by Western blotting with anti-V5 antibody, which confirmed the successful immunoprecipitation of the mature, catalytically active form of ADAMTS4 from supernatants of DOX-induced cells. Immunoprecipitated ADAMTS4 was then incubated with recombinant Aβ1–40 for 24 h, and the conversion of Aβ1–40 to Aβ4–40 peptides was measured by ELISA. An approximately fivefold increase in Aβ4–40 levels was observed with immunoprecipitated material from ADAMTS4 expressing versus non-induced control cells. The Aβ4–40 signal in the control condition was likely due to slight background expression of ADAMTS4 in non-induced cells and further enrichment by immunoprecipitation. Three independent biological experiments (n = 3) with two technical replicates for each experimental condition were performed. Unpaired t test was used to compare the means *p < 0.05. c Schematic representation of APP and the antibody epitopes that were analyzed to investigate its proteolytic processing by ADAMTS4. d Levels of the soluble APP ectodomain APPs were reduced in ADAMTS4 expressing cells. HEK–APP695sw/TetOn–ADAMTS4 cells were induced with DOX, and different epitopes spanning the APP ectodomain were analyzed in culture supernatants. Western blotting with antibody 22C11 against the N-terminus of APP (upper panel) revealed a decrease in full-length APPs levels and an additional APPs fragment of approximately 70 kDa (marked with *) in all three ADAMTS4 expressing cell clones versus non-induced control cells. Western blotting with antibodies IG7/5A3 (middle panel) and 6A1 (lower panel), which recognize the mid-region of APP or the C-terminal neoepitope generated by β-secretase cleavage of APP, confirmed a substantial reduction in full-length APPs and APPs-β levels after induction of ADAMTS4 expression. Three independent biological experiments were performed (n = 3), and one representative experiment is shown
Proteolytic processing of APP by ADAMTS4 affects levels of its soluble ectodomain APPs
To assess whether ADAMTS4 might cleave APP outside of the Aβ region and affect the levels of other APP metabolites, cell lysates of DOX-induced HEK-APP695sw/TetOn-ADAMTS4 cells and non-induced control cells were analyzed by Western blotting with antibody CT-15 against the last 15 C-terminal amino acids of APP (antibody epitopes are illustrated in Fig. 2c). No overt differences in the levels of full-length APP or the APP C-terminal fragments (APP-CTFs) generated by β- or α-secretase were observed (Supplementary Fig. S2a). However, in ADAMTS4 expressing cells a novel APP-CTF with a molecular weight between 25-35 kDa was observed (Supplementary Fig. S2a). Moreover, closer inspection of APP levels showed a significant, greater than 50% reduction in the mature, glycosylated forms of APP localized to late compartments of the secretory pathway and the cell surface [35], while the immature forms of APP remained unchanged (Supplementary Fig. S2b, c). Immunofluorescence stainings demonstrated co-localization of ADAMTS4 and APP with the Golgi marker TGN46 but not with the ER marker calnexin or the late endosome marker Rab11 (Supplementary Fig. S2d-e). Taken together, these findings suggested that ADAMTS4 might cleave APP in the secretory pathway presumably in the Golgi compartment, with an additional cleavage site located in the APP ectodomain further N-terminal to the β-secretase cleavage site. Accordingly, we examined the levels of the soluble APP ectodomain (APPs) in culture supernatants with a set of antibodies spanning epitopes along the extracellular domain (Fig. 2c). Western blotting with antibody 22C11, which detects the N-terminus of APP (amino acids 66-81), showed reduced levels of full-length APPs in ADAMTS4 expressing cells (Fig. 2d). In addition, an approximately 20–30 kDa smaller APPs fragment was prominently detected in supernatants of DOX-induced cells, which appeared to correspond to the 25–35 kDa APP-CTF detected in cell lysates (Fig. 2d and Supplementary Fig. S2a). Antibodies IG7/5A3 against the mid-region of the APP ectodomain (amino acids 380–665) and 6A1, which detects the C-terminal neo-epitope generated by β-secretase cleavage of APP, confirmed a strong reduction in APPs and APPs-β levels after induction of ADAMTS4 expression (Fig. 2d). However, no APPs band with faster mobility was detected likely because the antibody epitopes were removed from this C-terminally truncated APPs fragment (Fig. 2d). Very similar results were obtained in HEK293 cell lines with either stable overexpression of wild-type APP695 or endogenous APP expression, indicating that the Swedish mutation located just three amino acids N-terminal to the first ADAMTS4 recognition site in the Aβ domain did not interfere with the proteolytic cleavage of APP by ADAMTS4 (Supplementary Fig. S3). Overall, in ADAMTS4 expressing cells, a substantial reduction in APPs levels was observed, suggesting that ADAMTS4 might not only facilitate the generation of N-truncated Aβ4–x peptides but might also exert partial control over the levels of the soluble ectodomain APPs.
Genetic deletion of ADAMTS4 lowers Aβ4–x and increases APPs levels in vivo
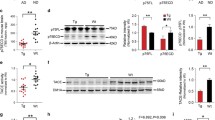
To validate the relevance of ADAMTS4 for Aβ generation and APP processing in vivo, ADAMTS4−/− knockout (KO) mice were crossed to the 5xFAD model of AD [54, 61]. Consistent with previous results [62], mass spectrometry analysis of Aβ peptide species in the brains of 12-month-old 5xFAD mice showed clearly detectable Aβ4–40 and Aβ4–42 peptides, demonstrating the suitability of the 5xFAD model and the age group (Supplementary Fig. S4). Hence, 5xFAD/ADAMTS4−/− and 5xFAD/ADAMTS4+/+ mice were aged to 12 months and TBS- and SDS-soluble brain fractions were analyzed by ELISA. Measurements of Aβ4–40 peptides in the SDS brain fractions demonstrated a significant 50% reduction in 5xFAD/ADAMTS4−/− mice corroborating that ADAMTS4 is critical for Aβ4–x levels in vivo (Fig. 3a). In contrast, Aβ1–40 and Aβ1–42 levels in the SDS fractions and Aβ1–42 levels in the TBS fractions were not different between the genotypes (Fig. 3b, d). As demonstrated in our previous study [79], the levels of the N-truncated Aβ4–40 peptides were low compared to full-length Aβ peptides in 5xFAD mice. In the SDS-soluble brain fractions of 12-month-old animals, Aβ4–40 levels were approximately 70-fold and 1500-fold lower than Aβ1–40 and Aβ1–42 levels (Fig. 3a–c). Consistent with these results, no obvious differences in the overall amyloid plaque load as determined by immunohistochemistry with antibodies against Aβ1–x or Aβx–42 peptides were observed between the genotypes (Supplementary Fig. S5a). Importantly, these results were confirmed in a second independent cohort of animals aged to 15 months (Supplementary Fig. S5b-e). In addition, soluble and membrane-bound APP fragments were analyzed by Western blotting in the brains of 12-month-old mice. The levels of full-length APP in the SDS fractions were unchanged between the genotypes (Fig. 3e). However, a significant 20–25% increase in APPs and APPs-β levels was observed in the TBS fractions of 5xFAD/ADAMTS4−/− mice as compared to 5xFAD/ADAMTS4+/+ control animals (Fig. 3f, g). These results were consistent with the tissue culture experiments, which had shown reduced APPs levels with overexpression of ADAMTS4 (Fig. 2d), and indicated that ADAMTS4 might also partially control APPs levels in the brain.
Genetic deletion of ADAMTS4 lowers Aβ4–x and increases APPs levels in vivo. a Aβ4–40 levels were significantly reduced in the SDS-soluble brain fractions of 12-month-old 5xFAD/ADAMTS4−/− mice (n = 3) as compared to 5xFAD/ADAMTS4+/+ control animals (n = 3). b–d In contrast, Aβ1–40 (b) and Aβ1–42 (c) in the SDS fractions and Aβ1–42 (d) in the TBS fractions were not different between the genotypes. Unpaired t test was used to compare the means between the genotypes. **p < 0.01. e–g Full-length APP levels (e) in the SDS brain fraction as measured by Western blotting with antibody CT-15 were not different between the genotypes. Conversely, APPs (f) and APPs-β (g) levels in the TBS fractions as measured by Western blotting with antibodies 22C11 and 6A1 were significantly increased in 5xFAD/ADAMTS4−/− mice as compared to 5xFAD/ADAMTS4+/+ control animals. Each Western blot was repeated three times and one representative blot is shown. For quantification, APP signal intensities were measured, normalized to actin levels, and averaged for the three technical replicates. Means were calculated for each genotype, and the values of the 5xFAD/ADAMTS4+/+ control animals were set to 100%. 5xFAD/ADAMTS4−/− (n = 3), 5xFAD/ADAMTS4+/+ (n = 3-4). Unpaired t test was used to compare the means between the genotypes. *p < 0.05. nTg, non-transgenic C57Bl/6 control mice
ADAMTS4 generated Aβ4–x peptides in white matter fiber tracts and neuronal processes
Previously, Aβ4–x peptides have been localized to amyloid plaque cores and vascular amyloid deposits in the brains of AD patients and different AD mouse models including 5xFAD [11, 79]. Intriguingly, using the Aβ4–x-specific antibody 029-2, we discovered an additional pool of Aβ4–x peptides in white matter fiber tracts and neuronal processes of aged 5xFAD mice, which was dependent on the expression of ADAMTS4. Immunohistochemical analyses of 12-month-old 5xFAD mice revealed abundant Aβ4–x peptides within axon-rich fiber tracts such as the dorsal hippocampal commissure and the dorsal fornix that co-localized with the axonal marker APP (Supplementary Fig. S6). In the corpus callosum, abundant dot-like immunoreactivity was detected, which was significantly reduced in age-matched 5xFAD/ADAMTS4−/− mice (Supplementary Fig. S7). Aged 5xFAD mice also showed abundant Aβ4–x positive fibers in cortical areas together with Aβ4–x positive extracellular amyloid deposits (Fig. 4a, b). In contrast, no such Aβ4–x positive cellular processes were detected in age-matched 5xFAD/ADAMTS4−/− mice (Fig. 4c, d). Further analysis of the dorsal fornix demonstrated abundant co-localization of Aβ4–x peptides within fibers positive for the oligodendrocyte marker 2′, 3′-cyclic nucleotide 3′-phosphodiesterase (CNPase), an early myelin protein that continues to be expressed at high levels in the adult CNS [5] (Fig. 4e, f). This Aβ4–x immunoreactivity was completely absent in age-matched 5xFAD/ADAMTS4−/− mice (Fig. 4g, h). In summary, these results indicated that ADAMTS4 contributes to a novel pool of Aβ4–x peptides localized to white matter structures and neuronal processes.
Aβ4–x peptides in neuronal processes and white matter fiber tracts. a, b Aβ4–x positive fibers (arrows) were abundantly present in the cortex of 12-month-old 5xFAD mice. c, d Aβ4–x immunoreactive fibers were not detected in age-matched 5xFAD/ADAMTS4−/− mice. e, f Prominent Aβ4–x immunoreactivity was present in CNPase-positive fibers in the dorsal fornix (df) of 12-month-old 5XFAD mice. g, h This immunoreactivity was absent in age-matched 5xFAD/ADAMTS4−/− mice. The images in the left panel show high-power magnifications of the immunofluorescence stainings on the right. Scale bars: a, c 50 µm; b, d 15 µm; e, g 100 µm; f, h 33 µm
ADAMTS4 is exclusively expressed in oligodendrocytes and is essential for Aβ4–x peptide generation in primary oligodendrocyte cultures
The remarkable, ADAMTS4-dependent localization of Aβ4–x peptides to white matter fiber tracts and neuronal processes prompted us to investigate the expression of ADAMTS4 in the adult murine brain. Prior studies had provided conflicting results and evidence for ADAMTS4 expression in both neurons and all glial cell types of the central nervous system (CNS) [19, 39, 43, 61, 69, 82]. We took advantage of the ADAMTS4−/− KO mice, in which the ADAMTS4 gene locus was disrupted by insertion of a bacterial lacZ gene such that the endogenous ADAMTS4 gene promotor would drive the expression of β-galactosidase (β-gal). This enabled us to examine the ADAMTS4 expression pattern by β-gal immunohistochemistry. In cortical brain sections of 6.5-month-old 5xFAD/ADAMTS4−/− mice, expression of β-gal did not co-localize with the neuronal marker MAP2, the astrocytic marker GFAP, or the microglia marker Iba1 (Supplementary Fig. S8a-c). Instead, clear co-localization of β-gal positive cell nuclei with CNPase was observed, indicating that ADAMTS4 was exclusively expressed in the oligodendrocytic cell lineage (Supplementary Fig. S8d-f). Next, we studied the role of ADAMTS4 in APP processing and Aβ generation specifically in oligodendrocytes. Initially, to confirm ADAMTS4 expression in oligodendrocytes, we extracted oligodendrocyte progenitor cells (OPCs) from whole brains of newborn wild-type mice and differentiated them in culture to mature oligodendrocytes over a period of 5 days. ADAMTS4 mRNA expression was detected by qPCR both in OPCs and mature oligodendrocytes with a clear trend for increased expression in the course of the differentiation process (Supplementary Fig. S9a-c). Endogenous APP processing was then compared in OPC cultures from ADAMTS4−/− KO and wild-type control mice. Full-length APP levels in cell lysates were unchanged between the genotypes (Fig. 5a). However, total APPs levels were more than threefold higher in conditioned media of ADAMTS4−/− KO oligodendrocytes compared to wild-type cells (Fig. 5b), reflecting the increased APPs levels in the brains of 5xFAD/ADAMTS4−/− mice. To enable Aβ measurements, OPC cultures from ADAMTS4−/− KO and wild-type control mice were infected with an adenovirus expressing human APP695sw, and conditioned media were analyzed by mass spectrometry. In the supernatants of wild-type oligodendrocytes, Aβ4–40 peptides were clearly detected alongside the more common species like Aβ1–40, Aβ1–42 and Aβ1–38. In contrast, the mass spectra acquired from ADAMTS4−/− KO cells showed no peak corresponding to Aβ4–40 while the other Aβ species remained unchanged (Fig. 5c). To quantify these differences, conditioned media were analyzed with Aβ1–40 and Aβ4–40 specific ELISAs. Both wild-type oligodendrocytes and ADAMTS4−/− KO cells secreted comparable amounts of Aβ1–40 peptides (Fig. 5d). However, in sharp contrast, Aβ4–40 peptides were only detectable in conditioned media of ADAMTS4 expressing wild-type oligodendrocytes, indicating that the enzyme was essential for the generation of Aβ4–x peptides in this cell type (Fig. 5e).
ADAMTS4 is essential for Aβ4–x peptide generation and controls APPs levels in primary oligodendrocyte cultures. a-b Endogenous, full-length APP levels (a) were unchanged in oligodendrocyte progenitor cells (OPCs) cultured from wild type versus ADAMTS4−/− KO mice as measured by Western blotting with antibody CT-15. In contrast, APPs levels (b) were significantly increased in conditioned media of ADAMTS4−/− KO cultures compared to wild-type OPC cultures as measured by Western blotting with antibody 22C11. The Western blots and the quantifications show the results of three independent biological experiments (n = 3). APP signal intensities were normalized to actin levels, while APPs signals were normalized to APP levels in the corresponding cell lysates. Means were calculated for each genotype, and the values of the ADAMTS4+/+ control cells were set to 100%. Unpaired t test was used to compare the means between the genotypes. **p < 0.01. c Mass spectrometry analysis of Aβ peptide species in OPC cultures. The cells were infected with an adenovirus expressing APP695sw, and conditioned media were immunoprecipitated with anti-Aβ antibody 4G8 and analyzed by MALDI-TOF mass spectrometry. The mass spectra acquired from ADAMTS4+/+ control cells displayed a peak corresponding to Aβ4–40 peptides (enlargement, black box), which was absent in the spectra generated from ADAMTS4−/− KO cultures. Other Aβ species appeared unchanged between the genotypes. Three independent biological experiments (n = 3) were performed and representative mass spectra are shown. The measured monoisotopic masses and the expected masses, and the primary amino acid sequences of the detected peptides are shown in Supplementary Table 2. d, e OPCs cultured from wild type or ADAMTS4−/− KO mice were infected with the APP695sw adenovirus and conditioned media were analyzed with Aβ1–40 or Aβ4–40 specific ELISA assays. Cells from both genotypes secreted comparable amounts of Aβ1–40 peptides (d). In contrast, Aβ4–40 peptides (e) were only detectable in conditioned media of ADAMTS4 expressing wild-type OPCs but not in ADAMTS4−/− KO cultures. The graphs depict the results of three independent biological experiments (Exp) (n = 3)
Discussion
N-truncated Aβ peptides, in particular Aβ4–x and pyroglutamated AβpE3-42, are abundantly present in AD brains [45, 51, 58], yet the molecular mechanisms of N-terminal residue removal are largely unknown. In this study, using cellular and in vivo models, we showed that Aβ4–x peptides can be generated by the secreted metalloprotease ADAMTS4. Intriguingly, ADAMTS4 was recently identified as a potential novel AD risk locus by two large-scale genome-wide association studies available in preprint form [31, 47]. Our findings connect ADAMTS4 to one of the core pathological pathways in AD, APP processing and Aβ generation. Stable co-expression of APP and ADAMTS4 in HEK293 cells facilitated the secretion of Aβ4–x and Aβ12–x peptides, while Aβ1–x peptides with an intact N-terminus were not affected. Importantly, this was confirmed in model systems with endogenous ADAMTS4 expression and through genetic controls. In primary oligodendrocytes, ADAMTS4 was essential for Aβ4–40 secretion, and Aβ4–40 levels were reduced in the brains of aged 5xFAD/ADAMTS4−/− mice. However, Aβ12–x peptides were neither secreted by oligodendrocytes nor detected in the brain tissue of 5xFAD mice. This indicated that the second putative ADAMTS4 cleavage site in the Aβ sequence, which deviates from the proposed consensus sequence [27], was not recognized to any measurable degree in cells with endogenous ADAMTS4 expression, in agreement with the fact that Aβ12–x peptides have not been detected in the human brain [51, 58]. APP is a type-1 transmembrane protein located in the secretory pathway and at the cell surface, whereas ADAMTS4 is a secreted soluble metalloprotease, which becomes activated in the trans-Golgi network but has its main substrates in the extracellular matrix [53, 76]. Hence, an intriguing question is the cellular compartment(s) in which enzyme and substrate meet to produce Aβ4–x peptides. Our data indicate that Aβ4–x peptide generation might occur both within the cell and in the extracellular space. Treatment of HEK293 cells co-expressing APP and ADAMTS4 with a BACE1 inhibitor reduced Aβ1–40 in culture supernatants to undetectable levels but had no effect on Aβ4–40 levels. Under these conditions, Aβ4–40 could not have been generated through truncation of Aβ1–40 in the extracellular space. Instead, these Aβ4–40 peptides must have been generated intracellularly by ADAMTS4 cleavage of APP within the Aβ domain followed by γ-secretase cleavage, which in its catalytically active form is also enriched in late compartments of the secretory pathway and at the cell surface [35]. However, in cell-free assays, ADAMTS4 was also able to generate Aβ4–40 and Aβ4–42 peptides by truncation of recombinant Aβ1–40/Aβ1–42, indicating that ADAMTS4 might also use Aβ1–x peptides generated by BACE1 as a substrate in the extracellular space. In vivo, both of these mechanisms could apply. Co-expression of APP and endogenous ADAMTS4 in oligodendrocytes was sufficient to generate Aβ4–x peptides demonstrating that these cells could produce a fraction of the Aβ4–x peptides. Alternatively, ADAMTS4 might be secreted by oligodendrocytes and could process Aβ1–x in the extracellular space, or neuronal APP, which has been localized to the surface of axonal membranes [15]. Yet another possibility is that ADAMTS4 is secreted by oligodendrocytes and taken up by neurons through endocytosis, leading to the processing of APP by ADAMTS4 in intracellular vesicular compartments. Indeed, in cartilage, it has been demonstrated that extracellular ADAMTS4 can be internalized via low density lipoprotein receptor-related protein (LRP1), which is abundantly expressed in neurons [57, 81]. Further studies are required to clarify this issue, but all three of these possibilities conform to our immunohistochemical detection of Aβ4–x peptides in white matter structures and their co-localization with APP in fiber tracts of 5xFAD mice (see below).
In addition, ADAMTS4 consistently affected the levels of the secreted ectodomain APPs, which has neurotrophic properties and was shown to rescue most of the phenotypic deficits in APP−/− KO mice [53, 63]. ADAMTS4 overexpression strongly reduced APPs levels while conditioned media of ADAMTS4−/− KO oligodendrocytes showed a substantial increase in APPs levels compared to wild-type cells. A less prominent but consistent increase in APPs levels was also detected in brain tissue of 5xFAD/ADAMTS4−/− mice, with the smaller effect size likely reflecting the restricted expression of ADAMTS4 in the mouse brain. However, other APP processing changes seen in ADAMTS4 overexpressing cells such as changes in mature APP levels and the appearance of larger APP-CTF and smaller APPs fragments were not confirmed in oligodendrocyte cultures or in vivo, indicating that they were likely overexpression artifacts. These results demonstrated that the effects of ADAMTS4 are not limited to the generation of Aβ4–x peptides and that ADAMTS4 might partially control APPs turnover. This was also reminiscent of findings with the membrane-bound metalloprotease meprin β, which has been implicated in the generation of N-truncated Aβ2–x peptides. Like ADAMTS4, meprin β has been shown to cleave in the Aβ domain and further N-terminal in the APP ectodomain at multiple sites, resulting in higher APPs levels in brain tissue of meprin β−/− KO mice [33, 66]. Taken together, these studies provide further evidence that the mechanisms controlling APPs levels in the brain are more complex than previously thought and involve multiple proteases and cell types.
In the SDS-soluble brain fractions of 12-month-old 5xFAD mice, we observed a 50% reduction in Aβ4–40 and unchanged Aβ1–x levels in the ADAMTS4 KO background. While these results confirmed our tissue culture findings and proved the relevance of ADAMTS4 for Aβ4–x peptide generation in vivo, they further indicated that other proteases might participate in the production of these N-truncated Aβ species. One candidate protease could be the zinc metalloendopeptidase neprilysin (NEP), which is localized to the plasma membrane and functions to degrade extracellular peptides. NEP primarily cleaves at sites that feature a hydrophobic or aromatic amino acid residue C-terminal to the scissile bond in the P1′ position [26]. NEP is a major Aβ degrading enzyme, and until now has been the only protease demonstrated to cleave the Glu3–Phe4 bond in vitro [28]. However, in vivo studies have indicated that the rate-limiting step in the degradation of Aβ by neprilysin is cleavage of the Gly9–Tyr10 bond, which would prevent the formation of full-length Aβ4–40 and Aβ4–42 peptides [30]. Alternatively, other ADAMTS proteases with similar substrate specificity to ADAMTS4 might also contribute to Aβ4–x peptide generation, but little is known about their protein expression levels and functions in the brain [22]. The Aβ ELISA measurements further revealed that Aβ4–40 peptides have low abundance in the brain of 5xFAD mice compared to full-length Aβ1–x peptides. In 5-month-old 5xFAD mice, we previously reported Aβ4–40 levels to be approximately 75-fold and 200-fold lower compared to Aβ1–40 and Aβ1–42 levels [79]. In the 12-month-old 5xFAD animals in this study, the ratio of Aβ1–40/Aβ4–40 peptides remained similar while the Aβ1–42/Aβ4–40 ratio increased to around 1500-fold, likely due to the substantial progress of the amyloid pathology in the 5xFAD model between 5 and 12 months of age. A similar predominance of Aβ1–x over N-truncated Aβ peptides has been observed in other APP-transgenic mouse models [36, 41]. The low abundance of Aβ4–x peptides in the 5xFAD model does provide a straightforward explanation for the observation that the overall amyloid plaque load was not affected at 12-months of age in the ADAMTS4 KO background. In addition, the low Aβ4–x peptide levels in 5xFAD mice point to major differences in the composition of amyloid deposits between common AD mouse models and the human disease as studies have supported a high prevalence of Aβ4–x peptides in human AD brains [45, 48, 50, 51, 58, 59, 68]. Why Aβ4–x peptides accumulate to a much lower extent in 5xFAD mice as compared to human AD brain tissue is unclear at this point. Species specific differences in the brain biochemistry and the much shorter life span of mice might account for the differences, but the reasons might also at least in part relate to the design of APP-transgenic mouse models. In 5xFAD as in other AD mouse models the APP transgene is driven by promotors that direct expression mainly to neurons. Therefore, the co-expression of ADAMTS4 and the APP substrate in oligodendrocytes might be insufficient to generate more substantial amounts of Aβ4–x peptides. However, further studies are evidently required to fully understand the low abundance of Aβ4–x and other N-truncated Aβ peptides in AD animal models [36, 41].
Neurons are thought to be the main source of Aβ peptides in the brain, but glial cells also produce Aβ [55]. Pertinent to our study, oligodendrocytes have been suggested to produce Aβ peptides in similar amounts to neurons [70]. Previous studies had detected ADAMTS4 protein expression by immunohistochemistry in the human, mouse and rat CNS in neurons, astrocytes and oligodendrocytes, but none of these studies had included ADAMTS4 KO tissue sections as genetic controls to validate the specificity of the employed commercial antibodies [19, 23, 39, 43]. Surprisingly, our studies using β-galactosidase reporter mice demonstrated that ADAMTS4 was exclusively expressed by oligodendrocytes in the adult murine brain. However, several previous studies had also strongly suggested that ADAMTS4 could be an oligodendrocyte-specific gene. First, an in situ hybridization study had detected ADAMTS4 mRNA specifically in oligodendrocytes of the mouse hippocampus and cortex at P21, and in co-localization studies the riboprobe only bound to cells positive for the oligodendrocyte marker Olig2 [44]. Second, a state-of-the-art transcriptome study of all major brain cell types had indicated that ADAMTS4 is specifically expressed in oligodendrocytes [82]. In his study, brain cell populations were acutely isolated from postnatal mice with a combination of immunopanning and fluorescence-activated cell sorting and analyzed by RNA-sequencing. In the resulting cell type-specific transcriptomes, ADAMTS4 was highly expressed in myelinating and mature oligodendrocytes but not in neurons, astrocytes, microglia, endothelial cells or pericytes [82]. This was further confirmed in a proteome study of murine brain cell types, in which ADAMTS4 belonged to the 10 most abundant proteins specifically expressed in cultured murine oligodendrocytes [69]. Most importantly, during the preparation of this manuscript, another research group had reported the use of the same ADAMTS4−/− KO reporter mouse strain to study the expression of ADAMTS4 in the brain, and had also detected ADAMTS4 in oligodendrocytes [61]. In the adult mouse brain, expression of β-galactosidase was exclusively localized to Olig2-positive oligodendrocytes. Further studies demonstrated that ADAMTS4 expression was restricted to cells co-expressing adenomatous-polyposis-coli protein, a marker for mature oligodendrocytes [61]. Finally, the exclusive expression of ADAMTS4 in oligodendrocytes is consistent with our immunohistochemical detection of Aβ4–x peptides in white matter fiber tracts and neuronal processes in cortical areas. This novel pool of Aβ4–x peptides was dependent on the expression of ADAMTS4, in contrast to the previously reported Aβ4–x peptides in amyloid plaque cores [11, 79], which appeared unchanged in the ADAMTS4−/− KO background. Aβ4–x peptides have displayed aggregation properties and toxicity comparable to Aβ1–42 peptides, which are regarded as a key trigger in the pathogenesis of AD [7, 11, 14, 56]. Consequently, the presence of Aβ4–x peptides in white matter structures might contribute to demyelination and white matter damage. Neuropathological, biochemical, and imaging studies have all provided evidence for white matter abnormalities and oligodendrocyte dysfunction in the brains of AD patients [10, 13, 52, 65]. Most intriguingly, magnetic resonance and diffusion tensor imaging have documented white matter abnormalities already in pre-symptomatic carriers of early-onset familial AD mutations [42, 64]. However, whether mouse models of AD display white matter abnormalities has been controversial and contradictory findings have been reported for commonly used mouse lines [17, 25, 37, 72], in line with the fact that these models mainly replicate the amyloidosis but not the degenerative hallmarks of AD [34]. The etiology of white matter lesions in AD is unknown and it has been proposed that oligodendrocyte degeneration occurs mainly secondary to the loss of neurons and axons (so called Wallerian degeneration) [49]. On the other hand, studies have shown that Aβ peptides are cytotoxic to oligodendrocytes and that they accumulate in white matter structures [12, 16, 32, 65, 80], raising the possibility that N-truncated Aβ4–x peptides produced by oligodendrocytes or in myelinated axons could be uniquely harmful. This putative pathogenic mechanism should be investigated further, which will likely require new animal models that account for a pro-amyloidogenic role of oligodendrocytes in the pathogenesis of AD.
References
Association Alzheimer’s (2016) 2016 Alzheimer’s disease facts and figures. Alzheimers Dement 12:459–509
Antonios G, Saiepour N, Bouter Y, Richard BC, Paetau A, Verkkoniemi-Ahola A et al (2013) N-truncated Abeta starting with position four: early intraneuronal accumulation and rescue of toxicity using NT4X-167, a novel monoclonal antibody. Acta Neuropathol Commun 1:56. https://doi.org/10.1186/2051-5960-1-56
Apte SS (2009) A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem 284:31493–31497. https://doi.org/10.1074/jbc.R109.052340
Bateman RJ, Aisen PS, De Strooper B, Fox NC, Lemere CA, Ringman JM et al (2011) Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res Ther 2:35. https://doi.org/10.1186/alzrt59
Baumann N, Pham-Dinh D (2001) Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev 81:871–927
Bayer TA, Wirths O (2014) Focusing the amyloid cascade hypothesis on N-truncated Abeta peptides as drug targets against Alzheimer’s disease. Acta Neuropathol 127:787–801. https://doi.org/10.1007/s00401-014-1287-x
Bouter Y, Dietrich K, Wittnam JL, Rezaei-Ghaleh N, Pillot T, Papot-Couturier S et al (2013) N-truncated amyloid beta (Abeta) 4-42 forms stable aggregates and induces acute and long-lasting behavioral deficits. Acta Neuropathol 126:189–205. https://doi.org/10.1007/s00401-013-1129-2
Brockhaus M, Grunberg J, Rohrig S, Loetscher H, Wittenburg N, Baumeister R et al (1998) Caspase-mediated cleavage is not required for the activity of presenilins in amyloidogenesis and NOTCH signaling. NeuroReport 9:1481–1486
Bros P, Delatour V, Vialaret J, Lalere B, Barthelemy N, Gabelle A et al (2015) Quantitative detection of amyloid-beta peptides by mass spectrometry: state of the art and clinical applications. Clin Chem Lab Med 53:1483–1493. https://doi.org/10.1515/cclm-2014-1048
Brun A, Englund E (1986) A white matter disorder in dementia of the Alzheimer type: a pathoanatomical study. Ann Neurol 19:253–262. https://doi.org/10.1002/ana.410190306
Cabrera E, Mathews P, Mezhericher E, Beach TG, Deng J, Neubert TA et al (2018) Abeta truncated species: implications for brain clearance mechanisms and amyloid plaque deposition. Biochim Biophys Acta 1864:208–225. https://doi.org/10.1016/j.bbadis.2017.07.005
Collins-Praino LE, Francis YI, Griffith EY, Wiegman AF, Urbach J, Lawton A et al (2014) Soluble amyloid beta levels are elevated in the white matter of Alzheimer’s patients, independent of cortical plaque severity. Acta Neuropathol Commun 2:83. https://doi.org/10.1186/s40478-014-0083-010.1186/preaccept-3091772881321882
De Strooper B, Karran E (2016) The Cellular Phase of Alzheimer’s Disease. Cell 164:603–615. https://doi.org/10.1016/j.cell.2015.12.056
De Strooper B, Vassar R, Golde T (2010) The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol 6:99–107. https://doi.org/10.1038/nrneurol.2009.218
DeBoer SR, Dolios G, Wang R, Sisodia SS (2014) Differential release of beta-amyloid from dendrite- versus axon-targeted APP. J Neurosci 34:12313–12327. https://doi.org/10.1523/jneurosci.2255-14.2014
Desai MK, Mastrangelo MA, Ryan DA, Sudol KL, Narrow WC, Bowers WJ (2010) Early oligodendrocyte/myelin pathology in Alzheimer’s disease mice constitutes a novel therapeutic target. Am J Pathol 177:1422–1435. https://doi.org/10.2353/ajpath.2010.100087
Desai MK, Sudol KL, Janelsins MC, Mastrangelo MA, Frazer ME, Bowers WJ (2009) Triple-transgenic Alzheimer’s disease mice exhibit region-specific abnormalities in brain myelination patterns prior to appearance of amyloid and tau pathology. Glia 57:54–65. https://doi.org/10.1002/glia.20734
Dimitrov M, Alattia JR, Lemmin T, Lehal R, Fligier A, Houacine J et al (2013) Alzheimer’s disease mutations in APP but not gamma-secretase modulators affect epsilon-cleavage-dependent AICD production. Nat Commun 4:2246. https://doi.org/10.1038/ncomms3246
Dubey D, McRae PA, Rankin-Gee EK, Baranov E, Wandrey L, Rogers S et al (2017) Increased metalloproteinase activity in the hippocampus following status epilepticus. Epilepsy Res 132:50–58. https://doi.org/10.1016/j.eplepsyres.2017.02.021
Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D et al (1998) A third-generation lentivirus vector with a conditional packaging system. J Virol 72:8463–8471
Gerber H, Wu F, Dimitrov M, Garcia Osuna GM, Fraering PC (2017) Zinc and Copper Differentially Modulate Amyloid Precursor Protein Processing by gamma-Secretase and Amyloid-beta Peptide Production. J Biol Chem 292:3751–3767. https://doi.org/10.1074/jbc.M116.754101
Gottschall PE, Howell MD (2015) ADAMTS expression and function in central nervous system injury and disorders. Matrix Biol 44–46:70–76. https://doi.org/10.1016/j.matbio.2015.01.014
Haddock G, Cross AK, Plumb J, Surr J, Buttle DJ, Bunning RA et al (2006) Expression of ADAMTS-1, -4, -5 and TIMP-3 in normal and multiple sclerosis CNS white matter. Mult Scler 12:386–396
Hahn S, Bruning T, Ness J, Czirr E, Baches S, Gijsen H et al (2011) Presenilin-1 but not amyloid precursor protein mutations present in mouse models of Alzheimer’s disease attenuate the response of cultured cells to gamma-secretase modulators regardless of their potency and structure. J Neurochem 116:385–395. https://doi.org/10.1111/j.1471-4159.2010.07118.x
Harms MP, Kotyk JJ, Merchant KM (2006) Evaluation of white matter integrity in ex vivo brains of amyloid plaque-bearing APPsw transgenic mice using magnetic resonance diffusion tensor imaging. Exp Neurol 199:408–415. https://doi.org/10.1016/j.expneurol.2006.01.002
Hersh LB, Rodgers DW (2008) Neprilysin and amyloid beta peptide degradation. Curr Alzheimer Res 5:225–231
Hills R, Mazzarella R, Fok K, Liu M, Nemirovskiy O, Leone J et al (2007) Identification of an ADAMTS-4 cleavage motif using phage display leads to the development of fluorogenic peptide substrates and reveals matrilin-3 as a novel substrate. J Biol Chem 282:11101–11109. https://doi.org/10.1074/jbc.M611588200
Howell S, Nalbantoglu J, Crine P (1995) Neutral endopeptidase can hydrolyze beta-amyloid(1-40) but shows no effect on beta-amyloid precursor protein metabolism. Peptides 16:647–652
Huttenrauch M, Baches S, Gerth J, Bayer TA, Weggen S, Wirths O (2015) Neprilysin deficiency alters the neuropathological and behavioral phenotype in the 5XFAD mouse model of Alzheimer’s disease. J Alzheimers Dis 44:1291–1302. https://doi.org/10.3233/JAD-142463
Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E et al (2000) Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat Med 6:143–150. https://doi.org/10.1038/72237
Jansen I, Savage J, Watanabe K, Bryois J, Williams D, Steinberg S et al (2018) Genetic meta-analysis identifies 9 novel loci and functional pathways for Alzheimers disease risk. bioRxiv 258533; doi: https://doi.org/10.1101/258533
Jantaratnotai N, Ryu JK, Kim SU, McLarnon JG (2003) Amyloid beta peptide-induced corpus callosum damage and glial activation in vivo. NeuroReport 14:1429–1433. https://doi.org/10.1097/01.wnr.0000086097.47480.a0
Jefferson T, Causevic M, Auf dem Keller U, Schilling O, Isbert S, Geyer R et al (2011) Metalloprotease Meprin beta Generates Nontoxic N-terminal Amyloid Precursor Protein Fragments in Vivo. J Biol Chem 286:27741–27750. https://doi.org/10.1074/jbc.M111.252718
Jucker M (2010) The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat Med 16:1210–1214. https://doi.org/10.1038/nm.2224
Kaether C, Lammich S, Edbauer D, Ertl M, Rietdorf J, Capell A et al (2002) Presenilin-1 affects trafficking and processing of betaAPP and is targeted in a complex with nicastrin to the plasma membrane. J Cell Biol 158:551–561. https://doi.org/10.1083/jcb.200201123
Kalback W, Watson MD, Kokjohn TA, Kuo YM, Weiss N, Luehrs DC et al (2002) APP transgenic mice Tg2576 accumulate Abeta peptides that are distinct from the chemically modified and insoluble peptides deposited in Alzheimer’s disease senile plaques. Biochemistry 41:922–928
Kastyak-Ibrahim MZ, Di Curzio DL, Buist R, Herrera SL, Albensi BC, Del Bigio MR et al (2013) Neurofibrillary tangles and plaques are not accompanied by white matter pathology in aged triple transgenic-Alzheimer disease mice. Magn Reson Imaging 31:1515–1521. https://doi.org/10.1016/j.mri.2013.06.013
Kelwick R, Desanlis I, Wheeler GN, Edwards DR (2015) The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol 16:113. https://doi.org/10.1186/s13059-015-0676-3
Krstic D, Rodriguez M, Knuesel I (2012) Regulated proteolytic processing of Reelin through interplay of tissue plasminogen activator (tPA), ADAMTS-4, ADAMTS-5, and their modulators. PLoS ONE 7:e47793. https://doi.org/10.1371/journal.pone.0047793
Kummer MP, Heneka MT (2014) Truncated and modified amyloid-beta species. Alzheimers Res Ther 6:28. https://doi.org/10.1186/alzrt258
Kuo YM, Kokjohn TA, Beach TG, Sue LI, Brune D, Lopez JC et al (2001) Comparative analysis of amyloid-beta chemical structure and amyloid plaque morphology of transgenic mouse and Alzheimer’s disease brains. J Biol Chem 276:12991–12998. https://doi.org/10.1074/jbc.M007859200
Lee S, Viqar F, Zimmerman ME, Narkhede A, Tosto G, Benzinger TL et al (2016) White matter hyperintensities are a core feature of Alzheimer’s disease: evidence from the dominantly inherited Alzheimer network. Ann Neurol 79:929–939. https://doi.org/10.1002/ana.24647
Lemarchant S, Pomeshchik Y, Kidin I, Karkkainen V, Valonen P, Lehtonen S et al (2016) ADAMTS-4 promotes neurodegeneration in a mouse model of amyotrophic lateral sclerosis. Mol Neurodegener 11:10. https://doi.org/10.1186/s13024-016-0078-3
Levy C, Brooks JM, Chen J, Su J, Fox MA (2015) Cell-specific and developmental expression of lectican-cleaving proteases in mouse hippocampus and neocortex. J Comp Neurol 523:629–648. https://doi.org/10.1002/cne.23701
Lewis H, Beher D, Cookson N, Oakley A, Piggott M, Morris CM et al (2006) Quantification of Alzheimer pathology in ageing and dementia: age-related accumulation of amyloid-beta(42) peptide in vascular dementia. Neuropathol Appl Neurobiol 32:103–118. https://doi.org/10.1111/j.1365-2990.2006.00696.x
Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J et al (2007) A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2:1236–1247. https://doi.org/10.1038/nprot.2007.135
Marioni R, Harris SE, McRae AF, Zhang Q, Hagenaars SP, Hill WD et al (2018) GWAS on family history of Alzheimer’s disease. Transl Psychiatry 8:99. https://doi.org/10.1038/s41398-018-0150-6
Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K (1985) Amyloid plaque core protein in Alzheimer disease and down syndrome. Proc Natl Acad Sci U S A 82:4245–4249
McAleese KE, Walker L, Graham S, Moya ELJ, Johnson M, Erskine D et al (2017) Parietal white matter lesions in Alzheimer’s disease are associated with cortical neurodegenerative pathology, but not with small vessel disease. Acta Neuropathol 134:459–473. https://doi.org/10.1007/s00401-017-1738-2
Miller DL, Papayannopoulos IA, Styles J, Bobin SA, Lin YY, Biemann K et al (1993) Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer’s disease. Arch Biochem Biophys 301:41–52. https://doi.org/10.1006/abbi.1993.1112
Moore BD, Chakrabarty P, Levites Y, Kukar TL, Baine AM, Moroni T et al (2012) Overlapping profiles of Abeta peptides in the Alzheimer’s disease and pathological aging brains. Alzheimers Res Ther 4:18. https://doi.org/10.1186/alzrt121
Nasrabady SE, Rizvi B, Goldman JE, Brickman AM (2018) White matter changes in Alzheimer’s disease: a focus on myelin and oligodendrocytes. Acta Neuropathol Commun 6:22. https://doi.org/10.1186/s40478-018-0515-3
Nhan HS, Chiang K, Koo EH (2015) The multifaceted nature of amyloid precursor protein and its proteolytic fragments: friends and foes. Acta Neuropathol 129:1–19. https://doi.org/10.1007/s00401-014-1347-2
Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J et al (2006) Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci 26:10129–10140
Oberstein TJ, Spitzer P, Klafki HW, Linning P, Neff F, Knolker HJ et al (2015) Astrocytes and microglia but not neurons preferentially generate N-terminally truncated Abeta peptides. Neurobiol Dis 73:24–35. https://doi.org/10.1016/j.nbd.2014.08.031
Pike CJ, Overman MJ, Cotman CW (1995) Amino-terminal deletions enhance aggregation of beta-amyloid peptides in vitro. J Biol Chem 270:23895–23898
Pohlkamp T, Wasser CR, Herz J (2017) Functional Roles of the Interaction of APP and Lipoprotein Receptors. Front Mol Neurosci 10:54. https://doi.org/10.3389/fnmol.2017.00054
Portelius E, Bogdanovic N, Gustavsson MK, Volkmann I, Brinkmalm G, Zetterberg H et al (2010) Mass spectrometric characterization of brain amyloid beta isoform signatures in familial and sporadic Alzheimer’s disease. Acta Neuropathol 120:185–193. https://doi.org/10.1007/s00401-010-0690-1
Portelius E, Lashley T, Westerlund A, Persson R, Fox NC, Blennow K et al (2015) Brain amyloid-beta fragment signatures in pathological ageing and Alzheimer’s disease by hybrid immunoprecipitation mass spectrometry. Neurodegener Dis 15:50–57. https://doi.org/10.1159/000369465
Pratta MA, Yao W, Decicco C, Tortorella MD, Liu RQ, Copeland RA et al (2003) Aggrecan protects cartilage collagen from proteolytic cleavage. J Biol Chem 278:45539–45545. https://doi.org/10.1074/jbc.M303737200
Pruvost M, Lepine M, Leonetti C, Etard O, Naveau M, Agin V et al (2017) ADAMTS-4 in oligodendrocytes contributes to myelination with an impact on motor function. Glia 65:1961–1975. https://doi.org/10.1002/glia.23207
Reinert J, Richard BC, Klafki HW, Friedrich B, Bayer TA, Wiltfang J et al (2016) Deposition of C-terminally truncated Abeta species Abeta37 and Abeta39 in Alzheimer’s disease and transgenic mouse models. Acta Neuropathol Commun 4:24. https://doi.org/10.1186/s40478-016-0294-7
Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA et al (2007) The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci 27:7817–7826. https://doi.org/10.1523/jneurosci.1026-07.2007
Ringman JM, O’Neill J, Geschwind D, Medina L, Apostolova LG, Rodriguez Y et al (2007) Diffusion tensor imaging in preclinical and presymptomatic carriers of familial Alzheimer’s disease mutations. Brain 130:1767–1776. https://doi.org/10.1093/brain/awm102
Roher AE, Weiss N, Kokjohn TA, Kuo YM, Kalback W, Anthony J et al (2002) Increased A beta peptides and reduced cholesterol and myelin proteins characterize white matter degeneration in Alzheimer’s disease. Biochemistry 41:11080–11090
Schonherr C, Bien J, Isbert S, Wichert R, Prox J, Altmeppen H et al (2016) Generation of aggregation prone N-terminally truncated amyloid beta peptides by meprin beta depends on the sequence specificity at the cleavage site. Mol Neurodegener 11:19. https://doi.org/10.1186/s13024-016-0084-5
Selkoe DJ, Hardy J (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8:595–608. https://doi.org/10.15252/emmm.201606210
Sergeant N, Bombois S, Ghestem A, Drobecq H, Kostanjevecki V, Missiaen C et al (2003) Truncated beta-amyloid peptide species in pre-clinical Alzheimer’s disease as new targets for the vaccination approach. J Neurochem 85:1581–1591
Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N et al (2015) Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci 18:1819–1831. https://doi.org/10.1038/nn.4160
Skaper SD, Evans NA, Evans NA, Rosin C, Facci L, Richardson JC (2009) Oligodendrocytes are a novel source of amyloid peptide generation. Neurochem Res 34:2243–2250. https://doi.org/10.1007/s11064-009-0022-9
Song RH, Tortorella MD, Malfait AM, Alston JT, Yang Z, Arner EC et al (2007) Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis Rheum 56:575–585. https://doi.org/10.1002/art.22334
Sun SW, Song SK, Harms MP, Lin SJ, Holtzman DM, Merchant KM et al (2005) Detection of age-dependent brain injury in a mouse model of brain amyloidosis associated with Alzheimer’s disease using magnetic resonance diffusion tensor imaging. Exp Neurol 191:77–85. https://doi.org/10.1016/j.expneurol.2004.09.006
Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R et al (1999) Purification and cloning of aggrecanase-1: a member of the ADAMTS family of proteins. Science 284:1664–1666
Tortorella MD, Malfait F, Barve RA, Shieh HS, Malfait AM (2009) A review of the ADAMTS family, pharmaceutical targets of the future. Curr Pharm Des 15:2359–2374
Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I et al (2000) Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4). J Biol Chem 275:18566–18573. https://doi.org/10.1074/jbc.M909383199
Wang P, Tortorella M, England K, Malfait AM, Thomas G, Arner EC et al (2004) Proprotein convertase furin interacts with and cleaves pro-ADAMTS4 (Aggrecanase-1) in the trans-Golgi network. J Biol Chem 279:15434–15440. https://doi.org/10.1074/jbc.M312797200
Weggen S, Eriksen JL, Das P, Sagi SA, Wang R, Pietrzik CU et al (2001) A subset of NSAIDs lower amyloidogenic Abeta42 independently of cyclooxygenase activity. Nature 414:212–216
Willem M, Lammich S, Haass C (2009) Function, regulation and therapeutic properties of beta-secretase (BACE1). Semin Cell Dev Biol 20:175–182. https://doi.org/10.1016/j.semcdb.2009.01.003
Wirths O, Walter S, Kraus I, Klafki HW, Stazi M, Oberstein TJ et al (2017) N-truncated Abeta4-x peptides in sporadic Alzheimer’s disease cases and transgenic Alzheimer mouse models. Alzheimers Res Ther 9:80. https://doi.org/10.1186/s13195-017-0309-z
Xu J, Chen S, Ahmed SH, Chen H, Ku G, Goldberg MP et al (2001) Amyloid-beta peptides are cytotoxic to oligodendrocytes. J Neurosci 21:Rc118
Yamamoto K, Owen K, Parker AE, Scilabra SD, Dudhia J, Strickland DK et al (2014) Low density lipoprotein receptor-related protein 1 (LRP1)-mediated endocytic clearance of a disintegrin and metalloproteinase with thrombospondin motifs-4 (ADAMTS-4): functional differences of non-catalytic domains of ADAMTS-4 and ADAMTS-5 in LRP1 binding. J Biol Chem 289:6462–6474. https://doi.org/10.1074/jbc.M113.545376
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S et al (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. https://doi.org/10.1523/jneurosci.1860-14.2014
Acknowledgements
We thank Karlheinz Baumann and Manfred Brockhaus (F. Hoffmann-La Roche Ltd., Basel, Switzerland) for carboxyl terminus-specific Aβ antibodies, and Guido Reifenberger (Heinrich-Heine-University Duesseldorf, Germany) for encouragement and support.
Funding
This work was supported by grants from the Stiftung VERUM (to S.W.) and the Forschungskommission of the Medical Faculty of the Heinrich-Heine-University Duesseldorf (grant 9772513 to T.J. and S.W.), the Alzheimer Forschung Initiative (grant 16013 to O.W.), and the foundations Strauss, Eclosion and SFNTF (to H.G. and P.C.F.).
Author information
Authors and Affiliations
Contributions
SW, TJ, DB, OW and SW designed the study. SW, TJ, MH, HG, MD, IO, SL, KL, SZ, SES, and OW designed and performed experiments. SW, TJ, MH, HG., MD, IO, SL, CB, JW, CB-P, CUP, PCF, OW and SW analyzed data, discussed results and provided scientific input throughout the study. SW, OW and SW wrote the paper with input and approval from all authors.
Corresponding authors
Ethics declarations
Conflict of interest
D.B. is the Chief Executive Officer of Asceneuron SA. All other authors declare that they have no conflict of interest.
Ethical approval
All animal experiments were carried out in accordance with German guidelines for animal care and have been approved by the local responsible committee.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Walter, S., Jumpertz, T., Hüttenrauch, M. et al. The metalloprotease ADAMTS4 generates N-truncated Aβ4–x species and marks oligodendrocytes as a source of amyloidogenic peptides in Alzheimer’s disease. Acta Neuropathol 137, 239–257 (2019). https://doi.org/10.1007/s00401-018-1929-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-018-1929-5