Abstract
Pilocytic astrocytomas (PAs) are the most common brain tumors in pediatric patients and can cause significant morbidity, including chronic neurological deficiencies. They are characterized by activating alterations in the mitogen-activated protein kinase pathway, but little else is known about their development. To map the global DNA methylation profiles of these tumors, we analyzed 62 PAs and 7 normal cerebellum samples using Illumina 450K microarrays. These data revealed two subgroups of PA that separate according to tumor location (infratentorial versus supratentorial), and identified key neural developmental genes that are differentially methylated between the two groups, including NR2E1 and EN2. Integration with transcriptome microarray data highlighted significant expression differences, which were unexpectedly associated with a strong positive correlation between methylation and expression. Differentially methylated probes were often identified within the gene body and/or regions up- or downstream of the gene, rather than at the transcription start site. We also identified a large number of differentially methylated genes between cerebellar PAs and normal cerebellum, which were again enriched for developmental genes. In addition, we found a significant association between differentially methylated genes and SUZ12 binding sites, indicating potential disruption of the polycomb repressor complex 2 (PRC2). Taken together, these data suggest that PA from different locations in the brain may arise from region-specific cells of origin, and highlight the potential disruption of key developmental regulators during tumorigenesis. These findings have implications for future basic research and clinical trials, as therapeutic targets and drug sensitivity may differ according to tumor location.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pilocytic astrocytoma (PA) is the most common central nervous system tumor in pediatric patients, with highest incidence between 5 and 15 years of age. They are classified as grade I by the World Health Organization (WHO), reflecting their slow growth and typically non-invasive behavior. Despite excellent overall survival of the patients, tumor recurrence is common. This is intrinsically linked to surgical accessibility, with complete resection associated with significantly longer progression-free survival than subtotal, partial or no resection [8, 30]. As such, PA is often considered as a chronic disease, and both the tumor and treatments can cause significant morbidity [1, 5]. PAs most frequently occur in the cerebellum, but are also found in the optic chiasm, hypothalamus, brain stem and, rarely, hemispheres or spinal cord. They are characterized by aberrations in the mitogen-activated protein kinase (MAPK) pathway, with approximately 60 % of cases demonstrating a KIAA1549:BRAF fusion gene as a result of tandem duplication at 7q34 [7, 15, 22, 28]. Alternate mechanisms of MAPK pathway activation include fusion of BRAF to the FAM131B gene, BRAF mutation, RAF1 fusion gene formation and NF1 mutation, as reviewed in Jones et al. [13]. The typically slow growth of PAs has been suggested to arise from oncogene-induced senescence in response to activation of the MAPK pathway [12, 23].
Besides MAPK pathway activation, little is known about the development and origin of PA. The majority of cases display a balanced karyotype as assessed by aCGH, with the remainder showing whole chromosomal gains, most commonly of chromosomes 5 and 7 [14, 22]. PAs arising in older patients (>15 years of age) show more frequent whole chromosomal changes, suggesting an age-based association [14]. Alterations in genes associated with higher-grade astrocytic brain tumors such as IDH1/2 mutation, TP53 mutation, MGMT methylation, EGFR amplification and PTEN loss are not typically seen in PA [6, 11, 14, 21, 24]. Transcriptome studies of PA to date have identified distinct gene expression signatures associated with PAs arising in patients with neurofibromatosis type 1 (NF1), as well as PAs arising in different locations within the brain [27, 33]. The overlap in gene signatures, however, is generally not consistent between studies, making it difficult to infer the importance of individual genes and pathways to PA development and prognosis.
To further define the molecular development of PA, we have used global DNA methylation profiling in a series of 62 tumors. The Illumina HumanMethylation450 platform contains in excess of 480,000 CpG probes, allowing comprehensive profiling of methylation patterns across the genome. We have identified distinct methylation profiles characterizing infratentorial and supratentorial subgroups of PA, with an excellent correlation to expression changes between the two groups. Our results provide an in-depth analysis of the molecular development underlying PAs and highlight new candidate genes in PA pathogenesis for future study.
Materials and methods
Tumor samples
DNA was extracted from a total of 129 tumors, including 39 PAs from the Cambridge cohort and 90 PAs from the Heidelberg cohort. Upon collection, tumors were bisected with one half taken for histopathological examination, and the second snap-frozen for molecular analysis. Only those samples with a tumor cell content estimated to be ≥70 % from histopathological assessment were included for molecular analysis. Full clinicopathological data was available for all cases, including location of the tumor, age of the patient at diagnosis and gender. All samples were collected with informed consent and ethical approval.
Illumina Infinium HumanMethylation450 BeadChips
Twenty-five PAs from the Cambridge cohort and 37 from the Heidelberg cohort were analyzed using the Illumina Infinium HumanMethylation450 BeadChips according to the manufacturer’s instructions, at the German Cancer Research Center (DKFZ) Genomics and Proteomics Core Facility. Seven normal cerebellum samples purchased from BioChain (US) were also analyzed: CP-D05-D1244039 (lot numbers: B405047, B405045, B405043), D1244039 (lot number: B207026) and CP-D05-D1234039 (lot numbers: B106087, B405048, A708134). Full clinicopathological data for tumors and normal cerebellum are listed in Supplemental Table 1. Two samples of whole-genome amplified DNA (unmethylated control) and two samples of M.SssI-treated DNA (100 % methylated control) were also included as further controls (data not shown). The following criteria were applied to filter the data: removal of probes targeting the X and Y chromosomes (n = 11,551), removal of probes containing a single-nucleotide polymorphism (dbSNP132 Common) within five base pairs of and including the targeted CpG-site (n = 24,536), and probes not mapping uniquely to the human reference genome (hg19) allowing for one mismatch (n = 9,993). In total 438,370 probes were kept for analysis. CpG methylation values for all samples are available through Gene Expression Omnibus (GEO: http://www.ncbi.nlm.nih.gov/geo/), accession number GSE44684.
Data analysis
Ensembl gene annotations (release 64) were obtained from BioMart (http://www.biomart.org/). Probes within ±10 kb of protein-coding genes were collapsed by taking the mean of the 25 % most variably methylated probes. The significance analysis of microarrays (SAM) method was used to identify genes which are differentially methylated. Correction for multiple testing was performed using the Benjamini–Hochberg method. Genes were considered significantly differentially methylated between groups when displaying an adjusted p value <0.05 and a methylation difference of ≥0.2. K-means consensus clustering (k = 2 to 4) was performed as previously described [31], using the 3,200 most variably methylated probes. For gene ontology analysis, the differentially methylated gene list was uploaded to the online bioinformatic tool DAVID, using a background of all genes included in the differential methylation analysis. For the SUZ12 analysis, binding site data was obtained from ENCODE via the UCSC Genome Browser, version hg19. A one-sided Wilcoxon rank sum test was used for each set of differentially methylated genes to determine whether the degree of differential methylation was higher in CpG probes at SUZ12 binding sites than in probes not at SUZ12 binding sites. The degree of differential methylation at individual CpG probes was assessed by one-sided Wilcoxon rank sum tests for hypermethylation and hypomethylation in tumor samples compared to normal tissue, while two-sided tests were used for non-directional differential methylation between supratentorial and infratentorial PA.
Expression profiling
Forty-nine PAs and 9 normal cerebellum samples were analyzed on the Affymetrix GeneChip® Human Genome U133 Plus 2.0 Array at the Microarray Department of the University of Amsterdam, the Netherlands. Sample library preparation, hybridization, and quality control were performed according to the manufacturer’s protocols. Expression data were normalized using the MAS5.0 algorithm of the GCOS program (Affymetrix Inc). Differences in gene expression for target genes were investigated using a t test between groups, with a Bonferroni correction for multiple testing (p ≤ 0.05). The raw cel files and array matrix data have been deposited in the GEO database, accession number GSE44971.
Pyrosequencing
To validate the methylation microarrays, DNA samples from 92 PAs were analyzed by pyrosequencing for the genes EN2, NR2E1, PAX3/CCDC140, VAX1, and FEZF2. These included 39 PAs from the Cambridge series (24 of which had been analyzed using the Illumina 450K BeadChip) and 53 from an independent set of PAs in the Heidelberg series (none of which had been analyzed by the Illumina BeadChips). Briefly, 500 ng of DNA was bisulphite-modified using the Zymo EZ DNA Methylation Kit (Zymo, US) according to the manufacturer’s instructions. The bisulphite-modified DNA was amplified using a gene-specific primer pair with a biotinylated reverse primer (see supplementary information for primer sequences). Single-strand purification of PCR products was performed using the PyroMark Vacuum Prep Workstation (Qiagen, UK) and samples were pyrosequenced using PyroGold Q96 SQA reagents with the PyroMark ID pyrosequencer according to the manufacturer’s recommendations. Data were analyzed using the Pyro-Q-CpG software (Qiagen, UK). Each assay was designed to include at least one CpG-site that was represented by a differentially methylated probe on the Illumina BeadChip. Results for each sample are expressed as an average value across all CpGs analyzed by any given assay (CpG count range 3–6). Statistical significance was determined by a t test performed in Microsoft Excel.
qRT-PCR
RNA was available for 14 tumors from the Cambridge PA series (7 of which had also been analyzed by the Illumina methylation array) and 42 PAs from the Heidelberg cohort. For the Cambridge series, 2 μg of RNA was reverse transcribed in a 40-μl reaction volume using SuperScript III (Invitrogen, UK) according to the manufacturer’s instructions. For the Heidelberg cohort, 500 ng of RNA was reverse transcribed in a 20-μl reaction volume. qPCRs were performed using the Light Cycler 480 Sybr Green Master Mix (Roche, UK) and the LightCycler 480 machine (Roche). Melting-curve analysis was used to determine the optimum conditions for each reaction. The housekeeping gene H6PD was used for relative quantitation; samples that showed outlier H6PD values were excluded (n = 3). Samples with assay replicates that showed >0.5 Ct difference were also excluded. All primer sequences are listed in the supplemental information.
Results
A series of 62 PAs was subjected to global DNA methylation profiling using Illumina Infinium HumanMethylation450 BeadChips, including 40 tumors from the cerebellum and 22 PAs from other locations within the brain (2 fourth ventricle, 2 brain stem, 8 diencephalon, 5 optic nerve, 4 cerebral and 1 third ventricle, Supplemental Table 1). In addition, 5 fetal and 2 adult normal cerebellum DNA samples from commercial sources were analyzed. Outlier detection excluded one cerebellar PA that clustered close to the normal cerebellum samples and was deemed to contain a significant component of non-tumor cell contamination, and one adult normal cerebellum. These two samples were removed from all further analyses, leaving 61 PAs and 6 normal cerebellums for downstream analysis (Supplemental Fig. S1).
Methylation profiling supports two distinct subgroups of PA that separate by tumor location
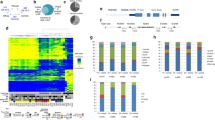
K-means consensus clustering was used to investigate the overall pattern of methylation in the tumor samples. This revealed two major subgroups that largely separated by tumor location, with 95 % (37/39) of cerebellar tumors in one cluster, and 91 % (20/22) of all other brain locations in the second cluster (Fisher’s exact test: p < 0.0001, Fig. 1, Supplemental Fig. 2). The two non-cerebellar tumors that clustered within the cerebellum group were both infratentorial tumors (either brain stem or fourth ventricle), as per the cerebellum itself. The remaining two brain stem/fourth ventricle PAs clustered with two outlying cerebellar samples in the non-cerebellum cluster. Thus, our data clearly indicate a difference in methylation profiles between infratentorial PAs (cerebellar, brain stem, and fourth ventricle) and supratentorial PAs (optic chiasm, diencephalon, third ventricle, and cerebral).
K-means consensus clustering of the top 3,200 most variant probes reveals two subgroups of PA based on tumor location. Clinicopathological and molecular details of MAPK alteration are shown beneath the heatmap. The normal cerebellum samples and control DNAs (100 % methylation and no methylation) are shown to the right of the main heatmap
Developmental genes are differentially regulated between infratentorial and supratentorial PA
We next analyzed the differences between the subgroups to identify those probes that were differentially methylated between the two clusters. A gene-centric approach was utilized using the top 25 % most variant probes for each gene (±10 kb of the gene body). Using a median β value difference of 0.2 or greater and an adjusted p value <0.05, we identified a total of 268 probes that showed significant differences in methylation between the two clusters. In total these probes covered 21 genes (Table 1), with an average of 13 differentially methylated probes per gene (range 3–24). As the four outlying infratentorial tumors in the supratentorial cluster showed some variation compared with the supratentorial tumors in the same cluster (Fig. 1), we also performed the differential methylation analysis with these four tumors excluded. This identified an additional 59 probes encompassing a further 5 genes that showed differential methylation between the two clusters (Table 1; Fig. 2a). All genes identified from the previous comparison were also identified as significantly differentially methylated in the second analysis. A volcano plot of the data revealed a general shift towards lower methylation levels (hypomethylation) in the infratentorial PAs as compared to the supratentorial PAs, further underlining the differences between these two subgroups (Fig. 2a).
Infratentorial versus supratentorial cluster analysis (excluding infratentorial outliers). a Volcano plot showing genes significantly differentially methylated (in red). Infratentorial tumors also show an overall pattern of hypomethylation compared with supratentorial PA, as evidenced by the large number of genes between −0.2 and 0.0 compared with 0.0 and 0.2. b IGV plot of the MASP1 gene (±10 kb), revealing differential methylation at the transcription start site (gene is encoded on the reverse strand). c Boxplot of MASP1 methylation values from the Illumina BeadChip by subgroup showing statistically significant differences between the two groups of PA. d Boxplot of MASP1 expression values from the Affymetrix microarrays, again showing a significant difference between infratentorial and supratentorial PA. e Correlation between methylation and expression in the 37 overlapping samples for which data was available from both microarray platforms
We used the IGV genome browser to visualize differentially methylated candidates on a gene-by-gene basis. Of the 26 genes, only 4 showed differential methylation of probes at the transcription start site (TSS: MASP1, PRKCDBP, GP6, and ACSBG2). The pattern of the remaining differentially methylated probes was either within the gene body and/or in regions upstream/downstream (within 10 kb) of the gene. To assess the impact of differential methylation on gene expression, we examined these genes in a dataset of 49 PAs analyzed by the Affymetrix HG U133 Plus 2.0 microarrays. This dataset comprised 12 supratentorial PAs and 37 infratentorial PAs (including 37 samples that had also been analyzed in the methylation study and 12 independent samples). Of those genes that showed differential methylation at the TSS, only MASP1 showed a concordant difference in gene expression (i.e., hypermethylation in the supratentorial tumors and decreased expression in comparison to infratentorial tumors; Fig. 2b–e). PRKCDBP, GP6, and ACSBG2 that also showed TSS methylation differences did not show any difference in expression between the two tumor groups (p > 0.05).
Significant expression changes were seen for the majority of the remaining genes that showed differential methylation within the gene body and/or upstream/downstream; however, there was a striking pattern of positive correlation between methylation and expression. For example, hypermethylation of EN2, PAX3, CCDC140, MSX2, and IRX3 was observed in the infratentorial PAs, but these tumors also showed significantly higher expression of these genes than their supratentorial counterparts. Similarly, NR2E1, SIX3, and FEZF2 showed hypermethylation and higher expression levels in supratentorial PAs when compared to infratentorial PAs (Fig. 3). Although the number of identified genes was small, gene ontology analysis revealed a significant enrichment of terms related to transcription, neuron development/differentiation and regional brain development (adjusted p < 0.05, Supplemental Table 2). Consistent with this, the only Interpro protein domains that were significantly enriched amongst the genes were either homeobox or homeodomain, both of which are associated with development and transcription factor binding (Benjamini adjusted p < 0.05, Supplemental Table 2).
Gene body methylation at the NR2E1 locus corresponds to a positive correlation between methylation and expression. a IGV plot of the NR2E1 gene (±10 kb) showing a block of differentially methylated probes between exons 2–5, compared to compete hypomethylation at the transcription start site for all PA. b Boxplot of NR2E1 methylation values from the Illumina Beadchip (infratentorial outliers excluded). c Boxplot of expression values from the Affymetrix microarrays. d Correlation between methylation and expression reveals a positive correlation (as methylation values increase, so do expression levels)
To validate the methylation and transcriptome findings, we designed pyrosequencing and qRT-PCR assays for a selection of 5 genes (EN2, NR2E1, PAX3/CCDC140, VAX1, and FEZF2). All pyrosequencing assays were designed to include at least one CpG that was represented by a differentially methylated probe on the Illumina Infinium 450k BeadChip. We analyzed a total of 92 PAs, which included 24 that had also been analyzed by the Infinium BeadChip and 68 independent cases. For all five genes, pyrosequencing confirmed the findings from the methylation array analysis and revealed statistically significant differences between supratentorial and infratentorial tumors (Fig. 4, Supplemental Fig. S3). In addition, a direct comparison of the BeadChip and pyrosequencing values for the 24 cases for which data were available from both methods revealed an excellent correlation between the two technologies, with an R 2 value of ≥0.9 across these samples (Supplemental Table 3). We also examined the expression levels for EN2, NR2E1, PAX3, FEZF2, and VAX1 using qRT-PCR assays and confirmed the correlation between non-TSS-based hypermethylation and overexpression of these genes (Fig. 4, Supplemental Fig. S3). Overall, these orthogonal techniques showed an excellent correlation with the array findings.
Developmental genes are amongst those differentially methylated between cerebellar PA and normal cerebellum
We next compared the cerebellar PA cluster with the normal cerebellum samples to identify differences between tumor and normal. A similar analysis for the supratentorial tumors was not possible because of the range of locations included and the paucity of frozen control material from regions such as the optic chiasm and diencephalon. Using a β value difference of 0.25 or greater, we identified 3,007 differentially methylated probes encompassing 408 genes. Of these, 228 genes were hypermethylated in the PA and 180 were hypomethylated (Supplemental Table 4). Gene ontology analysis revealed an enrichment of terms associated with neuron differentiation and development, immune response, cell death and the regulation of phosphorylation, although these categories did not reach statistical significance after correction for multiple testing (Supplemental Table 5). The enrichment of terms associated with neuronal features may be partly due to the large neuronal component of normal cerebellum, an inherent limitation of using this tissue for comparison. Affymetrix expression microarray data were used to investigate transcriptome differences for these genes, with 358/408 genes represented by probes on the array. Significant differential expression was identified for 212 genes (adjusted p ≤ 0.05). In contrast to the subgroup-specific differential methylation, the majority of genes in this comparison (86 %) were negatively correlated (i.e., hypermethylated and underexpressed, or hypomethylated and overexpressed), as expected by the classical model of gene expression regulation by DNA methylation.
Differential methylation is enriched for probes at SUZ12 binding sites
We noted from visual inspection of the differentially methylated genes that there appeared to be frequent overlap between differentially methylated probes and SUZ12 binding sites (Supplemental Fig. S4). We therefore proposed to formally test whether differential methylation was indeed enriched at these sites. An analysis of the probes across the 26 genes differentially methylated between the infratentorial PA cluster and supratentorial PA cluster revealed there was a significant association between probes that were differentially methylated and localization within SUZ12 binding sites (p = 0.00015). For analysis of the differentially methylated gene list from the cerebellar PA cluster versus normal cerebellum comparison, the genes were split into those that were hypermethylated in PAs and those that were hypomethylated, and then analyzed separately. This revealed that probes that were hypermethylated in the PAs showed a significant association with SUZ12 sites (p = 0.047), whereas those that were hypomethylated did not (p = 1.0).
Discussion
DNA methylation is a form of epigenetic regulation that is essential to developmental processes and normal gene expression. Alterations in DNA methylation have been increasingly identified as pivotal to disease, and efforts are emerging to use epigenetic alterations as druggable targets in cancer [16]. The results presented above indicate that differential methylation is likely to be important in the development of PA arising at different locations in the brain. Differential gene expression of PA based on tumor location has previously been reported, and was suggested to arise from differences in their cell of origin [27, 33]. Sharma and colleagues identified a 6 gene-signature that distinguished infratentorial and supratentorial PA, as well as cerebellar (infratentorial) and neocortical (supratentorial) astrocytes and neural stem cells. Of these 6 genes, 4 were found to be differentially methylated in this study (SIX3, NR2E1, IRX2, and PAX3), suggesting regional specificity may be largely defined by patterns of methylation that reflect local development and environmental cues at that particular site in the brain. In addition, Tchoghandjian and colleagues identified a large number of genes that were differentially expressed between PA from the hypothalamic/optic chiasm and cerebellar PA. This included a subset involved in brain development—a gene ontology category also identified by this study—however, there were no specific genes in common between our study and the study by Tchoghandjian and colleagues.
The developmental genes identified as differentially methylated between the PA subgroups support a pattern of regional development. For example, the Engrailed Homeobox 2 (EN2) gene encodes a protein that is pivotal to patterning of the cerebellum during development [4], and was found to show gene body hypermethylation with increased expression in the infratentorial cluster of tumors. Similarly, six homeobox 3 (SIX3), ventral anterior homeobox 1 (VAX1) and nuclear receptor subfamily 2, group E, member 1 (NR2E1, also known as TLX, or tailless) showed non-TSS based hypermethylation with overexpression in the supratentorial tumors. All three genes are important for development of the diencephalon and optic chiasm region, and NR2E1 in particular has a demonstrated role in neural stem cell expansion and glioma formation [3, 19, 34, 35]. Our data indicates the role of these genes warrants further investigation to determine their functional contribution to PA development. It would be interesting to investigate their role in regional specificity using tumors derived from different regions of the brain in the recently described mouse model of PA [9]. In addition, we identified global levels of hypomethylation in infratentorial PA compared to supratentorial PA. A similar distribution has been reported in ependymoma [26], further highlighting the intrinsic differences between tumors arising at different locations within the brain.
Evidence of regional specificity was provided in a recent study by Lee and colleagues [17], who demonstrated that mouse neural stem cells from different brain regions show distinct growth responses to the KIAA1549:BRAF fusion gene (the most common genetic aberration in PA). They found that neural stem cells from the third ventricle (where PA can arise) show increased proliferation in response to the KIAA1549:BRAF fusion gene, whereas proliferation rates were unchanged in neural stem cells from the lateral ventricle, where PAs are rare [17]. They suggested the differential response to genetic aberrations is a direct result of innate signatures reflecting the regional identity of progenitor cells, which, from the evidence presented in this study as well as previous expression studies of PA, could be linked to the distinct epigenetic programs of region-specific cells of origin [27, 33].
The positive association identified between DNA methylation and expression frequently involved gene body methylation, which has previously been linked to high levels of gene expression in a variety of contexts. Hellman and Chess [10] identified gene body-specific methylation on the active X chromosome that contrasted to promoter methylation and gene silencing on the inactive X chromosome. Similarly, Rauch and colleagues [25] used a technique for global enrichment of methylated DNA and identified that approximately 12 % of genes were methylated in the gene body near the 3′ end, and this correlated with increased expression levels. Gene body methylation has been suggested to increase levels of transcription by repressing internal gene promoters, suppressing repetitive elements and switching promoter usage within a gene [2, 10]. In addition, a recent study by Szulwach and colleagues [32] identified 5-hydroxymethylcytosine (5-hmC, a derivative of 5-methylcytosine) within the gene bodies of more highly expressed genes in the development of the mouse cerebellum, which was both age and location-specific.
We have also identified an association between SUZ12 binding sites and hypermethylation of genes in PA. SUZ12 is a member of the polycomb repressive complex 2 (PRC2) that plays an important role in controlling gene expression during embryogenesis and development. The core PRC2 complex is made up of four components (EZH1/2, SUZ12, EED, and RbAp46/48) that collectively bind to target sites and methylate histone H3 lysine 27 (a repressive mark that results in gene silencing), reviewed in Margueron and Reinberg [20]. Aberrant methylation patterns at SUZ12 binding sites could result in disruption of SUZ12 binding, with subsequent alteration in the activity of the PRC2 complex, as has been demonstrated at the imprinted Rasgrf1 locus in mice [18]. This would alter the transcriptional status of target genes and potentially enhance tumorigenesis through their disrupted expression. The targets of SUZ12 binding have been shown to be cell type specific, with transcription factors and homeobox genes targeted in embryogenesis compared to glycoproteins and immunoglobulin-related genes in adult tumors [29]. The enrichment of developmental transcription factors and homeobox proteins identified in this study is in keeping with the pattern of SUZ12 binding in progenitor cell populations.
In summary, we have identified distinct subgroups of PA that can be distinguished by the differential methylation of key brain developmental regulators, pointing towards distinct cells of origin in these subgroups. This study significantly increases our current knowledge of the biology underlying PA, and suggests that tumor location should be considered in future clinical trials, since the clear biological differences identified here may also have an effect on drug response and sensitivity. This is particularly important for supratentorial PA, where surgical resection is often challenging and for which new targeted therapeutics would be particularly advantageous.
References
Armstrong GT, Conklin HM, Huang S et al (2011) Survival and long-term health and cognitive outcomes after low-grade glioma. Neuro Oncol 13:223–234. doi:10.1093/neuonc/noq178
Bert SA, Robinson MD, Strbenac D et al (2013) Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell 23:9–22. doi:10.1016/j.ccr.2012.11.006
Bertuzzi S, Hindges R, Mui SH, O’Leary DD, Lemke G (1999) The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev 13:3092–3105
Cheng Y, Sudarov A, Szulc KU et al (2010) The Engrailed homeobox genes determine the different foliation patterns in the vermis and hemispheres of the mammalian cerebellum. Development 137:519–529. doi:10.1242/dev.027045
Dirven CM, Mooij JJ, Molenaar WM (1997) Cerebellar pilocytic astrocytoma: a treatment protocol based upon analysis of 73 cases and a review of the literature. Childs Nerv Syst 13:17–23
Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG (1999) Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 59:793–797
Forshew T, Tatevossian RG, Lawson AR et al (2009) Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol 218:172–181. doi:10.1002/path.2558
Gnekow AK, Falkenstein F, von Hornstein S et al (2012) Long-term follow-up of the multicenter, multidisciplinary treatment study HIT-LGG-1996 for low-grade glioma in children and adolescents of the German Speaking Society of Pediatric Oncology and Hematology. Neuro Oncol 14:1265–1284. doi:10.1093/neuonc/nos202
Gronych J, Korshunov A, Bageritz J et al (2011) An activated mutant BRAF kinase domain is sufficient to induce pilocytic astrocytoma in mice. J Clin Invest 121:1344–1348. doi:10.1172/JCI44656
Hellman A, Chess A (2007) Gene body-specific methylation on the active X chromosome. Science 315:1141–1143. doi:10.1126/science.1136352
Ichimura K, Pearson DM, Kocialkowski S et al (2009) IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol 11:341–347. doi:10.1215/15228517-2009-025
Jacob K, Quang-Khuong DA, Jones DTW et al (2011) Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas. Clin Cancer Res 17:4650–4660. doi:10.1158/1078-0432.CCR-11-0127
Jones DTW, Gronych J, Lichter P, Witt O, Pfister SM (2012) MAPK pathway activation in pilocytic astrocytoma. Cell Mol Life Sci 69:1799–1811. doi:10.1007/s00018-011-0898-9
Jones DTW, Ichimura K, Liu L, Pearson DM, Plant K, Collins VP (2006) Genomic analysis of pilocytic astrocytomas at 0.97 Mb resolution shows an increasing tendency toward chromosomal copy number change with age. J Neuropathol Exp Neurol 65:1049–1058. doi:10.1097/01.jnen.0000240465.33628.87
Jones DTW, Kocialkowski S, Liu L et al (2008) Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68:8673–8677. doi:10.1158/0008-5472.CAN-08-2097
Kelly TK, de Carvalho DD, Jones PA (2010) Epigenetic modifications as therapeutic targets. Nat Biotechnol 28:1069–1078. doi:10.1038/nbt.1678
Lee DY, Gianino SM, Gutmann DH (2012) Innate neural stem cell heterogeneity determines the patterning of glioma formation in children. Cancer Cell 22:131–138. doi:10.1016/j.ccr.2012.05.036
Lindroth AM, Park YJ, McLean CM et al (2008) Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet 4:e1000145. doi:10.1371/journal.pgen.1000145
Liu HK, Wang Y, Belz T et al (2010) The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev 24:683–695. doi:10.1101/gad.560310
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349. doi:10.1038/nature09784
Ohgaki H, Eibl RH, Schwab M et al (1993) Mutations of the p53 tumor suppressor gene in neoplasms of the human nervous system. Mol Carcinog 8:74–80
Pfister S, Janzarik WG, Remke M et al (2008) BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest 118:1739–1749. doi:10.1172/JCI33656
Raabe EH, Lim KS, Kim JM et al (2011) BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clin Cancer Res 17:3590–3599. doi:10.1158/1078-0432.CCR-10-3349
Rasheed BK, McLendon RE, Herndon JE et al (1994) Alterations of the TP53 gene in human gliomas. Cancer Res 54:1324–1330
Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP (2009) A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci USA 106:671–678. doi:10.1073/pnas.0812399106
Rogers HA, Kilday JP, Mayne C et al (2012) Supratentorial and spinal pediatric ependymomas display a hypermethylated phenotype which includes the loss of tumor suppressor genes involved in the control of cell growth and death. Acta Neuropathol 123:711–725. doi:10.1007/s00401-011-0904-1
Sharma MK, Mansur DB, Reifenberger G et al (2007) Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res 67:890–900. doi:10.1158/0008-5472.CAN-06-0973
Sievert AJ, Jackson EM, Gai X et al (2009) Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol 19:449–458. doi:10.1111/j.1750-3639.2008.00225.x
Squazzo SL, O’Geen H, Komashko VM et al (2006) Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res 16:890–900. doi:10.1101/gr.5306606
Stokland T, Liu JF, Ironside JW et al (2010) A multivariate analysis of factors determining tumor progression in childhood low-grade glioma: a population-based cohort study (CCLG CNS9702). Neuro Oncol 12:1257–1268. doi:10.1093/neuonc/noq092
Sturm D, Witt H, Hovestadt V et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437. doi:10.1016/j.ccr.2012.08.024
Szulwach KE, Li X, Li Y et al (2011) 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci 14:1607–1616. doi:10.1038/nn.2959
Tchoghandjian A, Fernandez C, Colin C et al (2009) Pilocytic astrocytoma of the optic pathway: a tumour deriving from radial glia cells with a specific gene signature. Brain 132:1523–1535. doi:10.1093/brain/awp048
Yu RT, Chiang MY, Tanabe T et al (2000) The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci USA 97:2621–2625. doi:10.1073/pnas.050566897
Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G (2002) Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development 129:2835–2849
Acknowledgments
This project was supported by grants from The Brain Tumour Charity, UK. Additional support was provided by the PedBrain Tumor Project contributing to the International Cancer Genome Consortium, funded by German Cancer Aid (109252) and by the German Federal Ministry of Education and Research (BMBF, grants #01KU1201A, MedSys #0315416C and NGFNplus #01GS0883), and the Dutch Cancer Foundations KWF (2010-4713) and KIKA to M. Kool. We thank Matthias Schick, Roger Fischer and Melanie Bewerunge-Hudler of the German Cancer Research Center (DKFZ) Genomics and Proteomics Core Facility for excellent technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sally R. Lambert and Hendrik Witt contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lambert, S.R., Witt, H., Hovestadt, V. et al. Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol 126, 291–301 (2013). https://doi.org/10.1007/s00401-013-1124-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-013-1124-7