Abstract
Purpose
Previous observational studies have shown that alcohol and coffee were associated with colorectal cancer (CRC) risk, but the causal relationships have not been adequately explored. This study aimed to assess the potential causal associations of alcohol and coffee with CRC risk using Mendelian randomization (MR) analyses in an East Asian population.
Methods
Publicly available summary-level genome-wide association studies data on ever/never alcohol drinker (n = 165,084), alcohol consumption (n = 58,610), coffee consumption (n = 152,634), and CRC (7062 cases and 195,745 controls) were obtained from the BioBank Japan (BBJ). Single-nucleotide polymorphisms (SNPs) that were significantly related to the exposures were identified as instrumental variables. Five, two, and six SNPs were used for ever/never alcohol drinkers, alcohol consumption, and coffee consumption, respectively. The inverse variance weighted method was used as the main MR method to calculate the odds ratios (ORs) and 95% confidence intervals (95% CIs) of CRC risk per one-unit change in exposures.
Results
Genetically predicted ever/never alcohol drinkers (OR: 1.08; 95% CI 1.06, 1.11; P < 0.001) and alcohol consumption (OR: 1.39; 95% CI 1.21, 1.60; P < 0.001) were positively associated with CRC risk. Conversely, genetically predicted coffee consumption was inversely related to CRC risk, with an OR (95% CI) of 0.80 (0.64, 0.99) (P = 0.037).
Conclusion
Genetically predicted alcohol use and consumption were risk factors for CRC while genetically predicted coffee consumption was a protective factor. Our findings highlight the effectiveness of keeping healthy dietary habits to prevent CRC. Further studies with more valid SNPs and CRC cases are needed. Validation of our findings is also recommended.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) ranked third for cancer incidence and second for cancer mortality worldwide in 2020 [1]. It is a multi-aetiological disease which involves many risk factors, such as old age, male sex, family history, smoking, alcohol, obesity, and unhealthy diet [2]. Therefore, identifying its causative factors is critical for its prevention [2].
Previous observational studies have shown that alcohol and coffee were associated with CRC risk. For example, a meta-analysis of 61 studies reported that both moderate and heavy alcohol drinkers had higher CRC risk compared with non-drinkers or occasional drinkers [3]. A recent umbrella review suggested that coffee consumption was inversely associated with CRC risk [4]. However, observational studies could not be used to infer causal association due to the inevitably uncontrolled confounders and possible reverse causality [5,6,7]. Randomized controlled trial (RCT), though is the gold standard to infer causality, is infeasible and unethical when it comes to some dietary habits such as alcohol [5,6,7]. Thus far, it remains unclear whether alcohol and coffee were causally associated with CRC risk.
Mendelian randomization (MR) is a study design for inferring causality [5,6,7]. It usually uses single-nucleotide polymorphisms (SNPs) as instrumental variables (IVs) to predict the causal effects of exposures on outcomes [5,6,7]. Since SNPs are naturally randomly assigned to individuals at meiosis (like a natural RCT), MR may overcome the limitations of observational studies and RCTs [5,6,7]. However, limited MR studies were performed to explore the associations of alcohol and coffee with CRC risk, and existing MR studies had inconsistent results. For example, Alex and colleagues selected three alcohol-related SNPs from a meta-analysis and four coffee-related SNPs from the Coffee and Caffeine Genetics Consortium (CCGC). But they found that both alcohol and coffee consumption were not associated with CRC risk in European populations based on MR analyses (26,397 CRC cases and 41,481 controls) [8]. In another MR study in European populations (46,155 CRC cases and 270,342 controls), coffee consumption had different associations with CRC risk based on adoption of SNPs from different sources [9]. More specifically, when the SNPs from the United Kingdom Biobank (UKB) were used, coffee was a risk factor for CRC; while data on SNPs extracted from the CCGC indicated that coffee consumption was not related to CRC risk.
In addition, since most related MR studies were conducted in European populations, the findings could not be directly applied to East Asian populations due to the different characteristics of alcohol consumption, coffee intake, and CRC between East Asian and European populations. For example, the alcohol per capita consumption (litres) of Central Europe (11.64; 95% uncertainty intervals (UI): 10.59, 12.68), Eastern Europe (11.55; 95% UI 9.64, 13.46), and Western Europe (11.13; 95% UI 10.46, 10.81) were all much higher than that of East Asia (7.14; 95% UI 5.34, 8.94) in 2015 [10]. People in European regions also consumed more coffee than those in East Asian regions such as Japan, China, and India in 2017 [11]. Additionally, in 2020, for both males and females, the incidence of both colon cancer and rectal cancer were higher in Europe than in East Asia [1].
In this study, we aimed to investigate the potential causal associations of ever/never alcohol drinkers, alcohol consumption, and coffee consumption with CRC risk using MR analyses in an East Asian population.
Methods
This MR study was performed and reported according to the strengthening the reporting of observational studies in epidemiology using Mendelian randomization (STROBE-MR) guideline [6]. Ethical approval and written informed consents were obtained in previous studies [12, 13]. This study did not exceed the scope of the original ethics committee approval.
Data sources
Summary-level genome-wide association studies (GWAS) data on ever/never alcohol drinker (n = 165,084), alcohol consumption (n = 58,610), coffee consumption (n = 152,634), and CRC (7062 cases and 195,745 controls) were obtained from the BioBank Japan (BBJ) [12, 13]. The BBJ is the largest East Asian biobank, which consists of over 200,000 East Asian individuals aged 20–89 years old [12, 13]. Participants were recruited and followed up between 2003 and 2018 in 12 medical institutions in the BBJ. Written informed consents were obtained from all participants, and related studies were approved by the ethical committees of RIKEN Yokohama Institute and the Institute of Medical Science, the University of Tokyo [12, 13]. All GWAS data are publicly available at website (JENGER; http://jenger.riken.jp/en/) and the National Bioscience Database Center Human Database (https://humandbs.biosciencedbc.jp/en/) without access restriction [12, 13].
For the exposures [12], in the BBJ, the information of alcohol and coffee were collected by a standardized questionnaire. For alcohol, the participants were first asked whether they had current or past alcohol drinking (yes or no). All alcohol drinkers were those who answered “yes”, and never drinkers were those who answered “no”. Next, for participants who drink alcohol currently, the information of the type, volume (ml/day), and frequency (days/week) of alcohol drinks were collected. The alcohol consumption (g/week) was then calculated by multiplying the percentage of alcohol in the drinks by the volume and frequency. For coffee [12], a four-point scale (1 = almost every day, 2 = 3–4 days per week, 3 = 1–2 days per week, and 4 = rarely) was used to evaluate the consumption frequency. SNPs with call rate <0.99, minor allele frequency (MAF) <0.005, or P-value of Hardy–Weinberg equilibrium (PHWE) <1.0 × 10−6 were excluded during quality control of genotypes. GWAS were performed to identify SNPs associated with the three dietary habits using the following covariates: age, age2, sex, and the status of diseases in the BBJ [12].
For the outcome, during the same period of the exposures, 7062 CRC cases (4496 males and 2566 females, aged 67.0 ± 10.2 years old) and 195,745 controls (97,655 males and 98,090 females, aged 61.6 ± 13.9 years old) were included in the BBJ [13]. Clinical data on CRC such as diagnosis, surgery, therapy, and tumor markers were provided by cooperating institutions. CRC was diagnosed by experienced physicians at each cooperating institution. Participants in the control group were recruited from other four Japanese population-based cohorts and the BBJ itself. Subjects with CRC and CRC-related cancers (esophageal cancer, gastric cancer, biliary tract cancer, hepatocellular carcinoma, and pancreatic cancer) were excluded from the control group. The genotype quality control criteria for the outcome are the same as those for the exposures. Age, sex, and the top five principal components were included as covariates in the GWAS of CRC [13].
Instrumental variables selection
SNPs significantly (P-value <5 × 10–8) associated with alcohol and coffee were selected as IVs [5,6,7]. SNPs with an effect allele frequency (EAF) ≤0.05 and palindromic SNPs (SNPs with A/T or C/G) with an EAF between 0.4 and 0.7 were further excluded [5,6,7]. We subsequently clumped the SNPs in the window of 10,000 kilobases and set a threshold of linkage disequilibrium (LD)-r2 > 0.01 to remove SNPs in high LD [5,6,7]. Finally, five (rs1260326, rs1229984, rs3043, rs8187929, and rs671), two (rs1229984 and rs671), and six (rs6681426, rs1260326, rs4410790, rs671, rs58806801, and rs5760444) SNPs were identified as IVs forever/never alcohol drinkers, alcohol consumption, and coffee consumption, respectively (Table 1).
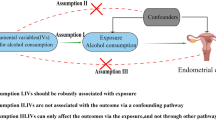
An IV should satisfy three basic MR assumptions: (1) the IV should be associated with the exposure (relevance assumption); (2) the IV should not be associated with confounders (independence assumption); and (3) the IV should affect the outcome only via the exposure (exclusion restriction assumption) (Fig. 1) [5,6,7]. The first assumption was tested by calculating the explained variance (R2) and the F-statistics of IVs in this study [5,6,7]. The R2 was calculated by: \({\beta }^{2}\times 2\times \mathrm{MAF}\times (1-\mathrm{MAF})]/\mathrm{Var}(Y)\), where β refers to the regression coefficient for one copy of the allele; MAF is the minor allele frequency; and Var(Y) indicates the variance of the exposure [14]. The F-statistic was calculated by: \(F=\frac{n-k-1}{k}\times \frac{{R}^{2}}{1-{R}^{2}}\), where n refers to the sample size of the exposure database; k is the number of IV; and R2 indicates the explained variance [15]. A F-statistic >10 was regarded as strong IVs [5,6,7]. The second and third assumptions cannot be tested directly, but can be partially satisfied if horizontal pleiotropy was not present [5,6,7]. Horizontal pleiotropy indicated that an IV affects the outcome via another trait other than the one of interest [5,6,7]. A P-value of MR Egger intercept <0.05 suggested that MR Egger intercept was significantly different from zero, and implied that horizontal pleiotropy is present [5,6,7].
Statistical analyses
The GWAS data on CRC were extracted according to the SNPs identified for the exposures. No SNP identified from the exposure dataset is absent in the outcome database. All GWAS data were then harmonized to ensure that the allele associated with higher levels of exposures was coded as the effect allele.
In the main MR analyses, the inverse variance weighted (IVW) method was used. The heterogeneity of SNPs in an IV was assessed by Cochran’s Q test. A P-value of heterogeneity <0.05 was considered as high heterogeneity and the random-effects (RE) IVW model was used, otherwise the fixed-effects (FE) IVW model was used. The odds ratios (ORs) and 95% confidence intervals (95% CIs) of CRC risk per one-unit change in exposures were calculated.
The following sensitivity analyses were conducted. First, to test the robustness of the associations, we performed MR analyses using three other MR methods (MR Egger, weighted median, and weighted mode) [7]. The MR Egger method could return unbiased results even if the exclusion restriction assumption is violated. In addition, the MR Egger intercept and corresponding P-value (P-value of pleiotropy) could be used to test the horizontal pleiotropy. A P-value of pleiotropy <0.05 was regarded as the presence of horizontal pleiotropy. The weighted median method takes the median effect of SNPs in an IV and allows up to 50% SNPs violate the three MR assumptions. The weighted mode method clusters SNPs in an IV into several groups according to the SNP-exposure associations, and computes the MR estimates based on the group with the largest number of SNPs. Therefore, the results will be unbiased if the SNPs in the largest group do not violate the three MR assumptions. Second, scatter plots were performed to visualize the association between SNP-exposure and SNP-outcome effects. Third, leave-one-out analyses were used to evaluate the effect of each SNP on CRC. The above sensitivity analyses were only performed for ever/never alcohol drinkers and coffee consumption, because the number of SNPs used in alcohol consumption were inadequate for the other three MR methods (SNPs number = 2).
Power calculations were conducted using an online tool (http://cnsgenomics.com/shiny/mRnd/) [16, 17], assuming a type-I error rate of 0.05. The needed data (R2 and sample size and cases proportion of outcome) were obtained from the BBJ [12, 13]. The minimum/maximum ORs to reach a power of 80% were 1.18/0.83 for ever/never alcohol drinkers, 1.33/0.68 for alcohol consumption, and 1.63/0.40 for coffee consumption. In addition, to detect an OR of 0.90 or 1.10, the statistical powers were 37% for ever/never alcohol drinkers, 14% for alcohol consumption, and 7% for coffee consumption (Table 2).
All MR analyses were performed using the TwoSampleMR package (version 0.5.6) in R (version 4.1.0). The details on R codes were shown in Online Resource 1. A two-sided P-value <0.05 was considered statistically significant.
Results
The characteristics of SNPs used in this study are shown in Table 1. Briefly, the IVs could explain 3.97% variance for ever/never alcohol drinkers, 1.11% variance for alcohol consumption, and 0.30% variance for coffee consumption. The F-statistics of IVs were 6827 for ever/never alcohol drinkers, 6603 for alcohol consumption, and 451 for coffee consumption. For the main analyses (Table 3), compared with never alcohol drinkers, ever alcohol drinkers had higher CRC risk (IVW-FE OR 1.08; 95% CI 1.06, 1.11; P < 0.001). Alcohol consumption was also positively associated with CRC risk (IVW-FE OR 1.39; 95% CI 1.21, 1.60; P < 0.001; per standard deviation (SD) increase). Conversely, coffee consumption was inversely related to CRC risk (IVW-RE OR: 0.80; 95% CI 0.64, 0.99; P = 0.037; higher versus lower consumption frequency).
In sensitivity analyses, all results of ever/never alcohol drinkers and coffee consumption were robust when three other MR methods were used (Table 3, Online Resources 2 and 3). The ORs (95% CIs) of MR Egger, weighted median, and weighted mode were 1.08 (1.04, 1.12), 1.08 (1.06, 1.11), and 1.08 (1.05, 1.10) for the association between ever/never alcohol drinkers and CRC risk. For coffee consumption, the ORs (95% CIs) of MR Egger, weighted median, and weighted mode were 0.58 (0.47, 0.71), 0.78 (0.69, 0.89), and 0.67 (0.58, 0.76). In addition, no horizontal pleiotropy was found for the SNPs of ever/never alcohol drinkers (P-value of pleiotropy = 0.848) and coffee consumption (P-value of pleiotropy = 0.059) (Table 3). Furthermore, the leave-one-out analyses showed that rs671 played the most significant role in the associations of ever/never alcohol drinkers and coffee consumption with CRC risk (Online Resources 4 and 5).
Discussion
In this study, MR analyses were performed to investigate the potential causal associations of alcohol and coffee with CRC risk in an East Asian population. We found that ever alcohol drinkers (versus never drinkers) and higher alcohol consumption (versus lower) were associated with increased CRC risk. In contrast, higher coffee consumption (versus lower) was associated with reduced CRC risk.
Consistent with our study, alcohol is an established risk factor for CRC in most previous observational studies. A meta-analysis pooled 16 studies revealed that each 10 g increase of alcohol intake per day was associated with a 7% increase in CRC risk (relative risk (RR): 1.07; 95% CI 1.05, 1.09) [18]. Another meta-analysis of 27 cohort and 34 case–control studies also suggested that compared to never or occasional drinkers, moderate (RR: 1.21; 95% CI 1.13, 1.28) and heavy drinkers (RR: 1.52; 95% CI 1.27, 1.81) had higher CRC risk [3]. However, a J-shape relationship was found in a meta-analysis, suggesting that CRC was inversely associated with light to moderate alcohol intake (≤2 drinks/day), but was positively related to heavy intake (≥3 drinks/day), whereas the inverse association may be explained by potential residual confounding or chance [19]. To our knowledge, only one MR study reported that alcohol consumption was not related to CRC (OR: 1.60; 95% CI 0.85, 3.04; P = 0.15) in European populations [8]. While the relative low number of SNPs (three SNPs compared to six SNPs in our study) with a limited explained variance (0.20% compared to 3.97% in our study) may be the main drivers for the null association [8]. Details on the comparisons between SNPs used in the current MR study and previous MR studies were shown in Online Resource 6. The carcinogenicity of alcohol has been attributed to the production of acetaldehyde, which could disrupt deoxyribonucleic acid (DNA) replication and repair [20]. The reactive oxygen species generated from alcohol may also play a carcinogenic role [19]. In addition, alcohol has been reported as a solvent that could promote the penetration of carcinogens into cells [21].
Coffee consumption was found to be inversely associated with CRC in this study, whilst previous observational studies presented mixed results. In line with our study, an umbrella review reported that coffee intake was inversely associated with CRC incidence (RR: 0.88; 95% CI 0.81, 0.96; the highest versus lowest intake) [4]. Nevertheless, the umbrella review failed to find a significant association between per 1 cup/day coffee intake and CRC (RR: 0.99; 95% CI 0.98, 1.01) [4]. A dose–response meta-analysis of cohort studies also showed that with the increase of the amount of coffee intake (from 1 cup/day to 6 cups/day), the CRC risk did not change significantly [22]. Similarly, some other meta-analyses reported an absence of relationship between coffee consumption and CRC risk [16, 23].
Previous MR studies also had inconclusive results for the potential causal association between coffee intake and CRC risk. A MR study based on European populations (26,397 CRC cases and 41,481 controls) identified four SNPs from the CCGC for coffee intake. But the MR analyses showed that coffee consumption (per SD increase) was not related to CRC risk (OR: 1.17; 95% CI 0.88, 1.55; P = 0.27) [8]. In another MR study conducted in European populations (46,155 CRC cases and 270,342 controls), Ong et al. [9] identified 36 SNPs from the UKB and six SNPs from the CCGC. However, different results were observed when the two sets of SNPs were used in MR analyses. One additional cup of coffee intake per day was associated with a 43% increase in CRC risk (OR: 1.43; 95% CI 1.22, 1.67; P < 0.001) when SNPs were used from the UKB, while a null association was reported when SNPs were adopted from the CCGC (OR: 1.15; 95% CI 0.92, 1.46; P = 0.22).
There were several potential explanations for the complicated relationship between coffee consumption and CRC risk. First, the inconclusive results may be partly explained by the carcinogenic and anticarcinogenic properties of coffee. On one hand, several components in coffee (such as caffeine and acrylamide) may act as carcinogens and promote carcinogenesis [22]. On the other hand, some other components may play anticarcinogenic roles. For example, diterpenes cafestol and kahweol could reduce the formation of bile acids and some DNA adducts, which were important CRC promoters [22, 24, 25]. Polyphenols, such as flavonoids and chlorogenic acids, may also play antitumor roles due to their antioxidant and insulin resistance reduction abilities [26, 27]. Moreover, coffee could increase colonic motility and hence decrease intestinal transport time [28], thereby reducing the exposure of epithelial cells to potential carcinogens [3]. These results suggested that future MR studies may need to separately evaluate the relationships between caffeinated and decaffeinated coffee and CRC risk.
Second, the differences in the used SNPs between the current MR study and the previous MR studies may play roles in the different results. In our study, six SNPs were identified for coffee consumption. While Alex et al. [8] selected four SNPs from the CCGC in their MR study, and Ong et al. [9] used 36 SNPs from the UKB and six SNPs from the CCGC. Details on these SNPs were shown in Online Resource 6. Briefly, all SNPs used in MR studies can explain only a limited variance in coffee intake (from 0.30 to 0.83%). Among the six SNPs used in our study, two SNPs (rs4410790 and rs1260326) overlapped with the two European MR studies [8, 9]. The overlap suggested that there may be shared biological effects of some SNPs on coffee consumption in different ethnicities. While the use of other different SNPs may be a contributor to the different results between our study and previous MR studies. Further internal and external validation are needed to confirm the association between genetically predicted coffee intake and CRC risk.
Three assumptions for an IV must be fulfilled for a MR study to be valid (relevance, independence, and exclusion restriction assumption) [5,6,7]. In this study, the first assumption was met because the F-statistics of all IVs were >10. The associations between the used SNPs and exposures (alcohol and coffee) may have potential biological plausibility. For alcohol, most selected SNPs are located in aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH) genes, which are known alcohol-related genes in East Asian populations. For example, people who carry the A allele of rs671 (ALDH2 gene) may drink less alcohol than those who do not carry it, because the A allele can slow the acetaldehyde metabolism and then cause flush response [5]. For coffee, the used SNPs are located in genes that encode factors related to the metabolism and biological functions of coffee. For example, cytochrome P450 1A2 (CYP1A2) is an enzyme involved in caffeine metabolism [29]. Aryl hydrocarbon receptor (AHR) exerts as an upstream inducer for CYP1A2 transcription [29]. Caffeine is able to block the adenosine A2a receptor (ADORA2A) [12]. Cafestol could induce cell apoptosis by decreasing the level of MCL1 [30]. Thus, coffee consumption could be affected by the polymorphisms of these genes. For instance, people with the C allele of rs4410790 (AHR gene) could drink more coffee than those without [29]. In addition, two SNPs were used for both alcohol and coffee (rs1260326 and rs671). The former one is located in glucokinase regulator (GCKR) gene which is significantly associated with the level of plasma glucose and multiple metabolic traits [31]. Hence, variation in GCKR gene may influence the glucose sensing in the brain, thereby affecting the central pathways that respond to alcohol and coffee [31]. Similarly, since another SNP (rs671) is significantly related to alcohol and alcohol drinking could influence other drinking behaviors like coffee, the association between the SNP and coffee may reflect a vertical effect [10]. In addition, the second and third assumptions were partly tested by the MR Egger intercept test in the current study. We found no evidence of horizontal pleiotropy for the SNPs used for ever/never alcohol drinkers and coffee consumption. These sensitivity analyses suggested that the two assumptions were not violated.
This study explored the potential causal associations of alcohol and coffee with CRC risk using MR analyses in an East Asian population. However, there are some limitations. First, for coffee consumption, the BBJ only collected the information of consumption frequency but not the actual consumption amount. In the BBJ, the participants were asked to clarify the consumption frequency based on a four-point scale (1 = almost every day, 2 = 3–4 days per week, 3 = 1–2 days per week, and 4 = rarely). Thus, the ORs only reflected the CRC risk change between higher versus lower consumption frequency, but not the actual amount of coffee intake. Further MR studies with more detailed coffee information in East Asian populations are needed. Second, since the participants of the exposures and outcome were derived from the same population in this study, MR estimates might be biased due to the sample overlap (known as weak instrument bias) [32]. This bias has been shown to inflate the Type 1 error rate and increase the false positive rate [32]. However, it was reported that for a categorical outcome involving both cases and controls, if the SNP-exposure association was only measured in controls, unbiased estimates could be obtained even in a one-sample setting [32]. In this study, none of the participants in the exposure dataset had CRC, which reduced the probability of bias [12]. Third, low statistical powers were found for most associations, suggesting that the explained variances of used SNPs were low and the number of CRC cases were not enough. Fourth, we could not perform stratified analyses based on important covariates (such as age, sex, and anatomic sites) due to the use of summary-level GWAS data. Last, since all GWAS data used in this study were based on East Asian populations, our findings should be interpreted with caution when generalizing to other ethnic populations.
Overall, this MR study found that CRC risk was positively associated with ever/never alcohol drinkers and alcohol consumption, but was inversely associated with coffee consumption. Our findings provided potential causal associations between the two beverages and CRC risk. People are encouraged to adopt healthy dietary habits to prevent CRC. Future MR studies with more valid SNPs, more detailed coffee information, and more CRC cases are needed. Internal and external validation are also recommended to confirm the findings.
The directed acyclic graph of the current study and three Mendelian randomization assumptions. A1: Relevance assumption: the instrumental variable is associated with the exposure. A2: Independence assumption: the instrumental variable is not to be associated with confounders. A3: Exclusion restriction assumption: the instrumental variable affects the outcome only via the exposure
Data availability statement
All data used in this study are publicly available at website (JENGER; http://jenger.riken.jp/en/) and the National Bioscience Database Center Human Database (https://humandbs.biosciencedbc.jp/en/) without any access restriction. One can also contact the corresponding author if he or she would like to assess them.
References
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Sung JJ, Ng SC, Chan FK et al (2015) An updated Asia Pacific consensus recommendations on colorectal cancer screening. Gut 64:121–132. https://doi.org/10.1136/gutjnl-2013-306503
Fedirko V, Tramacere I, Bagnardi V et al (2011) Alcohol drinking and colorectal cancer risk: an overall and dose–response meta-analysis of published studies. Ann Oncol 22:1958–1972. https://doi.org/10.1093/annonc/mdq653
Zhao L-G, Li Z-Y, Feng G-S et al (2020) Coffee drinking and cancer risk: an umbrella review of meta-analyses of observational studies. BMC Cancer 20:101. https://doi.org/10.1186/s12885-020-6561-9
Davies NM, Holmes MV, Davey Smith G (2018) Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362:k601. https://doi.org/10.1136/bmj.k601
Skrivankova VW, Richmond RC, Woolf BAR et al (2021) Strengthening the reporting of observational studies in epidemiology using Mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ 375:n2233. https://doi.org/10.1136/bmj.n2233
Hemani G, Zheng J, Elsworth B et al (2018) The MR-Base platform supports systematic causal inference across the human phenome. Elife. https://doi.org/10.7554/eLife.34408
Cornish AJ, Law PJ, Timofeeva M et al (2020) Modifiable pathways for colorectal cancer: a Mendelian randomisation analysis. Lancet Gastroenterol Hepatol 5:55–62. https://doi.org/10.1016/S2468-1253(19)30294-8
Ong J-S, Law MH, An J et al (2019) Association between coffee consumption and overall risk of being diagnosed with or dying from cancer among >300 000 UK Biobank participants in a large-scale Mendelian randomization study. Int J Epidemiol 48:1447–1456. https://doi.org/10.1093/ije/dyz144
Peacock A, Leung J, Larney S et al (2018) Global statistics on alcohol, tobacco and illicit drug use: 2017 status report. Addiction 113:1905–1926. https://doi.org/10.1111/add.14234
Reyes CM, Cornelis MC (2018) Caffeine in the diet: country-level consumption and guidelines. Nutrients 10:1772. https://doi.org/10.3390/nu10111772
Matoba N, Akiyama M, Ishigaki K et al (2020) GWAS of 165,084 Japanese individuals identified nine loci associated with dietary habits. Nat Hum Behav 4:308–316. https://doi.org/10.1038/s41562-019-0805-1
Ishigaki K, Akiyama M, Kanai M et al (2020) Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet 52:669–679. https://doi.org/10.1038/s41588-020-0640-3
Mozaffarian D, Kabagambe EK, Johnson CO et al (2015) Genetic loci associated with circulating phospholipid trans fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. Am J Clin Nutr 101:398–406. https://doi.org/10.3945/ajcn.114.094557
Xu L, Hao YT (2017) Effect of handgrip on coronary artery disease and myocardial infarction: a Mendelian randomization study. Sci Rep 7:954. https://doi.org/10.1038/s41598-017-01073-z
Burgess S (2014) Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 43:922–929. https://doi.org/10.1093/ije/dyu005
Brion MJ, Shakhbazov K, Visscher PM (2013) Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 42:1497–1501. https://doi.org/10.1093/ije/dyt179
Vieira AR, Abar L, Chan DSM et al (2017) Foods and beverages and colorectal cancer risk: a systematic review and meta-analysis of cohort studies, an update of the evidence of the WCRF-AICR continuous update project. Ann Oncol 28:1788–1802. https://doi.org/10.1093/annonc/mdx171
McNabb S, Harrison TA, Albanes D et al (2019) Meta-analysis of 16 studies of the association of alcohol with colorectal cancer. Int J Cancer 146:861–873. https://doi.org/10.1002/ijc.32377
Oyesanmi O, Snyder D, Sullivan N, et al (2010) Alcohol consumption and cancer risk: understanding possible causal mechanisms for breast and colorectal cancers. Evid Rep Technol Assess 1–151.
Papadimitriou N, Markozannes G, Kanellopoulou A et al (2021) An umbrella review of the evidence associating diet and cancer risk at 11 anatomical sites. Nat Commun 12:4579. https://doi.org/10.1038/s41467-021-24861-8
Micek A, Gniadek A, Kawalec P et al (2019) Coffee consumption and colorectal cancer risk: a dose-response meta-analysis on prospective cohort studies. Int J Food Sci Nutr 70:986–1006. https://doi.org/10.1080/09637486.2019.1591352
Sartini M, Bragazzi N, Spagnolo A et al (2019) Coffee consumption and risk of colorectal cancer: a systematic review and meta-analysis of prospective studies. Nutrients 11:694. https://doi.org/10.3390/nu11030694
Post SM, de Wit EC, Princen HM (1997) Cafestol, the cholesterol-raising factor in boiled coffee, suppresses bile acid synthesis by downregulation of cholesterol 7 alpha-hydroxylase and sterol 27-hydroxylase in rat hepatocytes. Arterioscler Thromb Vasc Biol 17:3064–3070. https://doi.org/10.1161/01.atv.17.11.3064
Higdon JV, Frei B (2006) Coffee and health: a review of recent human research. Crit Rev Food Sci Nutr 46:101–123. https://doi.org/10.1080/10408390500400009
Scalbert A, Johnson IT, Saltmarsh M (2005) Polyphenols: antioxidants and beyond. Am J Clin Nutr 81:215s–217s. https://doi.org/10.1093/ajcn/81.1.215S
Tunnicliffe JM, Shearer J (2008) Coffee, glucose homeostasis, and insulin resistance: physiological mechanisms and mediators. Appl Physiol Nutr Metab 33:1290–1300. https://doi.org/10.1139/H08-123
Brown SR, Cann PA, Read NW (1990) Effect of coffee on distal colon function. Gut 31:450–453. https://doi.org/10.1136/gut.31.4.450
Josse AR, Da Costa LA, Campos H et al (2012) Associations between polymorphisms in the AHR and CYP1A1-CYP1A2 gene regions and habitual caffeine consumption. Am J Clin Nutr 96:665–671. https://doi.org/10.3945/ajcn.112.038794
Choi MJ, Park EJ, Oh JH et al (2011) Cafestol, a coffee-specific diterpene, induces apoptosis in renal carcinoma Caki cells through down-regulation of anti-apoptotic proteins and Akt phosphorylation. Chem Biol Interact 190:102–108. https://doi.org/10.1016/j.cbi.2011.02.013
Coffee and Caffeine Genetics Consortium, Cornelis MC, Byrne EM et al (2015) Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol Psychiatry 20:647–656. https://doi.org/10.1038/mp.2014.107
Burgess S, Davies NM, Thompson SG (2016) Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 40:597–608. https://doi.org/10.1002/gepi.21998
Acknowledgements
We would like to express our gratitude to the BioBank Japan (BBJ) for providing publicly available summary-level genome-wide association studies (GWAS) data for dietary habits and colorectal cancer.
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
YYD: Conceived and designed this study, conducted the data acquisition, analyzed the data, and wrote the manuscript; JJH: Proofread the data and manuscript; MCSW: Critically revised the manuscript. All authors approved the final draft submitted.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics statement
Studies related to the BioBank Japan (BBJ) were approved by the ethical committees of RIKEN Yokohama Institute and the Institute of Medical Science, the University of Tokyo. Written informed consents were obtained from all participants. This study did not exceed the scope of the original ethics committee approval.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, Y., Huang, J. & Wong, M.C.S. Associations of alcohol and coffee with colorectal cancer risk in East Asian populations: a Mendelian randomization study. Eur J Nutr 62, 749–756 (2023). https://doi.org/10.1007/s00394-022-03002-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-022-03002-x