Abstract
Purpose
Colorectal cancer (CRC) is a clinically diverse disease whose molecular etiology remains poorly understood. The purpose of this study was to identify miRNA expression patterns predictive of CRC tumor status and to investigate associations between microRNA (miRNA) expression and clinicopathological parameters.
Methods
Expression profiling of 380 miRNAs was performed on 20 paired stage II tumor and normal tissues. Artificial neural network (ANN) analysis was applied to identify miRNAs predictive of tumor status. The validation of specific miRNAs was performed on 102 tissue specimens of varying stages.
Results
Thirty-three miRNAs were identified as differentially expressed in tumor versus normal tissues. ANN analysis identified three miRNAs (miR-139-5p, miR-31, and miR-17-92 cluster) predictive of tumor status in stage II disease. Elevated expression of miR-31 (p = 0.004) and miR-139-5p (p < 0.001) and reduced expression of miR-143 (p = 0.016) were associated with aggressive mucinous phenotype. Increased expression of miR-10b was also associated with mucinous tumors (p = 0.004). Furthermore, progressively increasing levels of miR-10b expression were observed from T1 to T4 lesions and from stage I to IV disease.
Conclusion
Association of specific miRNAs with clinicopathological features indicates their biological relevance and highlights the power of ANN to reliably predict clinically relevant miRNA biomarkers, which it is hoped will better stratify patients to guide adjuvant therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the fourth commonest cancer in men and the third in women worldwide [1]. Despite advances in surgical techniques and other treatment modalities, CRC remains a major cause of cancer mortality resulting in approximately 630,000 deaths worldwide each year. In the last decade, the emergence of novel molecular agents has heralded a new era of CRC treatment and has shown promising results [2, 3]. However, therapeutic responses to these agents have been unpredictable highlighting the underlying heterogeneity of the disease [4]. This stresses the need for phenotyping individual colorectal tumors in order to maximize therapeutic outcome, thus enable a better patient-tailored clinical management.
With a span from tumors that are barely starting to penetrate through the mucosa, through aggressive tumors involving the serosa or adjacent organs, stage II CRC presents a significant therapeutic challenge. Even though up to 25% of patients with stage II CRC develop disease recurrence during their lifetime, the role of adjuvant chemotherapy remains unclear based on under-powered studies [5–7]. Therefore, molecular dysregulation and interactions underlying colorectal tumorigenesis and progression warrant further investigations in order to improve prognostication to guide therapy and to optimize a long-postulated individualized targeted therapeutic strategy.
Microarray technology has provided a useful strategy to globally screen for potential molecular markers for disease classification, prognostication and prediction of response to therapy [8–10]. Breast cancer represents an example of successful translation of gene expression profiling into clinical practice with significant prognostic and therapeutic implications [11]. Thus far, this success has not been reproduced in the management of CRC.
Mi(cro)RNAs play critical roles in the regulation of gene expression by targeting mRNA sequences for cleavage and/or translational repression, and thereby govern cellular processes including cell differentiation, proliferation, and apoptosis [12–15]. Furthermore, accumulating evidence indicates that dysregulated miRNA expression is a feature of most malignancies examined to date, including CRC [16–18]. Aberrant miRNA expression in tumors indicates their potential causal roles for tumorigenesis, tumor growth, and invasion, which may have significant diagnostic, prognostic, and therapeutic implications. Moreover, miRNA profiles have been reported to be more accurate than genomic profiles in disease classification [16]. The interaction between miRNAs and mRNAs is complex: each miRNA can regulate up to 200 mRNAs, and in turn, each mRNA can have multiple conserved target sites for miRNA binding [19]. At the time of writing, 721 miRNA sequences have been described in human, which have opened a new avenue to investigate CRC biological pathways [20].
Reliability and biological relevance of data generated from high throughput array technology depends heavily on bioinformatic and statistical analyses. Conventional hierarchical clustering methods have their limitations [21]. Artificial neural networks (ANN) are a form of artificial intelligence that can accurately analyze complex and non-linear datasets through training and testing predictive models that relate biological features such as genes, miRNAs or proteins to clinical characteristics of an individual [22, 23]. ANNs have been reliably and successfully applied in discriminating disease states, grades, and subgroups in different cancers [24–26].
Using microarray profiling and ANNs, the aim of this study was to identify miRNAs to differentiate stage II tumors from controls. Identification of reliable biomarkers of disease stratification will further current understanding of CRC etiology and foster improvement of current prognostic and therapeutic regimens.
Materials and methods
Tissue samples
Written informed consent from 58 consecutive patients undergoing surgical resection for CRC at Galway University Hospital, Ireland was included in this study. Ethical approval was sought and granted by Galway University Hospital research ethics committee. Tissue retrieval was carried out subsequent to review by a consultant pathologist. Samples were immediately snap-frozen in liquid nitrogen and stored at −80°C. MiRNA profiling was performed on 20 paired tumor and normal tissues from ten patients with stage II disease. RT-qPCR was performed on a cohort of 102 tissue specimens of varying stages. Patient demographics, clinicopathological, and survival data were collected and maintained prospectively. Tumors were staged according to the TNM staging system [27].
Clinicopathological parameters and follow-up data are summarized in Supplementary Data Tables 1 and 2. Risk of disease recurrence was stratified based on pathological parameters such as tumor size, adequacy of node sampling, mucinous phenotype, and the presence of perineural or lymphovascular invasion.
RNA extraction
Total RNA was extracted from 50 to 100 mg of snap-frozen colorectal tissue homogenized in QIAzol Lysis reagent (Qiagen, Crawley, UK) as previously described [28]. Concentration and integrity of extracted RNA were assessed using NanoDrop 1,000 spectrophotometry (NanoDrop Technologies, USA) and an Agilent 2,100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), respectively. Electropherograms and gel-like images were evaluated using Agilent 2,100 Expert software (Version B.02.03) which generated RNA integrity numbers (RIN). Only RNA of good quality was used in the array experiment with RIN range from 5.5 to 8.6.
MiRNA profiling
TaqMan Human MicroRNA arrays (Applied Biosystems) were used to simultaneously measure the expression of 380 miRNAs on a microfluidic PCR platform. Each array contained quadruplicate probes to a mammalian U6 target, and an Arabidopsis process control (ath-miR-159a). Simultaneous synthesis of cDNA for mature miRNAs was performed using Megaplex Reverse Transcription Human Pool A (Applied Biosystems), which is a set of pre-defined pools of stem-looped reverse transcription primers. In accordance to the manufacturer’s instructions, miRNAs with low concentration (below 350 ng/μL) underwent pre-amplification to increase the quantity of desired cDNA to allow better detection. RT-qPCR was performed using the Applied Biosystems 7900HT Fast Real-Time PCR System.
Relative quantification of profiling data and artificial neural network analysis
Raw fluorescence data generated from the microarray experiment were normalized using qBasePlus software (Biogazelle NV, Belgium). Using the lowest expressed sample as a calibrator, the relative quantity (RQ) of miRNA expression was calculated using the comparative cycle threshold (ΔΔCT) method, normalized to two reference miRNAs (miR-16 and miR-345) [28].
ANN-based algorithms coupled with an additive stepwise approach were applied to profiling data in order to generate miRNA signatures predictive of tumor status as previously described [26]. In brief, a three-layer multi-layer perceptron with a feedforward back-propagation algorithm and a sigmoidal transfer function was employed. The relative expression of miRNA was presented in the input layer, the tumor status was represented in the output layer coded as 0 for control and 1 for tumor, with two hidden nodes within the hidden layer. An extensive Monte Carlo cross-validation strategy was applied to overcome the issue of over-fitting, which is particularly at risk considering the low number of samples [26]. A subset of cases (60% of the cases) was randomly assigned to a training dataset. A separate testing dataset (20% of the cases) monitored and stopped the training once the model was optimized. Finally, the remaining 20% of samples was used as validation to test the finalized trained ANN independently from training. The random selection process was repeated over 50 times reshuffling all the cases in different subsets, so that all the cases have fallen in each subset during the model development.
An additive stepwise approach was coupled in order to identify reliable signature for the question investigated. This consists of testing each miRNA as a single predictor. The miRNAs could therefore be ranked according to their predictive ability (by predictive error on validation set). This ranking gave the best single miRNA to predict cancer versus control tissue on its own. Subsequently, in a second step, the selected miRNA was used in a two-input model, iteratively using each of the remaining miRNA as a second input, the ANN was built and trained. The ranking of the 2-miRNA signature was once again ranked giving the best predictive pair. This was resumed in subsequent steps until no further improvement in prediction accuracy and error was achieved, thereby generating an optimal model with the best predictive performance.
RT-qPCR validation
The primer sequences of miR-10b, miR-16, and miR-21 have been previously described and were obtained from MWG Biotech (Ebersberg, Germany) [29]. For all other miRNAs, assays containing stem-looped primers were sourced from Applied Biosystems (USA). Each reaction was primed using sequence-specific stem-loop primers. All reagents were included in the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT-negative controls were included in each batch of reactions. PCR reactions were carried out in final volumes of 10 μL. Inter-assay controls were included on each plate. All reactions were performed in triplicate. The threshold standard deviation for intra- and inter-assay replicates was 0.3. Percent PCR amplification efficiencies (E) for each assay were calculated as E = (10−1/slope − 1) × 100, using the slope of the semi-log regression plot of Ct versus log input of cDNA (10-fold dilution series of five points). A threshold of 10% above or below 100% efficiency was applied. Similar methods were applied to calculate the RQ of miRNA in both RT-qPCR and profiling datasets.
Statistical analysis
Statistical analysis was performed using SPSS 14.0 (Chicago, USA). Values were expressed as mean ± s.e.m. Distribution of continuous data was determined using the Kolmogorov–Smirnov Z test. Differential miRNA expression was expressed as fold change, calculated by log2 (RQ in tumor/RQ in normal). Differences between groups were calculated using analysis of variance and Student’s t tests. Correlation between profiling and RT-qPCR expression data was calculated using Pearson’s correlation. p values of less than 0.05 were considered statistically significant for all tests.
Results
MiRNAs predictive of tumor status
ANN analysis of miRNA profiling data identified a distinct miRNA signature predictive of tumor status in stage II CRC samples. This signature consisted of three miRNA transcripts (miR-139-5p, miR-31, and miR-17-92) which distinguished tumor from normal correctly with a median accuracy of 100% (Table 1). Figure 1 shows the iterative improvement of predictive accuracy and drop of predictive error, for the cumulative building of the model at each step of the cumulative stepwise approach.
Artificial neural network model performance analysis. Bars represent the median model accuracy with lines indicating the mean squared error for the predictions at each step. After the addition of the third miRNA transcript, the accuracy of the predictive model has reached 100% without further improvement in the error in subsequent steps. At this point, the model is considered to contain the transcripts which most accurately model the data and 3-miRNA signature predictive of tumor status in stage II colorectal cancer was generated
Identification of dysregulated miRNAs in CRC
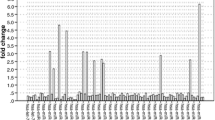
We compared the expression profiles of 380 miRNAs in stage II tumors and normal tissues using fold change. Thirty-three individual miRNAs were found to be differentially expressed in tumors (Table 2). Twenty miRNAs displayed higher levels of expression, with miR-31 being the most significantly upregulated in tumors at 5.7-fold, and miR-139-5p the most downregulated at −2.0-fold.
MiRNA selection and PCR validation
Seven candidate miRNAs were chosen for validation by RT-qPCR. Three miRNAs identified from the signature (miR-17, miR-31, and miR-139-5p) were chosen alongside four additional miRNAs (miR-10b, miR-21, miR-143, and miR-145) [18, 30]. In addition, miR-16 and miR-345 were used as internal controls. In most cases, miRNA expression data in the validation cohort correlated with the microarray profiling data (Fig. 2). MiRNAs, which were upregulated in tumors, were: miR-17 (p = 0.029), miR-21 (p = 0.002), and miR-31 (p < 0.001). MiRNAs that showed reduced levels of expression were: miR-10b (p < 0.001), miR-139-5p (p < 0.001), miR-143 (p = 0.003), and miR-145 (p = 0.001). These results are in keeping with previous published data.
Correlation between microarray and RT-qPCR data. Correlation of miRNA expression levels as determined by microarray (dashed lines) and RT-qPCR (solid lines) for a subset of 6 candidates. Both datasets were normalized to miR-16 and miR-345. Good correlation between both platforms was demonstrated in most cases; however, probe-specific differences were occasionally observed. r and p values were calculated using Pearson correlation
Association between miRNA expression and clinicopathological parameters
Comparison of expression data with clinicopathological variables identified differential expression of three miRNAs associated with mucinous phenotype in patients with stage II disease (n = 15). In this group, higher expression of miR-31 (p = 0.004) and miR-139-5p (p < 0.001) and lower expression of miR-143 (p = 0.016) were shown. Interestingly, miR-31 expression was found to be higher in T3 tumors compared with T4 lesions (p = 0.040).
When the analysis was expanded to the entire cohort of different tumor stages, miR-10b (p = 0.004) was significantly upregulated in mucinous tumors. miR-10b also displayed an increasing level of expression from T1 to T4 tumors in a progressive manner (p = 0.026) and from stage I to stage IV disease in a similar pattern albeit not statistically significant (p = 0.069; Fig. 3).
Discussion
In this study, miRNA expression profiling in stage II CRC was investigated using a high throughput microarray approach and ANN analysis. To our knowledge, this is the first study to generate distinctive miRNA signatures to reliably predict stage II colorectal tumor status from normal tissue. Subsequently, aberrantly expressed miRNAs that associated with specific clinicopathological parameters were identified in a larger heterogeneous cohort of colorectal tumors and adjacent normal tissues.
Up to 25% of patients with stage II colorectal tumors develop disease recurrence despite undergoing curative resection. Multiple clinical trials have shown conflicting results regarding the use of adjuvant chemotherapy in these patients [5–7]. Thus, there is a clear clinical need for improved stratification of patients with stage II CRC in order to predict their risk of disease recurrence and therefore to guide adjuvant therapy. The largest clinical trial to date involving approximately 3,000 patients with stage II CRC demonstrated a small (3.6%) absolute risk reduction in 5-year mortality in patients who received adjuvant therapy in comparison to patients who did not [7]. This result was subsequently reproduced in a Cochrane Systematic Review [31]. The decision to provide such therapies is largely based on traditional clinical and histological features of the tumor. With the lack of accurate prognostic tools, patients with resected stage II cancer are currently either needlessly exposed to the side effects of chemotherapy or left untreated with a significant risk of disease recurrence. Two patients in our stage II cohort developed disease recurrence: both underwent elective resections with clear margins, and histology revealed favorable features including adequate node sampling, and the lack of perineural or lymphovascular invasion, hence did not receive adjuvant therapy. This highlights the heterogeneity of the disease, and the need for novel biomarkers for classification and prognostication.
Frederiksen et al. investigated the genetic profiles in CRC and identified distinctive expression patterns which discriminate between Dukes’ B and C tumors [8]. This result suggests differing underlying genetic events leading to lymph node metastasis in Dukes’ C tumors. Wang et al. identified a 23-gene signature to predict recurrence in Dukes’ B colon cancer using DNA microarray technology [9]. Most genes in the reported signature are not inherently involved in carcinogenesis, and there has been a lack of translation of this prognostic assay into clinical practice which can be explained by the size of such genetic signature.
MiRNA expression profiles have been reported to be more accurate in cancer classification than mRNA profiles [16]. Furthermore, as less than a thousand human miRNA transcripts (721 at the time of writing) have been identified thus far in comparison to the ~30,000 genes in the human genome, this substantially decreases both throughput and complexity of data analysis algorithms [20]. As demonstrated by our study, a 3-miRNA signature generated by ANN analysis predicted tumor status in stage II CRC with 100% accuracy. As clinical data accrues with adequate patient follow-up, it is hoped that future analysis may reveal further miRNA signatures to predict disease recurrence.
The accuracy of miRNA expression profiles may be further strengthened when used in combination with mRNA profiles. Increasing understanding of the molecular mechanism underlying resistance to anti-epidermal growth factor receptor (EGFR) monoclonal antibodies have led to the concept of the “quadruple negative” CRC [4]. Mutations in downstream effectors of EGFR including KRAS, BRAF, PTEN, and PIK3CA have been shown to confer therapeutic resistance to Cetuximab and Panitumumab. Unresolved questions remain with regard to the underlying mechanism of resistance and the upstream regulation of these genes. Intriguingly, KRAS is a validated target of miR-143, and PTEN is known to be suppressed by miR-21 [32, 33]. Further research to elucidate the specific roles of these miRNAs in chemoresistance may provide insights into the molecular basis of these interactions.
The three miRNAs in the predictive signature identified in this study have been shown to be dysregulated in CRC and other cancer in previous studies, which indicates their biological relevance. miR-31 was found to be upregulated in CRC and was associated with advanced disease stage and deeper invasion of tumors [18, 34]. Increased expression of the miR-17-92 cluster which encompasses six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1) has been reported to be associated with microsatellite stable colon tumors, neoplastic transformation of colonic epithelium, and adenoma to carcinoma progression [35–37]. A single miRNA (miR-17) representative of the six transcripts in the miR-17-92 cluster was chosen for validation as co-expression of these polycistronic transcripts is a well-established feature of cancer regulatory networks [26, 37, 38]. Interestingly, fluorouracil (5-FU), which has been the cornerstone of CRC medical treatment since the late 1950s and remains to be the backbone of current treatment regime, was reported to downregulate the expression of the miR-17-92 cluster in a dose-dependent manner [39]. A better understanding of the interaction between these miRNAs and 5-FU may provide novel therapeutic insights. In contrast to both miR-31 and miR-17-92, miR-139-5p has not been previously shown to be dysregulated in CRC. Our finding of reduced miR-139-5p expression in colorectal tumors is in keeping with its reported tumor suppressor roles in gastric and endometrial adenocarcinomas [40, 41]. Despite the difference in our profiling methodology compared to previous studies, a number of miRNAs found to be dysregulated in our study overlap with previous reports, highlighting the consistency of the findings [42, 43]. Rather than using hybridization array platforms, the microarray chosen for use in this study was a PCR-based microfluidic system. The increased sensitivity, specificity and dynamic range facilitated by RT-qPCR made it an attractive alternative to more traditional hybridization methodologies.
The recent discovery of the presence of miRNAs in the circulation has generated much scientific and clinical interest. MiRNAs have been shown to be stable non-invasive biomarkers for early diagnosis and potentially postoperative monitoring of cancer patients [44–46]. Intriguingly, two separate studies to date have reported that plasma miR-92 is a sensitive marker that discriminates between CRC patients and normal controls [47, 48]. This has major clinical implications in the early diagnosis of CRC. Furthermore, circulating miRNAs levels have been demonstrated to correlate with tumor miRNAs expression [46, 47]. The signature predictive of tumor status generated in our study consists of the miR-17-92 cluster, which is in keeping with its predictive role in the plasma.
In conclusion, this study demonstrates that a RT-qPCR-based microarray platform and ANN analysis generated a miRNA signature predictive of tumor status in stage II CRC. Differential miRNA expression and association of specific miRNAs with clinicopathological features indicates their biological relevance in tumor development, growth, progression and invasion. This highlights their potential for development as novel biomarkers to better stratify patients at increased risk of disease recurrence to guide adjuvant therapy. Increasing understanding of the miRNA–mRNA regulatory mechanisms may provide a novel individual-based therapeutic strategy.
References
Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel RL, Thun MJ (2007) Global cancer facts & figures. American Cancer Society, Atlanta, GA
Hurwitz H, Fehrenbacher L, Novotny W et al (2004) Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 350(23):2335–2342
Cunningham D, Humblet Y, Siena S et al (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 351(4):337–345
Bardelli A, Siena S (2010) Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol 28(7):1254–1261
Mamounas E, Wieand S, Wolmark N et al (1999) Comparative efficacy of adjuvant chemotherapy in patients with Duke’s B versus Duke’s C colon cancer: Results from four National Surgical Adjuvant Breast and Bowel Project adjuvant studies (C-01, C-02, C-03, and C-04). J Clin Oncol 17(5):1349–1355
International Multicentre Pooled Analysis of Colon Cancer Trials (IMPACT) Investigators (1999) Efficacy of adjuvant fluorouracil and folinic acid in B2 colon cancer. J Clin Oncol 17(5):1356–1363
QUASAR Collaborative Group (2007) Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomized study. Lancet 370(9604):2020–2029
Frederiksen CM, Knudsen S, Laurberg S, Orntoft TF (2003) Classification of Duke’s B and C colorectal cancers using expression arrays. J Cancer Res Clin Oncol 129(5):263–271
Wang Y, Jatkoe T, Zhang Y et al (2004) Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J Clin Oncol 22(9):1564–1571
Ghadimi BM, Grade M, Difilippantonio MJ et al (2005) Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol 23(9):1826–1838
Sorlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98(19):10869–10874
Lai EC (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30(4):363–364
Engels BM, Hutvagner G (2006) Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene 25(46):6163–6169
Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303(5654):83–86
Croce CM, Calin GA (2005) miRNAs, cancer, and stem cell division. Cell 122(1):6–7
Lu J, Getz G, Miska EA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838
Volinia S, Calin GA, Liu CG et al (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103(7):2257–2261
Bandrés E, Cubedo E, Agirre X et al (2006) Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 5:29
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20
Griffiths-Jones S (2004) The microRNA registry. Nucleic Acid Res 32:D109–D111
Simon R, Radmacher MD, Dobbin K, McShane LM (2003) Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst 95(1):14–18
Khan J, Wei JS, Ringner M et al (2001) Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 7(6):673–679
Lancashire L, Schmid O, Shah H, Ball G (2005) Classification of bacterial species from proteomic data using combinatorial approaches incorporating artificial neural networks, cluster analysis and principle components analysis. Bioinformatics 21(10):2191–2199
Matharoo-Ball GR, Lancashire L, Ugurel S et al (2007) Diagnostic biomarkers differentiating metastatic melanoma patients from healthy controls identified by an integrated MALDI-ToF mass spectrometry/bioinformatic approach. Proteomics 1:605–620
Ball G, Mian S, Holding F et al (2002) An integrated approach utilizing artificial neural networks and SELDI mass spectrometry for the classification of human tumors and rapid identification of potential biomarkers. Bioinformatics 18(3):395–404
Lowery AJ, Miller N, Devaney A et al (2009) MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 11:R27
Sobin LH, Wittekind CH (2002) TNM classification of malignant tumors, 6th edn. John Wiley & Sons, New York, pp 170–173
Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N (2010) MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer 10:173
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25(4):402–408
Akao Y, Nakagawa Y, Naoe T (2006) MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep 16(4):845–850
Figueredo A, Coombes ME, Mukherjee S (2008) Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev 16(3):CD005390
Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133(2):647–658
Chen X, Guo X, Zhang H et al (2009) Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28(10):1385–1392
Wang CJ, Zhou ZG, Wang L et al (2009) Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers 26(1):27–34
Lanza G, Ferracin M, Gafa R et al (2007) mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 6:54
Monzo M, Navarro A, Bandres E et al (2008) Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res 18(8):823–833
Diosdado B, van de Wiel MA, Terhaar Sive Droste JS et al (2009) MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer 101(4):707–714
Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB (2008) MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA 105(50):19678–19683
Zhao HY, Ooyama A, Yamamoto M et al (2008) Down regulation of c-Myc and induction of an angiogenesis inhibitor, thrombospondin-1, by 5-FU in human colon cancer KM12C cells. Cancer Lett 270(1):156–163
Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y (2009) Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 24(4):652–657
Hiroki E, Akahira J, Suzuki F et al (2009) Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci 101(1):241–249
Schetter AJ, Leung SY, Sohn JJ et al (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299(4):425–436
Schepeler T, Reinert JT, Ostenfeld MS et al (2008) Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 68(15):6416–6424
Chen X, Ba Y, Ma L et al (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18(10):997–1006
Mitchell PS, Parkin RK, Kroh EM et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518
Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ (2010) Circulating microRNAs novel minimally invasive biomarkers for breast cancer. Ann Surg 251(3):499–505
Ng EK, Chong WW, Jin H et al (2009) Differential expression of microRNAs in plasma of colorectal cancer patients: a potential marker for colorectal cancer screening. Gut 58(10):375–381
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X (2009) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127(1):118–126
Acknowledgments
We would like to acknowledge the National Breast Cancer Research Institute (NBCRI) for the continued financial support. The authors gratefully acknowledge Ms. Emer Hennessy and Ms. Catherine Curran for continued technical assistance and curation of the Department of Surgery BioBank.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 48 kb)
Rights and permissions
About this article
Cite this article
Chang, K.H., Miller, N., Kheirelseid, E.A.H. et al. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis 26, 1415–1422 (2011). https://doi.org/10.1007/s00384-011-1279-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00384-011-1279-4