Abstract
Purpose
The NOD2 gene is known to have a strong association with Crohn’s disease, but different trends were reported in occurrence of NOD2 variants in distinct ethnicities. The aim of this study was to assess all exonic sequences of the NOD2 gene in Iranian Crohn's disease patients and healthy controls to identify any existing variation and evaluate their association with Crohn's disease.
Methods
A total of 90 non-related Crohn's disease patients and 120 sex- and age-matched healthy controls of Iranian origin were enrolled in this study. The participants were referred to a tertiary center in a 2-year period (2006–2008). The exonic regions of the NOD2 gene were amplified by polymerase chain reaction and evaluated by direct sequencing.
Results
A total of 21 sequence variations were identified among all exonic regions of the NOD2 gene, of which eight had an allele frequency of more than 5%. Eight new mutations (one in exon 2 and seven in exon 4) were observed. The three main variants (R702W, G908R, and 1007fs) showed allele frequencies of 13.3%, 2.2%, and 1.7%, respectively. Three new variations (P371T, A794P, and Q908H) and R702W mutation were significantly more frequent in Crohn's disease patients compared to controls.
Conclusions
Eight novel mutations were identified in the NOD2 exons, but the pathophysiological importance of these variants remains unclear. Iranian patients with their different genetic reservoirs may demonstrate some novel characteristics for disease susceptibility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crohn's disease (CD), a major form of inflammatory bowel disease (IBD), is a chronic granulomatous inflammation of the gut. It may involve any portion of the gastrointestinal (GI) tract and could affect all layers of the digestive wall [1]. The exact pathogenesis of CD is still unknown; but it is thought to result from inappropriate activation of the mucosal immune system in response to the antigens of intestinal bacteria [2].
The role of genetic factors in CD was first suggested by epidemiologic data and confirmed by familial aggregation and monozygote twin studies [3]. Microsatellite markers were then used to identify the areas of linkage with CD in the whole genome. In these challenges, several loci have been marked to be linked with CD [4, 5]. One of the strongest associations was observed with a locus on chromosome 16q12 assigned as IBD1 [4]. This locus contains the first reported CD linked gene, NOD2 [6–8]. NOD2 protein, the product of this gene, is a cytosolic pattern recognition receptor responsible for regulation of innate immunity by intracellular recognition of the microorganisms’ particles such as lipopolysaccharides [9]. Moreover, NOD2 protein plays a role in apoptosis, nuclear factor kappa B (NF-kB) activation, and development of inflammatory response in epithelial cells. As NF-kB has a key role in inflammation, NOD2 can be considered as a pro-inflammatory protein [10]. Structurally, it contains two caspase recruitment domains (CARDs), a nucleotide-binding oligomerization domain, and a C-terminal leucine-rich repeat (LRR) [11].
Many studies were performed to clarify the NOD2 gene’s role in IBD pathogenesis. Different variations of this gene were reported to be associated with CD in Caucasian populations [12–17]. Three common mutations of the NOD2 gene (R702W, G908R, and 1007fsinsC) showed a significant association with CD in many European and American studies [18] while the lack of association was reported in other ethnicities such as Japanese and Koreans [19]. In a recent study, only one of the three common NOD2 variations (R702W) was reported to be associated with CD in Iranian population, and two other variations demonstrated a very low frequency (<5%) in both patients and controls [20]. These results raised the idea that other variations of the NOD2 gene might be responsible for susceptibility to CD among this ethnic group. Considering the rising number of affected individuals in Iran [21] and previously reported associations of other gene variants (such as MDR1 and VDR) with CD, as well as different trends of the NOD2 common variations in this ethnicity [22, 23], we evaluated all exonic regions of the NOD2 gene in Iranian CD patients and healthy controls to identify any existing variations and assess any probable association of them with CD.
Methods and materials
In this case–control designed study, a total of 90 unrelated CD patients as well as 120 age- and sex-matched healthy controls, all of Iranian origin, were recruited. Patients were referred to a tertiary GI center in Tehran (the capital of Iran) in a two-year period (2006–2008). CD was diagnosed based on the clinical, endoscopic, radiologic, and histopathologic findings [1, 24, 25] by an expert gastroenterologist. A complete clinical questionnaire was filled for each patient at admission. CD phenotype was categorized according to Vienna classification including disease location, behavior, and age at diagnosis [26]. Controls were selected from healthy individuals without any GI symptoms or positive familial or personal history of any significant disorders. The study protocol was approved by the Ethics Committee of Research Center for Gastroenterology and Liver Diseases, and a written informed consent was obtained from each participant.
Genomic DNA was extracted from peripheral blood leukocytes by standard phenol–chloroform method [27]. All exons and flanking intronic sequences of the NOD2 gene (HUGO Gene Nomenclature Committee accession number AF178930) were amplified by previously reported primers (Table 1) [12]. A pair of primers was used for each exon except for exon 4, for which five pairs of primers were utilized, resulting in five overlapping fragments.
Polymerase chain reactions (PCRs) were performed in a 25-μl volume containing 10 mM Tris–HCl, pH 8.8, 1.5 mM MgCl2, 50 mM KCl, 250 mM dNTPs, a 0.50-mM concentration of each primer, 2 U of Taq DNA polymerase (Fermentas, Germany), and 200 ng of genomic DNA. The PCR condition for all fragments was as follows: initial denaturation at 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min. Appropriate synthesis of PCR products was confirmed by agarose gel electrophoresis (1.5%) and visualized by ethidium bromide staining (0.5 μg/ml). The fragments were analyzed by an ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster, CA, USA) sequencer. The sequence data were aligned using Lasergene software, version 6.00 (DNASTAR Inc., Madison, WI, USA) and were compared to the published NOD2 gene sequence (GenBank accession number AF178930). In silico predictions were evaluated for novel coding variants using SNAP online software (http://cubic.bioc.columbia.edu/services/SNAP/). Statistical analysis was performed using X 2 test and the fisher exact test to compare variables among study groups. P values less than 0.05 were considered significant. The data were analyzed using SPSS software version 13 (SPSS, Chicago, IL, USA).
Results
The median age (range) of healthy controls and CD patients were 31 (17–62 years) and 32 years (16–65 years), respectively. The male to female ratio was 1.6:1.0 in both study groups. The clinical characteristics of patients based on Vienna classification [26] are summarized in Table 2.
A total of 420 chromosomes of the NOD2 gene from 90 CD patients and 120 healthy controls were screened for any exonic nucleotide changes by direct sequencing. In analyzed coding regions and intronic flanking sequences, 21 variations were detected in four exons including exons 2, 4, 8, and 11. The identified mutations are listed in Table 3 in comparison to previously reported variations in the NOD2 gene sequence by Lesage et al. [12]. Splice sites and other exons did not demonstrate any sequence change in our study population. The observed sequence changes were mostly concentrated in exon 4, which included higher than 70% (n = 16) of the detected variations. Among five remaining mutations, four were observed in exons 2 and 8, two in each and one in exon 11. No insertion or deletion mutations were detected.
Most variations were a transition type (61.9%), especially a C → T substitution occurred more frequently (33.3%). All observed variations were due to a single nucleotide substitution, except the insertion frameshift mutation, 1007fsinsC (SNP13). A novel nonsense mutation was detected in exon 4, which resulted from a C to A transversion forming a premature stop codon in the location 813 of the amino acid chain. More than 50% (n = 11) of the observed variations were missense mutations.
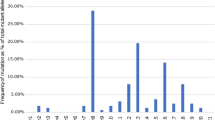
Over 85% (n = 18) of the observed variations had an allele frequency of more than 1% in CD patients, while only 47.6% (n = 10) of these mutations showed the same frequency in controls. Eight sequence changes had an allele frequency of more than 5% in CD patients including S178S, P268S, P371T, R459R, R587R, R702W, A794P, and Q809H (Figs. 1 and 2). Four of them had a frequency of more than 10% in healthy controls, including S178S in exon 2 and P268S (SNP5), R459R (SNP6), and R587R (SNP7) in exon 4. None of the four mentioned mutations showed a significant higher frequency in CD patients compared to controls.
Forward and reverse electropherograms of NOD2 gene. a V162I variation in a CD patient, with a heterozygote genotype; b P371T variation in a CD patient, with a heterozygote genotype; c T389T variation in a CD patient, with a heterozygote genotype; d E462K variation in a CD patient, with a heterozygote genotype; e A611A variation in a CD patient, with a heterozygote genotype; f A794P variation in a CD patient, with a heterozygote genotype; g Q809H variation in a CD patient, with a heterozygote genotype
Totally, eight novel variations were identified in Iranian CD patients, one in exon 2 and seven in exon 4 (Fig. 3). Five out of eight were missense mutations, of which three of them, including P371T, A794P, and Q809H, had a higher allele frequency in CD patients compared to controls (P < 0.001, with odds ratio (OR) = 9.26, 7.07, and 9.26, respectively). The related 95% confidence intervals were as follows: P371T, OR = 9.26, 95% CI, 2.06–41.59; A794P, OR = 7.71, 95% CI, 2.21–26.88; and Q809H, OR = 9.26, 95% CI, 2.06–41.59. None of these novel variations presented with an allele frequency of more than 10% in healthy controls.
Cumulative risk was performed for eight rare mutations including V162I, N289S, T389T, V432A, E462K, A611A, S732S, and R760C. The number of CD patients carrying rare variants was significantly higher compared to controls (17.8% in CD patients vs. 0.8% in controls, P < 0.001, OR = 25.73, 95% CI, 3.34–198.08).
In silico predictions for coding novel variants—using SNAP online software (http://cubic.bioc.columbia.edu/services/SNAP/)—are presented in Table 4. The power calculation was performed for variations without a significant p value. The power for these analyses varied from 5.5% in S178S to 61.7% in P792P, which could be due to the small number of patients included in the study.
Among the three common mutations of the NOD2 gene, only R702W (SNP8) showed a significantly higher allele frequency in CD patients compared to healthy controls (13.3% vs. 1.3%, P < 0.001, OR = 12.15, 95% CI, 3.60–41.05). The allele frequencies of SNP 12 (G908R) and SNP 13 (1007fsinsC) were lower than 3% in Iranian CD patients (2.2% and 1.7%, respectively).
Discussion
This study provides confirmation for the previously reported [20] association of the NOD2 R702W mutation with CD in Iranian population (p < 0.001). In addition, we observed eight novel variations in exonic regions of the NOD2 gene. Finally, allele frequency analysis among patients and healthy controls indicated the association of some of the NOD2 gene variations with CD in Iranian population.
The NOD2 gene is known to be involved in susceptibility to CD in many Caucasian studies. Three common variants of this gene (G908R, R702W, and 1007fsinsC) were documented to be strongly associated with CD in white American and European populations [28–30]. Analysis of non-Caucasian groups has failed to identify a significant contribution of these variations in CD susceptibility. In Japanese and Korean populations, as well as in Chilean patients, the NOD2 common variants were very rare [30–32]. Iranian population, in the Middle East, has a complete mixed genetic reservoir. During the past years, some ethnicities like Turkish and Arabs who were mixed with the inhabitant population previously immigrated to Iran from northern regions. This makes Iranians have a different genetic background in comparison to Caucasians as well as Asians. In a recent study in Iranian CD patients, only R702W (SNP8) was reported to be associated with CD (p < 0.001) [20]. The different distribution patterns of the NOD2 common variations raised an opinion whether any other sequence changes of this gene were present in CD patients in Iran.
In this study, 21 variations were detected, including eight novel mutations, comparing to dbSNP sequence. Many studies have attempted to discover other sequence variations of the NOD2 gene in different populations [12, 33]. In a multi-center study on European population, 67 variations were identified in 453 CD cases [12], while in another research in Scotland, 18 mutations were reported in 663 CD patients [33]. According to these results, the NOD2 sequence changes in Iran showed a comparable occurrence and frequency to European studies.
More than 70% (n = 16) of the observed variations occurred in exon 4, a 1,816-bp fragment, which encodes a large size of the NOD2 protein from NBD to LRR segment. LRR fragment of the NOD2 protein has a critical role in the detection of bacterial components [11]; thus, sequence changes influencing this part of the protein could probably result in an impaired function.
Three of the novel variations (P371T, A794P, and Q809H) showed a statistically significant higher frequency in CD patients compared to controls (P < 0.001). These variations were all in a conserved sequence of the NOD2 gene, assessed by UCSC website (http://genome.ucsc.edu). The observed associations could clarify the probable role of the NOD2 gene variations in susceptibility to CD in Iran. As a separate ethnicity, Iranian patients demonstrated a different occurrence of NOD2 variations.
Two common mutations of the NOD2 gene (G908R and 1007fsinsC) were previously reported with a frequency lower than 5% in Iranian CD patients without any significant association with disease [20]. These results were in line with our observations. The allele frequencies of SNP 12 (G908R) and SNP 13 (1007fsinsC) were presented, 2.2% and 1.7%, respectively in CD patients. None of these variations showed a significant higher frequency in CD patients compared to controls. These results are in contrast to the reports from other Caucasian populations with a high frequency of these variations [28–30].
The analysis method used in this study did not include the promoter or other potential regulatory sequences which may influence the gene expression; and also, the number of included cases was somehow restricted. Despite these limitations, a comparable number of variations were detected in exonic regions of the NOD2 gene in Iranian CD patients. The observed variations, especially missense mutations (n = 4), could theoretically change the expression and function of the NOD2 protein. Changes in the structure of this protein may account as a predisposing factor for CD.
In conclusion, our study demonstrated the association between some of the NOD2 variations with CD in Iranian patients and could propose this gene as a contributing genetic factor in disease susceptibility. Evaluation of exonic and non-exonic segments including promoter regions in a study with larger sample size would be helpful in establishing the exact role of the NOD2 gene in Iranian CD patients.
References
Podolsky DK (2002) Inflammatory bowel disease. N Engl J Med 347(6):417–429
Bonen DK, Cho JH (2003) The genetics of inflammatory bowel disease. Gastroenterology 124:521–536
Hugot JP, Cézard JP, Colombel JF et al (2003) Clustering of Crohn’s disease within affected sibships. Eur J Hum Genet 11:179–184
Hugot JP, Laurent-Puig P, Gower-Rousseau C et al (1996) Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature 379:821–823
Satsangi J, Parkes M, Louis E et al (1996) Two stage genomewide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet 14:199–202
Ogura Y, Bonen DK, Inohara N et al (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411:603–606
Hugot JP, Chamaillard M, Zouali H et al (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603
Ahmad T, Tamboli CP, Jewell DP et al (2004) Clinical relevance of advances in genetics and pharmacogenetics of IBD. Gastroenterology 126:1533–1549
Girardin SE, Boneca IG, Viala J et al (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278:8869–8872
Hugot JP (2004) Genetic origin of IBD. Inflamm Bowel Dis 10(Suppl 1):S11–S15
Lichtenberger GS, Flavell RA, Alexopoulou L (2004) Innate immunity and apoptosis in IBD. Inflamm Bowel Dis 10(Suppl 1):S58–S62
Lesage S, Zouali H, Cezard JP et al (2002) CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet 70:745–747
Economou M, Trikalinos TA, Loizou KT et al (2004) Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol 99:2393–2404
Cuthbert AP, Fisher SA, Mirza MM et al (2002) The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology 122:867–874
Vermiere S, Wild G, Kocher K et al (2002) CARD15 genetic variation in a Quebec population: prevalence, genotype–phenotype relationship, and haplotype structure. Am J Hum Genet 71:74–83
Radlmayr M, Torok HP, Martin K et al (2002) The c-insertion mutation of the NOD2 gene is associated with fistulizing and fibrostenotic phenotypes in Crohn’s disease. Gastroenterology 122:2091–2092
Tukel T, Shalata A, Present D et al (2004) Crohn disease: frequency and nature of CARD15 mutations in Ashkenazi and Sephardi/Oriental Jewish families. Am J Hum Genet 74(4):623–636
Van Heel D, Fisher S, Kirby A et al (2004) Inflammatory bowel disease susceptibility loci defined by genome scan meta-analysis of 1952 affected relative pairs. Hum Mol Genet 13(7):763–770
Ogura Y, Inohara N, Benito A et al (2001) Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF (kappa) B. J Biol Chem 276:4812–4818
Derakhshan F, Naderi N, Farnood A et al (2008) Frequency of three common mutations of CARD15/NOD2 gene in Iranian IBD patients. Indian J Gastroenterol 27:8–11
Aghazadeh R, Zali MR, Bahari A et al (2005) Inflammatory bowel disease in Iran: a review of 457 cases. J Gastroenterol Hepatol 20:1691–1695
Farnood A, Naderi N, Moghaddam SJ et al (2007) The frequency of C3435T MDR1 gene polymorphism in Iranian patients with ulcerative colitis. Int J Colorectal Dis 22:999–1003
Naderi N, Farnood A, Habibi M et al (2008) Association of vitamin D receptor gene polymorphisms in Iranian patients with inflammatory bowel disease. J Gastroenterol Hepatol 23(12):1816–1822
Lennard-Jones JE (1989) Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl 170:2–6
Malchow H, Ewe K, Brandes JW et al (1984) European Cooperative Crohn’s Disease Study (ECCDS): results of drug treatment. Gastroenterology 86:249–266
Monkholm P, Binder V (2004) Clinical features and national history of Crohn's disease. In: Sartor RB, Sandborn WJ (eds) Kirsner's inflammatory bowel disease, 6th edn. Saunders, Philadelphia, pp 289–300
Sambrooks J, Fritsch EF, Manitis T (1992) Molecular cloning, laboratory manual. Cold Spring Harbor Laboratory Press, New York
Büning C, Genschel J, Bühner S et al (2004) Mutations in the NOD2 gene in Crohn's disease are associated with ileocecal resection and are a risk factor for reoperation. Aliment Pharmacol Ther 19:1073–1078
Vavassori P, Borgiani P, Biancone L et al (2004) CARD15 mutation analysis in an Italian population: Leu1007fsinsC but neither Arg702Trp nor Gly908Arg mutations are associated with Crohn's disease. Inflamm Bowel Dis 10:116–121
Inoue N, Tamura K, Kinouchi Y et al (2002) Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology 123:86–91
Yamazaki K, Takazoe M, Tanaka T et al (2002) Absence of mutation in the NOD2 gene among 483 Japanese patients with Crohn’s disease. J Hum Genet 47:469–472
Figueroa C, Peralta A, Herrera L et al (2006) NOD2 and toll-like 4 receptor gene polymorphism in Chilean patients with inflammatory bowel disease. Eur Cytokine Netw 17:125–130
Russell RK, Drummond HE, Wilson DC et al (2008) Detailed assessment of NOD2 exonic variation in inflammatory bowel disease in Scotland: implications for disease pathogenesis. Genes Immun 9:556–560, Abstract
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Naderi, N., Farnood, A., Habibi, M. et al. NOD2 exonic variations in Iranian Crohn's disease patients. Int J Colorectal Dis 26, 775–781 (2011). https://doi.org/10.1007/s00384-011-1145-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00384-011-1145-4