Abstract
Background and aims
Hypoxia-inducible factor-1α (HIF-1α) is the main active subunit of HIF-1 that promoted tumor cells survival and critical steps in tumor progression and aggressiveness. The authors aimed to investigate the role of HIF-1α and Survivin in colorectal cancer (CRC) progression.
Materials and methods
Plasmid expressing small interfering RNA (siRNA) against HIF-1α was constructed and transfected into LS174T cells with Lipofectamine. The LS174T cells were incubated for 24 h under hypoxic condition. The inhibitory effects of siRNA on HIF-1α gene was determined by semiquantitative reverse transcriptase polymerase chain reaction and Western blot. Expression of HIF-1α and Survivin was investigated by immunohistochemistry in colorectal adenocarcinomas tissue microarrays.
Results
HIF-1α and Survivin expressions were markedly downregulated after the siRNA expression vector against HIF-1α was transfected into the LS174T cells. Of the eight adenoma lesions, one case (12.25%) and four cases (50%) were positive for HIF-1α and Survivin, respectively. Of the 69 cases of CRCs, 46 cases (66.7%) and 39 cases (56.5%) were positive for HIF-1α and Survivin, respectively. The positive rate of HIF-1α protein in CRCs was significantly higher than that in colorectal adenoma lesions (P < 0.05). HIF-1α protein expression was significantly higher in patients with stage III than in patients with stage I–II CRCs (P < 0.01). In addition, overexpression of HIF-1α in higher stages of CRCs was found to correlate positively with Survivin levels (P < 0.001).
Conclusions
Our data demonstrate that HIF-1α and Survivin are mostly expressed in invasive CRCs. Inhibition of HIF-1α may lead to exploration of its potential as a diagnostic tool and possibly a target for gene therapy for colorectal carcinoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypoxia inducible factor-1(HIF-1), a master transcription factor of oxygen-regulated genes, mediates a wide range of cellular and physiological adaptive responses to changes in oxygen tension [1]. HIF-1 is composed of two subunits, HIF-1α and HIF-1β [2, 3]. HIF-1α is the main active subunit. Hypoxia or genetic alterations of the hypoxia signaling cascade leading to the constitutive expression of HIF-1 could promote intense and chaotic neovascularization that facilitates tumor spread [4]. HIF-1 can induce a vast array of gene products controlling energy metabolism, neovascularization, survival, and cell migration and has become recognized as a strong promoter of tumor growth. It has now been firmly established that HIF-1 has important roles in tumor progression [5].
Survivin, a member of the inhibitor of apoptosis family, has been reported to be associated with hypoxia [6]. Tumor cannot grow in the absence of angiogenesis, because of the lack of oxygen in the center of tumors, which results in apoptosis and necrosis. Surprisingly, Survivin is only weakly or not expressed at all in normal tissues, but is overexpressed in many tumors. This overexpression is mainly correlated with poor prognosis [7–9]. In patients with rectal carcinoma treated with a combination of radiotherapy and chemotherapy, increased Survivin expression was inversely related to the levels of apoptosis and was also associated with a significantly higher risk of a local tumor recurrence [10]. Recently, it has been found that tumor-specific expression of Survivin is increased by hypoxia, and the addition of hypoxia-responsive elements (HREs) to the Survivin promoter results in a further increase in its expression [11]. A putative HRE, 5′-GCGTG-3′, located at −81 to −85 nucleotides of the 5′-flanking region of the Survivin gene. HIF-1α directly interacts with the Survivin promoter, upregulating the level of Survivin gene expression, which results in resistance to apoptosis in tumor cells [12].
We have recently found that HIF-1α expression level was higher in SW480 cells under hypoxic than under normoxic conditions. HIF-1α messenger ribonucleic acid (mRNA) expression level was found to be expressed at higher levels in adenocarcinoma than in adenoma tissues. We also found that HIF-1α protein expression was significantly correlated with angiogenic markers [5, 13]. Consistent with our results, the expression of HIF-1α has been reported in others human malignancies [14, 15]. In this study, we first examined the effect of small interfering RNA (siRNA) against HIF-1α mRNA on the expression of Survivin in human colonic cancer cell line LS174T. Then, to further confirm the observed preclinical data at the clinical level, we investigated the possible correlation of HIF-1α and Survivin expression levels in tissue microarrays of human colorectal carcinomas.
Materials and methods
Cell lines and condition of transfections
Human LS174T cells were maintained in Dulbecco’s modified Eagle medium (GIBCO/BRL) supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 IU/ml streptomycin. Cell lines were incubated at 37°C in an atmosphere of 5% CO2. For experiment, cancer cells were seeded at 4.0 × 105 cells per well in 24-well plates and grown to the confluence reaching approximately 60% at the time of transfection. The cells were harvested 24 h after transfection. For experiment, cancer cells were seeded at 4.0 × 105 cells per well in 24-well plates and grown to the confluence reaching approximately 60% at the time of transfection. siRNA (25 pmol per well) was delivered to the plated cells using Lipofectamine 2000 (Invitrogen, Basel, Switzerland) following the manufacturer’s instructions. Then, cells were set in different oxygen conditions. Under the normoxic condition, cells were cultured in 20% O2, 5% CO2, and 75% N2, whereas under the hypoxic condition, cells were cultured in 1% O2, 5% CO2, and 94% N2. To ensure the level of hypoxia was verified, PO2 of the culture medium was tested when the cells culture was done. The cells were harvested 24 h after transfection.
Plasmid construction and transfection
According to human HIF-1 α complementary deoxyribonucleic acid (cDNA) sequence in the gene bank (NM001530), targets for siRNA against HIF-1α mRNA were selected as described previously [16–18]. The sequences of three target sites in HIF-1α mRNA and the primers that were used to make the corresponding pSilence-U6-siRNA plasmids are shown in Table 1. The sequences of target sites in the primers were underlined.
RNA isolation and RT-PCR assay
LS174T cells were transfected with siRNA expression plasmids. Total RNA was then extracted from transfected cells by Trizol Reagent (Invitrogen) according to the method described in the manufacturer’s manual. Reverse transcription was performed with total RNA as the template. The cDNAs were synthesized with HIF-1α gene-specific primer. β-Actin was used as an internal control. The cDNA was amplified by 25 polymerase chain reaction (PCR) cycles of denaturation for 1 min at 94°C, annealing for 45 s at 60°C, and extension for 1 min at 72°C. The PCR primers were the following:
-
HIF-1α up: 5′-GCATCTCCATCTCCTACCCACA-3′
-
HIF-1α down: 5′-TGCATCCTTTTACACGTTTCCA-3′, 2,202–2,559, 358 bp
-
β-Actin up: 5′-CTTGCCATCCTAAAAGCCACC-3′
-
β-Actin down: 5′-GACCAAAAGCCTTCATACATCTC-3′, 1,485–1,715, 231 bp
For the expression of specific double-stranded RNA, each template contains sequences of 19 nucleotides from HIF-1α mRNA and their complement sequence separated by nine bases to facilitate the formation of the hairpin structure. BamHI and HindIII restriction enzyme sites were introduced into primers for the cloning purpose. Sense and antisense primers were annealed. Their products were digested with BamHI and HindIII, and inserted into pSilence-2.1-U6 plasmid downstream of U6 promoter. Plasmids containing the right insert were confirmed by sequencing. The corresponding plasmids were named pSilence-2.1-U6-siRNA-HIF-1α-1, 2 and 3. A control plasmid containing an insertion of unrelated sequence was used as the control. A cation-based transfecting reagent, sofastTM (Xiamen Taiyangma Biotech), was used for transfecting plasmids into LS174T cells. The intensities of HIF-1α mRNA were determined by using HPLAS-2000 analysis software (Qianping Biotechnology, Wuhan, China).
Western blot analysis
Western blots for HIF-1α and Survivin were performed as described previously [19]. Briefly, total protein was extracted from transfected LS174T cells. Cells transfected a plasmid containing an insertion of unrelated sequence were used as a control. A primary goat polyclonal anti-HIF-1α(C-19) and Survivin(C-19) antibody (Santa Cruz Biotechnology) was used according to the manufacturer’s protocol. Fifty micrograms of whole-cell extracts per lane were resolved using 6% (for HIF-1α) or 12% (for Survivin) sodium dodecyl sulfate polyacrylamide gel electrophoresis. The proteins were then transferred onto nitrocellulose membranes in the blotting buffer (5% [vol/vol] methanol, 25 mmol/L Tris, 120 mmol/L glycine). Membranes were blocked with 5% nonfat dried milk, 2% bovine serum albumin, and Tris-buffered saline–Tween 20. Endogenous HIF-1α or Survivin was detected with 1:600 dilution of anti-HIF-1α or anti-Survivin polyclonal antibody, followed by a secondary horseradish peroxidase-conjugated rabbit anti-goat IgG antibody. The optical density of HIF-1α and Survivin protein were compared with β-actin by using HPLAS-2000 analysis software (Qianping Biotechnology).
Patients and samples
Tissue microarrays used in this study were purchased from Pantomics (San Francisco, CA, USA), including 61 cases of colorectal adenocarcinomas and eight cases of mucinous adenocarcinomas, eight cases of normal tissues, and eight cases of adenoma in duplicates. Adenoma patients had a mean age of 45 years, ranged from 7 to 68 years, and colorectal carcinoma patients had a mean age of 56 years, ranged from 29 to 89 years. According to the TNM classification and International Union Against Cancer colorectal cancer stages, there were 39 patients with stage I–II and 30 with stage III colorectal adenocarcinomas. The clinicopathological data including grade, stage, histological type, and lymph node metastasis were provided by the producers. All tissues were fixed in 10% neutral buffered formalin for 24 h and processed using identical standard operating procedures and cut into 4-μm serial sections.
Immunostaining
Streptavidin/peroxidase kits (Fuzhou Maxim Biotech, China) were used for immunohistochemistry. The procedures for immunohistochemical detection were performed according to previous study [5]. Briefly, sections were incubated with anti-HIF-1α- or anti-Survivin-specific goat polyclonal antibodies (at a working dilution of 1:100; Santa Cruz Biotechnology). For a negative control, sections were incubated with phosphate-buffered saline (0.01 mol/L, pH 7.4) instead of the primary antibodies. For HIF-1α, brown-yellow staining of the cytoplasm indicated a positive result. Nuclear and cytoplasm Survivin expression was considered as positive.
Evaluation of HIF-1α and Survivin immunostaining
Staining for HIF-1α and Survivin was assessed in five high power fields, and they were believed to be representative of the average in tumors at ×400 magnification. HIF-1α and Survivin expression was determined by assessing semiquantitatively the percentage of decorated tumor cells and the staining intensity (SI). The percentage of positive cells (PP) was rated as follows: 1–10% PPs +, 11–50% PPs ++, and greater than 50% PPs +++. SI was scored as: weak +, moderate ++, and strong +++. Points for expression and percentage of PPs were calculated to an immunoreactive score (IRS). IRS = PP × SI. IRS scored 0 were taken as negative, scored less than 3 as low expression, scored 3 to 6 as moderate expression, and scored more than 6 as high expression. The score was determined by two authors (all slides were examined and scored independently by two investigators [Li-Fang Fan and Wei-Guo Dong] independently after the blind analysis principle).
Statistical analysis
All statistical analyses were performed by the SPSS 13.0 software package for Windows. Experimental in vitro data are presented as means ± SD. Levels of significance for these data were calculated using Student’s t test. To evaluate whether the frequency of cells with the elevated levels of HIF-1α protein and Survivin expression increased during colorectal carcinogenesis, we performed a Chi-square test for trend, grouping the results as adenoma, stage I–II, and stage III. Spearman rank correlation test was used to analyze the correlation between HIF-1α and Survivin proteins. P < 0.05 was considered to be statistically significant. No cases were excluded from statistical analysis.
Results
Inhibition of the expression of HIF-1α mRNA
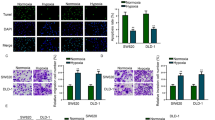
DNA sequence analysis confirmed the successful insertion of template as we designed. Figure 1 showed reverse transcriptase (RT)-PCR results analyzing mRNA levels of HIF-1α in LS174T cells transfected with siRNA plasmids. Using β-actin as the internal control, transfection of pSilence-2.1-U6-siRNA1 and 2 had minor inhibitory effects on the mRNA level of HIF-1α in LS174T cells, compared lanes 1 and 2 to 3. On the other hand, compared to the control group (lane 4), cells transfected with pSilence-2.1-U6-siRNA3 had a dramatic reduction in HIF-1α mRNA level (lane 3).
Inhibition of the expression of HIF-1α and Survivin proteins
As shown in Fig. 2, cells incubated 24 h under hypoxia after transient transfection with HIF-1α siRNA3 had HIF-1α and Survivin protein expressions significantly lower than in the control (P < 0.05). Furthermore, the HIF-1α and Survivin protein expression ratio was significantly lower than HIF-1α siRNA1 and 2 (P < 0.05).
a Western blot analysis of HIF-1α protein in LS174T cells transfected with siRNA plasmids and control. HIF-1α protein was inhibited by HIF-1αsiRNA3 significantly. b Western blot analysis of Survivin protein in LS174T cells transfected with siRNA plasmids and control. Survivin protein was inhibited by HIF-1αsiRNA3 significantly. Lane 1, HIF-1αsiRNA1; lane 2, HIF-1αsiRNA2; lane 3, HIF-1αsiRNA3; lane 4, control
Expression of HIF-1α and Survivin in normal colorectal mucosa
Specific immunoreactivity for HIF-1α was not observed in normal colorectal mucosa. In contrast, in HIF-1α expression, one case (12.5%) out of eight normal colorectal mucosa expressed Survivin, mainly located in the cytoplasm of epithelial cells of colonic crypts.
Expression of HIF-1α in colorectal adenomas and carcinomas
The expression of HIF-1α protein was observed mainly in the cytoplasm of tumor cells (Fig. 3). HIF-1α was predominantly expressed in cells that were located at the outer, invading edge of the tumors, adjacent to the necrotic areas and surrounding areas of neovascularization in colorectal cancers. There was only one case of colorectal adenoma expressed HIF-1α. The other seven cases were negative for HIF-1α. Among colorectal carcinoma tissue samples, the positive rate of HIF-1α expression was 66.7% (46 of 69). HIF-1α expressed differently in adenomas, carcinomas stages I–II and stage III (P = 0.016). The cases of negative, low, moderate, and high expression of HIF-1α were 23, 17, 22, and 7, respectively. HIF-1α immunoreactivity was found to be significantly higher in colorectal cancers compared to adenomas (P = 0.000). The expression of HIF-1α were significantly higher in tissue samples classified as stage III than those classified as stage I–II (P = 0.01). However, no significant difference was found between adenomas and pathological stages I–II (Table 2).
Representative slides showing the immunohistochemical staining of HIF-1α. a, b No HIF-1α. Immunoreactivity was detected in the normal colorectal mucosa (×40, ×200 magnification, respectively). c HIF-1α expressed negative in colorectal adenoma (×200 magnification). d, e By contrast, strong HIF-1α immunoreactivity was detected in the cytoplasm of tumor cells in carcinoma tissue (×40, ×200 magnification, respectively). f HIF-1α was expressed in cells located at the outer edge, adjacent to the necrotic areas in carcinoma tissue
Expression of Survivin in colorectal adenomas and carcinomas
The Survivin protein expression showed both cytoplasmic and nuclear staining in tumor cells (Fig. 4). Half of the adenoma samples positively expressed Survivin. Among colorectal carcinoma tissue samples, the rate of positive Survivin protein expression was 56.5% (39 of 69). Thirty cases were with negative expression, ten cases were with low expression, 18 cases were with moderate expression, and 11 cases were with high expression. There was no statistical difference between adenoma and carcinoma groups. In carcinoma samples, we also could not find a significant correlation between stage I–II and stage III groups (Table 2).
Representative slides showing the immunohistochemical staining of Survivin in normal colorectal mucosa and adenoma. a Survivin expressed negative in normal colorectal mucosal glands. (×200 magnification). b, c Survivin immunoreactivity was also detected in the nuclei of tumor cell in the same field of colorectal carcinoma tissue (×40, ×200magnification, respectively). d Survivin expressed strongly in nuclei of cells in adenoma (×200 magnification). e, f Survivin immunoreactivity was also detected in the cytoplasm of tumor cells in carcinoma tissue (×40, ×200 magnification, respectively)
Correlation between HIF-1α and Survivin expression in colorectal carcinoma
Table 3 shows the correlation between HIF-1α and Survivin immunoreactivity. Using the Spearman rank correlation test, a positive linear relationship was found between HIF-1α and Survivin (rho = 0.326, P = 0.001). HIF-1α protein expression correlated closely with Survivin protein expression in colorectal carcinoma.
Discussion
HIF-1 is the central transcription factor that is activated by hypoxia and modulates the expression of many genes involved in cell metabolism, proliferation, apoptosis, and angiogenesis. Recently, it has been reported that HIF-1 contributes to tumor radioresistance by upregulating Survivin expression under hypoxic conditions [12].
In this study, we used siRNA molecules to decrease HIF-1α in LS174T colorectal cancer cells. This showed a significant decrease in HIF-1α mRNA and HIF-1α protein. We also showed that HIF-1α protein expression was significantly higher in tissue samples classified as stage III than those classified as stages I–II, suggesting that HIF-1α protein expression correlated with tumor progression and advanced pathological stage in colorectal cancer.
We demonstrated previously that HIF-1α plays an important role in angiogenesis and tumor progression by regulating the expression of vascular endothelial growth factor (VEGF) in human colorectal carcinomas. The positive rate of HIF-1α mRNA expression was significantly higher in adenocarcinoma specimens than in adenomas. There exists an increase in the positive rate of HIF-1α expression from Dukes A to C + D stages in tumor development [5]. These reports all demonstrated that HIF-1α implicated in the progression of colorectal carcinoma.
Survivin is a bifunctional regulator of spindle microtubule function at mitosis and an inhibitor of apoptosis. It is overexpressed in nearly every human cancer, including colorectal cancer but rarely expressed in normal tissues [20, 21]. Survivin immunoreactivity expanded from the base crypt toward the surface when the normal crypts shift to adenoma [22]. Survivin expression between adenoma and different Dukes stages of colorectal carcinoma was different [23, 24]. In our study, Survivin immunoreactivity was observed in one case of normal tissue where 50% adenomas expressed Survivin, and we found no difference between adenomas and carcinomas. These different results were reasoned by a small sample size or difference of staining method or immunostaining evaluation. Although no difference of the Survivin proteins expression was found between pathological stages of colorectal carcinoma, the in vivo data show strong correlation between the expression of HIF-1α and Survivin (Table 3). Only HIF-1α is expressed differently in adenomas and carcinomas stages I–II and stage III. It may be credible that HIF-1α and Survivin have a positive role in tumor progression in colorectal cancer.
Predictive and prognostic detection of Survivin has been implemented in some types of cancer, and strategies for targeting Survivin as a new cancer treatment are underway [25, 26]. Recently, a few reports have shown a strong correlation between HIF-1α and Survivin in breast [27] and pancreatic [28] cancer cells. The expression of HIF-1α mRNA was closely correlated with the expression of Survivin proteins in pancreatic cancer [29]. Previous data also demonstrate that antisense HIF-1α inhibits expressions of Survivin in human pancreatic cancer BxPc-3 cells [28]. In breast cancer cells, overexpression of Survivin was mediated by oxygen-independent HIF-1α upregulation in endothelial growth factor-treated cancer cells. Furthermore, hypoxia alone via HIF-1α induces Survivin expression in the brain with a mouse model of stroke [29]. However, no relationship between Survivin protein expression and HIF-1α was found in a small sample size of advanced cervical carcinoma of patients treated by radiotherapy [6].
In this study, we used siRNA molecules to decrease HIF-1α in LS174T colorectal cancer cells, resulting in a significant attenuation of Survivin protein expression after transfection, demonstrating that HIF-1α regulates Survivin expression. Statistical analysis shows a significant association of HIF-1α with Survivin, but we could not prove a statistical significant relationship between Survivin expressions with pathological stage. Survivin protein expression has no statistical difference between the pathological stages of colorectal carcinoma. Possibly, this result is reasoned by the small sample size or the activation of Survivin through a parallel pathway.
The data implicate that HIF-1α might be a key transcription factor for Survivin gene expression, since the downregulation of HIF-1α using HIF-1α siRNA significantly reduced the level of Survivin expression in human colon cancer cells. For the first time, our study provides evidence of the correlation between HIF-1α and Survivin in cultured colon cancer cells and human colorectal cancers. Currently, the precise mechanism for HIF-1α-mediated transcriptional activation of the Survivin gene is under investigation. However, we believed it possible the transcriptional activation of Survivin gene expression by HIF-1α in colorectal cancer; whether this mechanism is universal need further studies.
In summary, our study highlights the importance of HIF-1α in tumor progression and the expression of Survivin in colorectal cancer.
Conclusion
In conclusion, we have shown that the inhibition of HIF-1α results in attenuation of Survivin expression. HIF-1α tends to increase with tumor progression and correlates with Survivin in human colorectal carcinoma. Inhibition of HIF-1α may be an important and approachable therapeutic target for colorectal carcinoma.
References
Hochachka PW, Buck LT, Doll CJ, Land SC (1996) Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA 93:9493–9498
Semenza GL, Wang GL (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol 12:5447–54543
Wang GL, Semenza GL (1995) Purification and characterization of hypoxia-inducible factor 1. J Biol Chem 270:1230–1237
Pouyssegur J, Dayan F, Mazure NM (2006) Hypoxia signalling in cancer and approaches to enforce tumor regression. Nature 441:437–443
Jiang CQ, Liu ZS, Fan LF, Liu ZS, Qian Q, Xia D, Diao LM, He YM, Ai ZL (2004) Expression levels and significance of hypoxia inducible factor-1 alpha and vascular endothelial growth factor in human colorectal adenocarcinoma. Chin Med J 117:1541–1546
Bache M, Holzapfel D, Kappler M, Holzhausen HJ, Taubert H, Dunst J, Hänsgen G (2007) Survivin protein expression and hypoxia in advanced cervical carcinoma of patients treated by radiotherapy. Gynecol Oncol 104:139–144
Haberler C, Slavc I, Czech T, Gelpi E, Heinzl H, Budka H, Urban C, Scarpatetti M, Ebetsberger-Dachs G, Schindler C, Jones N, Klein-Franke A, Maier H, Jauk B, Kiefer A, Hainfellner JA (2006) Histopathological prognostic factors in medulloblastoma: high expression of survivin is related to unfavourable outcome. Eur J Cancer 42:2996–3003
Sohn DM, Kim SY, Baek MJ, Lim CW, Lee MH, Cho MS, Kim TY (2006) Expression of survivin and clinical correlation in patients with breast cancer. Biomed Pharmacother 60:289–292
Akyürek N, Memiş L, Ekinci O, Köktürk N, Oztürk C (2006) Survivin expression in pre-invasive lesions and non-small cell lung carcinoma. Virchows Arch 449:164–170
Rödel F, Hoffmann J, Distel L, Herrmann M, Noisternig T, Papadopoulos T, Sauer R, Rödel C (2005) Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res 65:4881–4887
Yang L, Cao Z, Li F, Post DE, Van Meir EG, Zhong H, Wood WC (2004) Tumor specific gene expression using the Survivin promoter is further increased by hypoxia. Gene Ther 11:1215–1223
Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L (2006) Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem 281:25903–25914
Jiang YA, Fan LF, Jiang CQ, Zhang YY, Luo HS, Tang ZJ, Xia D, Wang M (2003) Expression and significance of PTEN, hypoxia-inducible factor-1 alpha in colorectal adenoma and adenocarcinoma. World J Gastroenterol 9:491–494
Matsuyama T, Nakanishi K, Hayashi T, Yoshizumi Y, Aiko S, Sugiura Y, Tanimoto T, Uenoyama M, Ozeki Y, Maehara T (2005) Expression of hypoxia-inducible factor-1alpha in esophageal squamous cell carcinoma. Cancer Sci 96:176–182
Vleugel MM, Greijer AE, Shvarts A, van der Groep P, van Berkel M, Aarbodem Y, van Tinteren H, Harris AL, van Diest PJ, van der Wall E (2005) Differential prognostic impact of hypoxia induced and diffuse HIF-1alpha expression in invasive breast cancer. J Clin Pathol 58:172–177
Zhang Q, Zhang ZF, Rao JY, Sato JD, Brown J, Messadi DV, Le AD (2004) Treatment with siRNA and antisense oligonucleotides targeted to HIF-1alpha induced apoptosis in human tongue squamous cell carcinomas. Int J Cancer 111:849–857
Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL (2003) Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res 63:6130–6134
Berchner-Pfannschmidt U, Petrat F, Doege K, Trinidad B, Freitag P, Metzen E, de Groot H, Fandrey J (2004) Chelation of cellular calcium modulates hypoxia-inducible gene expression through activation of hypoxia-inducible factor-1alpha. J Biol Chem 279:44976–44986
Itoh T, Namba T, Fukuda K, Semenza GL, Hirota K (2001) Reversible inhibition of hypoxia-inducible factor 1 activation by exposure of hypoxic cells to the volatile anesthetic halothane. FEBS Lett 509:225–229
Ponnelle T, Chapusot C, Martin L, Bouvier AM, Plenchette S, Faivre J, Solary E, Piard F (2005) Cellular localisation of Survivin: impact on the prognosis in colorectal cancer. Cancer Res Clin Oncol 131:504–510
Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N (2000) Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res 6:127–134
Boman BM, Walters R, Fields JZ, Kovatich AJ, Zhang T, Isenberg GA, Goldstein SD, Palazzo JP (2004) Colonic crypt changes during adenoma development in familial adenomatous polyposis: immunohistochemical evidence for expansion of the crypt base cell population. Am J Pathol 165:1489–1498
Tan HY, Liu J, Wu SM, Luo HS (2005) Expression of a novel apoptosis inhibitor—survivin in colorectal carcinoma. World J Gastroenterol 11:4689–4692
Abd El-Hameed A (2005) Survivin expression in colorectal adenocarcinoma using tissue microarray. J Egypt Natl Canc Inst 17:42–50
Altieri DC (2003) Validating Survivin as a cancer therapeutic target. Nat Rev Cancer 3:46–54
Smith SD, Wheeler MA, Plescia J, Colberg JW, Weiss RM, Altieri DC (2001) Urine detection of Survivin and diagnosis of bladder cancer. JAMA 285:324–328
Chang Q, Qin R, Huang T, Gao J, Feng Y (2006) Effect of antisense hypoxia-inducible factor 1alpha on progression, metastasis, and chemosensitivity of pancreatic cancer. Pancreas 32:297–305
Conway EM, Zwerts F, Van Eygen V, DeVriese A, Nagai N, Luo W, Collen D (2003) Survivin-dependent angiogenesis in ischemic brain. Am J Pathol 163:935–994
Wei H, Wang C, Chen L (2006) Proliferating cell nuclear antigen, Survivin, and CD34 expressions in pancreatic cancer and their correlation with hypoxia-inducible factor 1alpha. Pancreas 32:159–163
Acknowledgments
The authors thank Kai-Lang Wu and Hong-Lei Chen for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fan, LF., Dong, WG., Jiang, CQ. et al. Role of Hypoxia-inducible factor-1α and Survivin in colorectal carcinoma progression. Int J Colorectal Dis 23, 1057–1064 (2008). https://doi.org/10.1007/s00384-008-0511-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00384-008-0511-3